Abstract
E3-19K binds to and retains MHC class I molecules in the endoplasmic reticulum, suppressing anti-Adenovirus (Ad) activities of T-cells. We determined the structure of the Ad serotype 2 (Ad2, species C) E3-19K–HLA-A2 complex to 1.95 Å resolution. Ad2 E3-19K binds to the N-terminus of the HLA-A2 groove, contacting the α1-, α2-, and α3-domains and β2m. Ad2 E3-19K has a unique structure comprised of a large N-terminal domain, formed by two partially overlapping β-sheets arranged in a V-shape, a C-terminal α-helix and tail. The structure reveals determinants in E3-19K and HLA-A2 that are important for complex formation; conservation of some of these determinants in E3-19K proteins of different species and MHC I of different loci suggests a universal binding mode for all E3-19K proteins. Our structure is important for understanding the immunomodulatory function of E3-19K.
Adenoviruses (Ads) are widespread in the human population, with at least 51 known serotypes (Ad1 to Ad51) classified into six species (A to F)1. Ads cause a number of infections that are linked to respiratory, gastrointestinal, and ocular diseases2. While primary Ad infection elicits antiviral cytotoxic T-lymphocyte (CTL) immune responses, these responses are usually insufficient to clear the virus. Consequently, Ads cause lifelong asymptomatic infections in healthy individuals and can be fatal in children and immunocompromised patients.
Large deoxyribonucleic acid (DNA) viruses such as Ads have evolved strategies for evading antiviral immune responses3, 4. It was shown in the late 1980s that the E3-19K protein of Ad binds to and retains MHC class I molecules in the endoplasmic reticulum (ER) of infected cells, preventing their egress to the cell surface5, 6, 7. As a consequence, Ad-infected cells are less susceptible to lysis by Ad-specific CTLs6, 8, 9, 10. At that time, this represented the first example of a viral protein capable of interfering with MHC I antigen presentation. It was also shown that when the lungs of cotton rats are infected with wild-type Ad, immunopathological responses are less severe than when the lungs are infected with a mutant Ad containing a deletion of the E3-19K gene11. It was suggested that the absence of E3-19K in the mutant virus activated CTLs as part of the host inflammatory response to the virus11. It is thought that E3-19K, through its association with MHC I, enables Ads to establish persistence in host cells4, 11, 12.
E3-19K is a type I transmembrane glycoprotein that comprises a N-terminus ER-lumenal domain, a transmembrane domain, and a C-terminus cytosolic tail. The ER-lumenal domain of E3-19K associates with the ER-lumenal domain of MHC class I molecules8, 13, 14, 15, 16, while the dilysine motif in the cytosolic tail of E3-19K provides the signal for localization of the E3-19K–MHC I complex in the ER8, 17, 18. An analysis of immunoprecipitates from Ad2-, Ad5-, and Ad19-infected mammalian cells suggested that E3-19K displays differential interaction with MHC I19, 20, 21. Using recombinant, soluble E3-19K proteins of species B, C, D, and E and HLA-A, -B, and -C molecules, we showed that E3-19K proteins associate with MHC I in a locus-specific manner22, 23; E3-19K associates with HLA-A and HLA-B but displays essentially no affinity for HLA-C. We also showed that E3-19K proteins of species B, C, and E associate with HLA-A and -B in an allele-dependent manner22, 23.
The ER-lumenal domain of E3-19K has been subdivided into three regions24, 25: 1. residues 1 to ~78–81 are rather variable between E3-19K proteins of different species; 2. residues ~79–82 to ~98 are rather conserved between E3-19K proteins of different species; and 3. residues ~99 to 107 act as linker between the ER-lumenal domain and the transmembrane domain. Secondary structure predictions of Ad2 E3-19K have provided some insights into potential architectural elements26, 27, 28. Structure-function relationship studies have identified specific residues in both the variable and conserved regions of E3-19K that are critical for its MHC I-binding function 22, 23, 25, 27, 29. Evidence from several studies has suggested that E3-19K binds to the peptide-binding groove of MHC I13, 14, 22, 23. We proposed recently a model of interaction in which E3-19K has contact sites on MHC I at the N-terminal end of the α1-helix and C-terminal end of the α2-helix23. To date, more than 20 years after its discovery as the first viral immunomodulatory protein, our lack of knowledge of the three-dimensional structure of E3-19K alone or in complex with MHC I severely limits our understanding of its immunomodulatory function. Progress has been hampered largely by the fact that E3-19K is a protein recalcitrant to crystallization.
We set out to determine the x-ray three-dimensional structure of recombinant, soluble Ad2 E3-19K bound to HLA-A2–Tax peptide. We describe a novel rescue refolding strategy that allowed formation of Ad2 E3-19K–HLA-A2, from which we generated crystals diffracting to 1.95 Å resolution. Our structure offers precise explanations for the mechanism by which E3-19K modulates antiviral cellular immunity.
RESULTS
Structure of the Ad2 E3-19K–HLA-A2 complex
The ER-lumenal domain of Ad2 E3-19K has two N-linked glycans (Asn12 and Asn61)30. In previous studies, we showed that treatment of Ad2 E3-19K with PNGase F and Endo H31 lead to precipitation of Ad2 E3-19K. This suggested an important role for the glycans in stabilization of Ad2 E3-19K. When Ad2 E3-19K is bound to HLA-A2–Tax, it can more easily withstand hydrolysis of its glycans; crystallization trials of PNGase F- and Endo H-treated Ad2 E3-19K–HLA-A2 failed to produce crystals (our unpublished results).
The expression of Ad2 E3-19K in E. coli led to production of inclusion bodies from which we were unable to refold E3-19K (discussed in Ref. 31). In marked contrast, the addition of submolar amounts of folded HLA-A2–Tax in the oxidative refolding buffer containing urea-solubilized inclusion bodies of Ad2 E3-19K generated the Ad2 E3-19K–HLA-A2 complex in solution. Most likely, upon refolding, Ad2 E3-19K can bind immediately to HLA-A2 and form a stable Ad2 E3-19K–HLA-A2 complex before it can aggregate in solution.
Using this strategy, we determined the structure of the complex between the ER-lumenal domains of Ad2 E3-19K and HLA-A2–Tax to 1.95 Å resolution by molecular replacement (see METHODS) (Table 1). Only minor differences were observed between the two molecules of E3-19K in the asymmetric unit (rmsd of 0.52 Å for 97 equivalent Cα atoms) such that only chain G will be discussed further. Clear electron density was observed for nearly all residues of Ad2 E3-19K–HLA-A2; residues 2–100 of Ad2 E3-19K, residues 1–275 of HLA-A2 heavy chain, residues 0–99 of β2-microglobulin (β2m), and the HIV-1 Tax LLFGYPVYV peptide. The overall structure of the Ad2 E3-19K–HLA-A2 complex is shown in Fig. 1A.
Table 1.
Data collection and refinement statistics
| Ad2 E3-19K–HLA-A2 complex | |
|---|---|
| Data collectiona | |
| Space group | I212121 |
| Cell dimensions | |
| a, b, c (Å) | 113.19, 133.67, 196.72 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.00–1.95 (2.02–1.95)b |
| Rmerge | 0.148 (0.540) |
| I / σI | 15.97 (3.20) |
| Completeness (%) | 99.7 (97.4) |
| Redundancy | 11.0 (7.1) |
| Refinement | |
| Resolution (Å) | 37.06–1.95 |
| No. reflections | 102,933 |
| Rwork / Rfree | 0.210/0.241 |
| No. atoms | |
| Protein | 7,968 |
| Water | 472 |
| B-factors | |
| Protein | 35.6 |
| Water | 34.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.3 |
Data were collected from a single crystal.
Values in parentheses are for highest-resolution shell.
Figure 1.
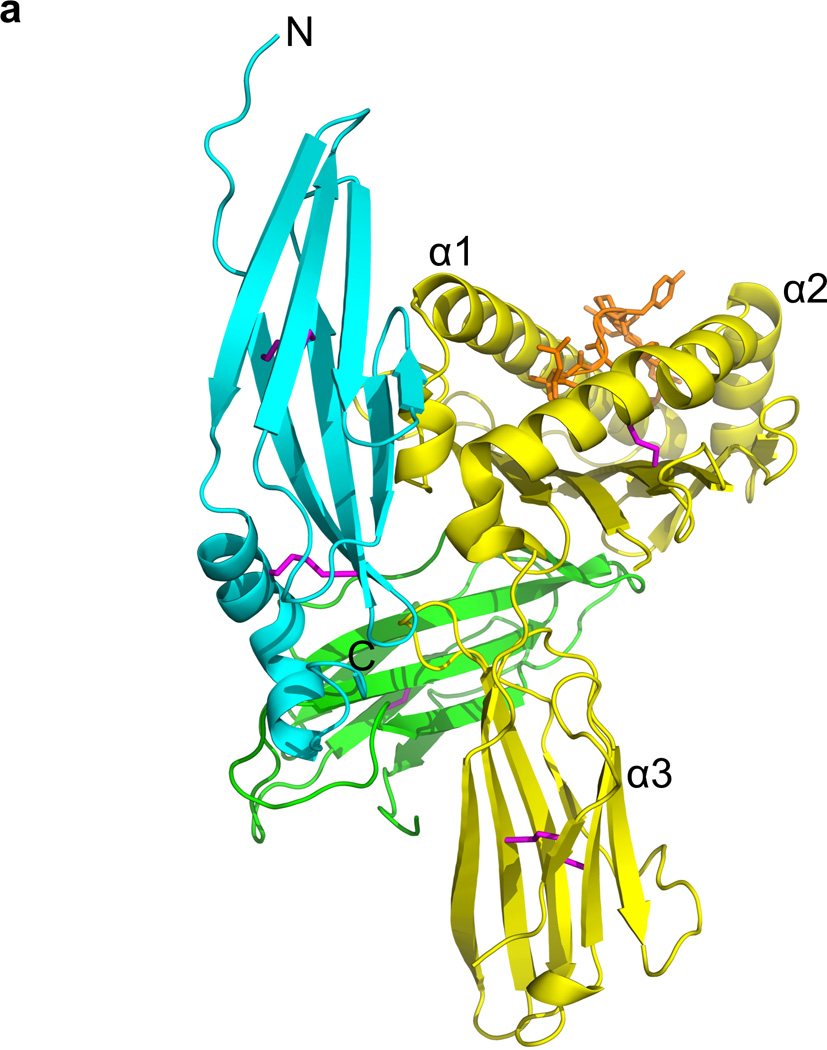
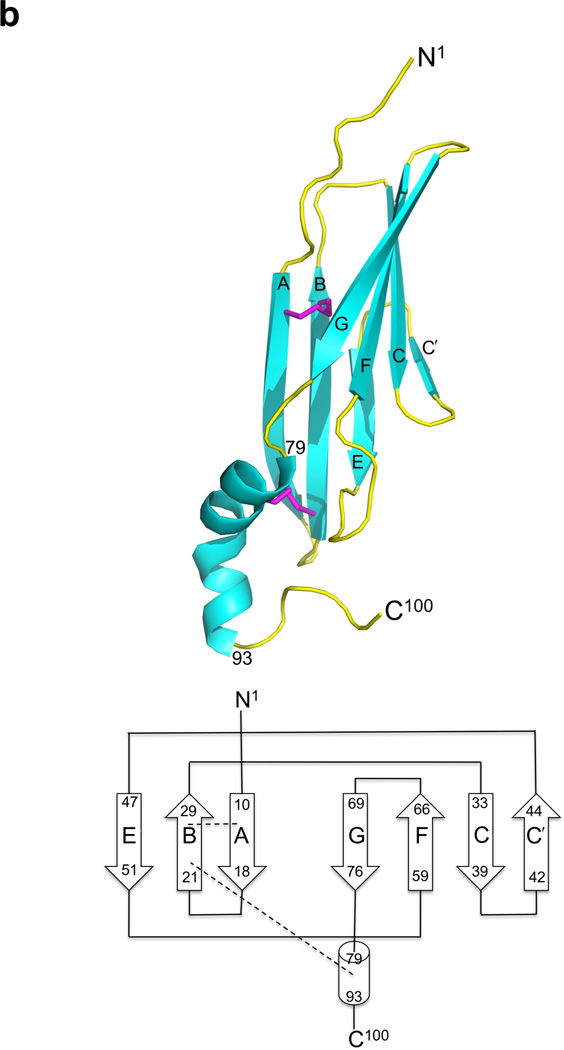
The structure of Ad2 E3-19K–HLA-A2. (a) Ribbon representation of the structure of Ad2 E3-19K–HLA-A2: E3-19K, cyan; heavy chain (α1-, α2-, and α3-domains), yellow; β2m, green; HIV-1 Tax (LLFGYPVYV) peptide, orange; and disulfide bond, magenta. The N- and C-termini of Ad2 E3-19K are labeled. (b) Ribbon representation of the structure of liganded Ad2 E3-19K (top) and its topology diagram (below). The N-terminal domain (residues 1–78), α-helix (residues 79–93), and tail (residue 94–100) are indicated. The disulfide bonds are shown in magenta. The β strands A, B, C, C′, E, F, and G, and N1- and C100-termini are labeled. The β strands are represented by arrows and the α-helix is shown as a cylinder. The two disulfide bonds are indicated by dashed lines.
The structure of Ad2 E3-19K contains a large N-terminal domain that forms two antiparallel β-sheets defined by β-strands A, B, and E and β-strands C, C′, F, and G (Fig. 1B). The C-terminus of this domain is connected to an α-helix and a tail (Fig. 1B). The two β-sheets of the N-terminal domain overlap with each other only partially, with an angle of approximately 30° between each other, such that overall this domains resembles a V-shaped structure. The apex of the V-shape is formed by contacting β-strands C′ and E and the opening is defined by β-strands A and G. This unusual relative arrangement of β-sheets confines the hydrophobic core to the deepest part of the V-shape. The hydrophobic core comprises residues of β-strands B (Cys22, Thr24, Ile26, and Cys28) and E (Val48 and Ala50) of the ABE β-sheet and β-strands C (Leu35, Ile37, and His39), C′ (Gly44), and F (Val62 and Val64) of the CC′FG β-sheet (Fig. 2); only one residue of the edge β-strand A (Val13), and none of the edge β-strand G, is part of the hydrophobic core. Other residues such as Pro9 and Trp52 of the large N-terminal domain, Phe79 and Met87 of the α-helix, and Trp96 of the tail also participate in forming the hydrophobic core (Fig. 2).
Figure 2.
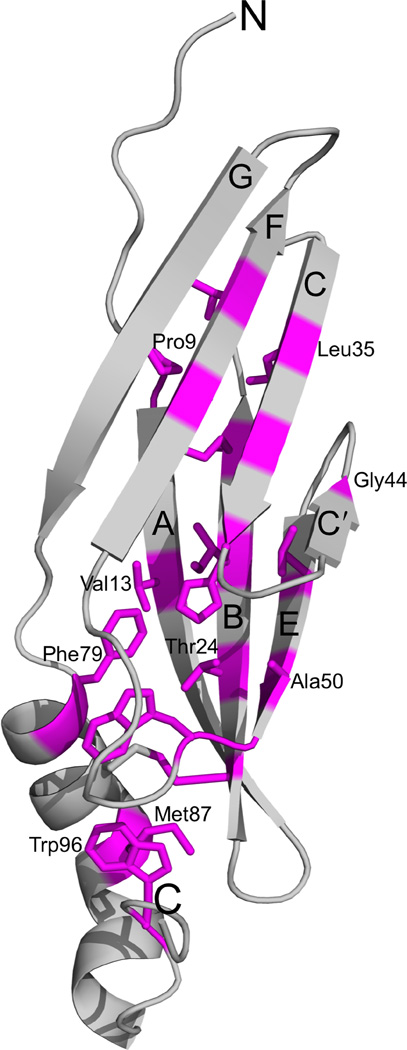
Hydrophobic core of Ad2 E3-19K. Ribbon representation of the structure of liganded Ad2 E3-19K showing the side chain of the core-forming residues (magenta) (some side chains are labeled for clarity). Core-forming residues (>90% buried) were determined by using SPDBV45. The β strands A, B, C, C′, E, F, and G, and N- and C-termini are labeled.
In addition to the hydrophobic core, a number of interactions keep the α-helix packed against the N-terminal domain. These include the disulfide bond between Cys83 and Cys22 and also several mostly water-mediated (water G271) hydrogen bonds between Ser90 (α-helix) and Ser17 (β-strand A) and Asn20 (AB-loop; loop connecting β-strands A and B). The well-ordered electron density of the N-terminal residues Lys2–Pro9 reveals that main chain atoms of Lys7, Glu8, and Pro9 form water-mediated hydrogen bonds (water G231, G224, and G237, respectively) with the side chains of Thr30 and Glu31 (BC-loop). Similarly, the side chain of Trp96 participates in a water-mediated (water G202) hydrogen-bond network that contributes to the well-defined backbone curvature of the tail. Overall, the presence of important hydrogen bonds at the N- and C-termini and of a hydrophobic core along the entire length of Ad2 E3-19K, appear to provide structural rigidity to the protein. Finally, it is clear that the strategically positioned disulfide bonds that link β-strands A to B (Cys11, Cys28) and the α-helix to the N-terminal domain (Cys22, Cys83) are critical structural features of Ad2 E3-19K, consistent with results from mutational analysis of cysteine residues in Ad2 E3-19K32.
The coordinates of Ad2 E3-19K were submitted to the DALI server33 to establish how E3-19K might be related to other proteins of known structures in the Protein Data Bank (PDB). The search revealed a relatively low degree of structural similarity with gp130. The closest structural relative is D2 of D2D3 gp130 (PDB 1pvh, rmsd of 3.4 Å) with a Z-score of 6.6. The next two best matches are D1 of D1D2 gp130 (PDB 1bqu, rmsd of 3.5 Å) and D2 of D1D2D3 gp130 (PDB 1i1r, rmsd of 3.7 Å) with Z-scores of 6.5, and 6.4, respectively. These domains adopt fibronectin type III folds. The superposition of these gp130-based structures onto the Ad2 E3-19K structure revealed that similarities are limited to 1 or 2 β-strands of the CC′FG β-sheet; no protein could be identified that aligned well with E3-19K in its entirety. No closely related structures were also found with the SSM server34. We conclude that the E3-19K protein has a unique tertiary structure.
The E3-19K–HLA-A2 binding interface
Ad2 E3-19K binds to the N-terminus of the α1-helix on HLA-A2, contacting also the C-terminus of the α2-helix and prominent loops of the α3-domain and β2m (Fig. 3). The E3-19K–HLA-A2 binding interface shields a molecular surface area of 1929 Å2 from solvent. The shape complementarity coefficient, Sc, of the complex was determined to be 0.68 (Sc = 1 for perfect geometrical fits 35).
Figure 3.
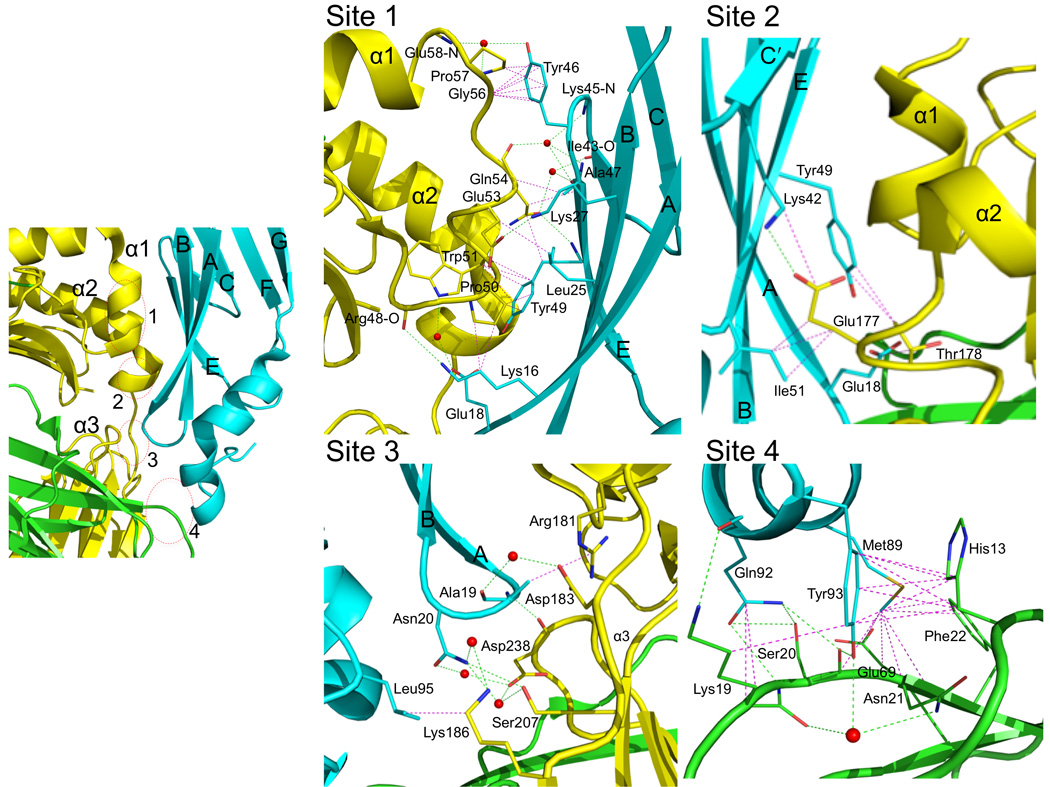
Interaction surface between Ad2 E3-19K and HLA-A2. Interaction sites 1, 2, 3 and 4 of Ad2 E3-19K–HLA-A2 are shown in separate panels using the same color code as in Figure 1(a). Nitrogen, oxygen and sulfur atoms are colored blue, red, and light orange, respectively. Hydrogen bonds (distance ≤ 3.5 Å) and salt bridges are indicated by dashed green lines, hydrophobic contacts (distance < 4.0 Å) are represented by dashed magenta lines. Water molecules are shown as red spheres. The β strands of Ad2 E3-19K and α1-, α2-, α3-domains of the heavy chain are labeled.
Each interaction site (referred to as sites 1, 2, 3, and 4) is discussed separately (Fig. 3). Site 1 is located at the N-terminus of the α1-helix on HLA-A2 and contributes 36% to the total buried surface area. The site features a salt bridge between Lys27 (β-strand B) of E3-19K and Glu53 of HLA-A2, as well as hydrogen bond interactions involving Lys16 and Tyr49 (β-strands A and E, respectively) of E3-19K and Arg48 and Gln54 of HLA-A2, respectively. These interactions are supplemented by a number of solvent-mediated hydrogen bonds and hydrophobic contacts. Site 2 is located at the C-terminus of the α2-helix on HLA-A2 and occupies 16% of the total buried surface area. The key interaction at site 2 is a salt bridge between Lys42 (β-strand C′) of E3-19K and Glu177 of HLA-A2. Site 3 involves prominent loops of E3-19K and the α3-domain of HLA-A2, contributing 17% of the total buried surface area. The interface at that site is defined by a number of mostly water-mediated hydrogen bonds and hydrophobic contacts. Site 4 involves the α-helix of E3-19K and β2m, representing 31% of the total buried surface area. Interactions at site 4 include mostly hydrogen bonds involving Gln92 and Tyr93 of E3-19K and several residues (Lys19, Ser20, and Asn21) of β2m, as well as a number of hydrophobic contacts. Overall, interactions at sites 1, 2, 3, and 4 define a novel binding surface on MHC I. No large conformational changes were observed in the polypeptide backbone of HLA-A2–Tax upon complex formation; the rmsd between free (PDB 1HHK) and bound HLA-A2–Tax is 0.66 Å for 384 equivalent Cα atoms.
The complementarity of charges at each interaction surface is shown in Fig. 4. The binding interface at site 1 includes a highly electropositive region contributed by E3-19K that targets an electronegative region on HLA-A2. Similarly, the interface region at site 2 is largely electropositive in nature for Ad2 E3-19K and complementarily electronegative for HLA-A2. Site 3, in contrast, is formed by areas of electronegativity and electropositivity on both E3-19K and HLA-A2. Finally, Site 4, which involves the α-helix of Ad2 E3-19K and β2m, is the most apolar region of the overall binding interface.
Figure 4.
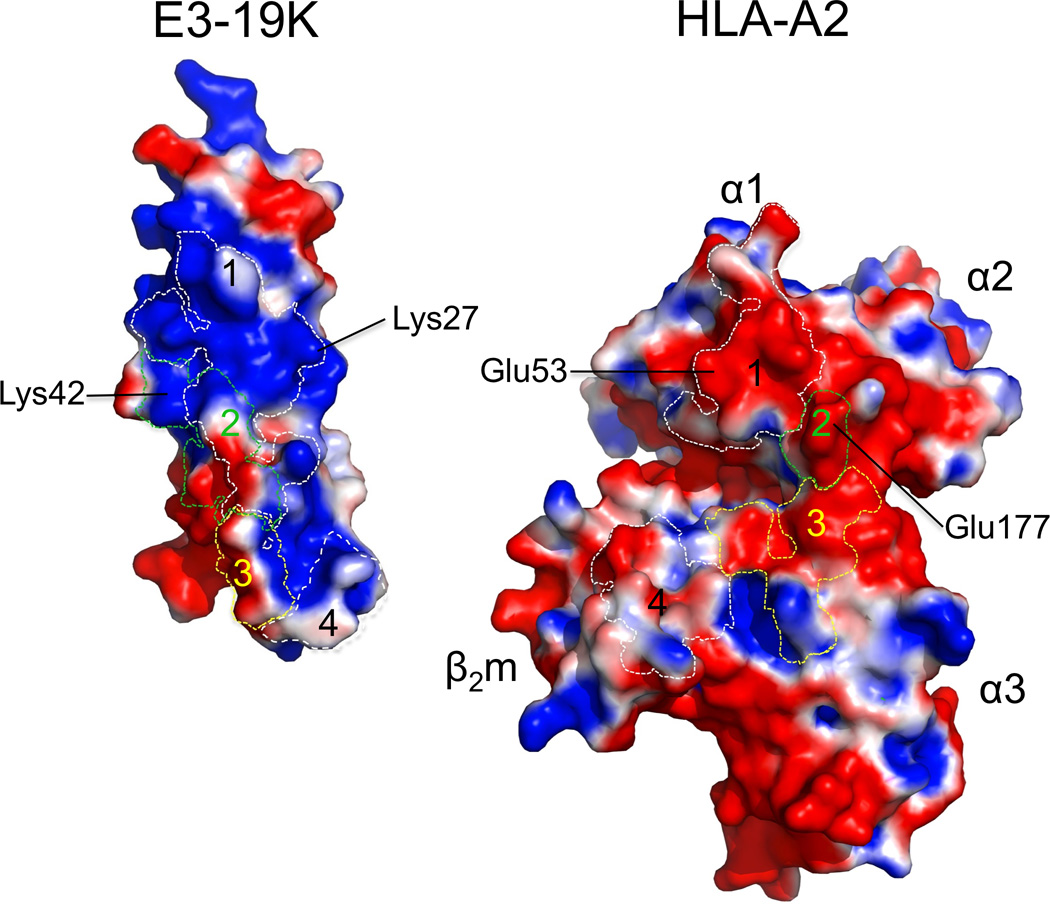
Electrostatic properties of interacting surfaces in Ad2 E3-19K and HLA-A2. Electrostatic surfaces of Ad2 E3-19K and HLA-A2 at interaction sites 1, 2, 3, and 4 (marked by dashed lines): electronegative (red, −1.000 kTe−1), electropositive (blue, +1.000 kTe−1), and apolar (white). Conserved contact residues in Ad2 E3-19K and HLA-A2 at site 1 (Lys27 and Glu53, respectively) and site 2 (Lys42 and Glu177, respectively) that form salt bridges are labeled. The α1-, α2-, and α3-domains of the heavy chain and β2m are labeled.
E3-19K proteins across Ad species; roles of conserved residues
A sequence alignment of E3-19K proteins of different species reveals residues that have been maintained by evolution as strictly and highly conserved (blue and cyan, respectively, in Supplementary Fig. 1). These residues were mapped in the Ad2 E3-19K structure (Fig. 5). The analysis shows that several strictly conserved residues are clustered in the N-terminal domain, particularly in the central β-strands B and F (Fig. 5). The N-terminus of the α-helix and tail both also contain several strictly conserved residues. Strict conservation is also observed for four cysteine residues that form the two disulfide bonds (Cys11, Cys28 and Cys22, Cys83).
Figure 5.
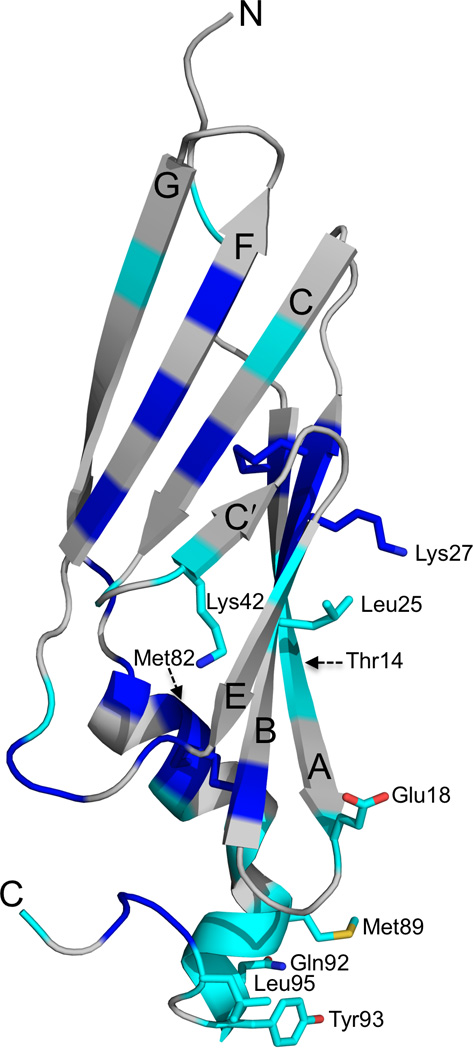
Conserved residues in Ad2 E3-19K. Strictly (blue) and highly (cyan) conserved residues are mapped in the structure of Ad2 E3-19K. The side chains of strictly and highly conserved residues at sites 1 (Glu18, Leu25, Lys27), 2 (Glu18, Lys42), 3 (Leu95), and 4 (Met89, Gln92, Tyr93) are indicated; nitrogen, oxygen and sulfur atoms are colored blue, red, and light orange, respectively. The positions of Thr14 and Met82 side chains, which are important for MICA and MICB interaction27, are indicated by dashed arrows. The β strands A, B, C, C′, E, F, and G, and N- and C-termini are labeled.
Interestingly, nine strictly (Cys22, Ile26, Cys28, Ile37, Trp52, Val62, Val64, Phe79, and Trp96) and four highly (Val13, Leu35, Val48, and Met87) conserved residues (Supplementary Fig. 1) are part of the hydrophobic core (compare Figs. 2 and 5). Thus, several residues have been maintained evolutionarily because of the key role that they play in the structural integrity of the E3-19K fold. Other strictly (Lys27) and highly (Glu18, Leu25, Lys42, Met89, Gln92, and Tyr93, and Leu95) conserved residues have been maintained because of the important role that they play in the MHC I-binding function of E3-19K (residues marked in red in Supplementary Fig. 1 and side chains indicated inFig. 5). These residues mediate hydrophilic and hydrophobic interactions with HLA-A2 at site 1 (Glu18, Leu25, Lys27), site 2 (Glu18 and Lys42), site 3 (Leu95), and site 4 (Met89, Gln92, and Tyr93). Importantly, sequence alignment reveals strict conservation of the WPP motif (residue 96–98) (Supplementary Fig. 1). Trp96 is part of the hydrophobic core (compare Figs. 2 and 5) and participates in a network of solvent-mediated hydrogen bonds, as mentioned above. This effect, together with the exceptional conformational rigidity of proline residues, suggests that the WPP motif plays an important structural role (see DISCUSSION).
Conserved MHC I residues at the binding interface
A sequence alignment of consensus HLA-A, -B, and -C heavy chains at each of the three interaction sites on HLA-A2 heavy chain is shown in Fig. 6. The analysis shows that Ad2 E3-19K binds to conserved residues of the HLA-A2 heavy chain at sites 1 and 2 but to polymorphic residues at site 3. Site 4, which involves β2m, is obviously strictly conserved in HLA molecules. Overall, our structure shows that Ad2 E3-19K interacts with largely conserved residues of HLA-A2.
Figure 6.
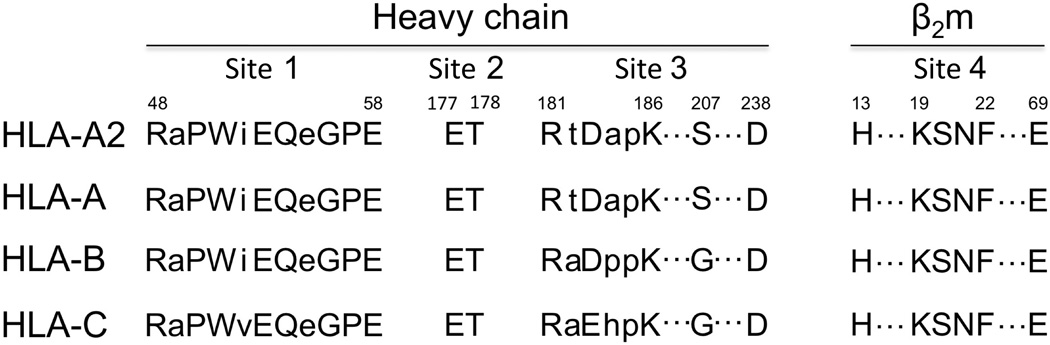
Amino acid sequence alignment at the four interaction sites. Alignment of HLA-A2 and consensus HLA-A, -B, and -C heavy chain sequences at sites 1, 2, and 3. Interaction site 4 on β2m is also indicated. Capital letters designate contact residues at each of the four sites (see legend of Figure 3). Consensus sequences were obtained from the IMGT-HLA Sequence Database (www.ebi.ac.uk/imgt/hla/align.html).
DISCUSSION
It was previously speculated that the E3-19K protein resembles an Ig-like domain26, our structure shows that this is not the case. Structural alignment searches against the DALI and SSM servers did not identify any structures that aligned well with Ad2 E3-19K in its entirety, indicating a unique tertiary structure.
E3-19K proteins share relatively low levels of sequence homology in their N-terminal 100 residues (Supplementary Fig. 1). In spite of this, several residues in this region that are part of the hydrophobic core are strictly or highly conserved (Supplementary Fig. 1). This includes residue Trp96, which together with Pro97 and Pro98 likely provide conformational rigidity to the tail region of Ad2 E3-19K. Such rigidity of the tail should restrict conformational freedom of the membrane-anchored ER-lumenal domain. The strictly conserved WPP motif may thus be important for correctly aligning E3-19K onto MHC I. It is of note that a mutation of Trp96 in Ad2 E3-19K decreased interaction with MHC I27. Thus, it is clear that the conservation of key structural determinants in E3-19K proteins reflects a strong evolutionary pressure to maintain a function-specific tertiary structure, consistent with our knowledge that all E3-19K proteins display an MHC I-binding function.
The characterization of interaction in E3-19K–MHC I pairs is important because of the role that this association plays in Ad pathogenesis. We suggest that Ad2 E3-19K residues Lys27 (site 1), which is strictly conserved (Supplementary Fig. 1), and Lys42 (site 2), which is conserved by charge, are key determinants of the MHC I-binding function of E3-19K proteins. We showed previously the importance of the electrostatic interaction mediated by Lys42 from a mutation of E177K in HLA-A*1101 which abolished interaction with E3-19K of species B, C, D, and E23. Furthermore, we suggest that the highly conserved Ad2 E3-19K residues Gln92 and Tyr93 (site 4) (Supplementary Fig. 1) are important determinants of interaction in E3-19K–MHC I pairs. Consistent with this, we showed that a Y93G mutation in Ad2 E3-19K abolished interaction with HLA-A molecules29. We, and others, have also shown that mutation of non-contact Ad2 E3-19K residues such as Met87 and Lys91 (site 4) destabilized Ad2 E3-19K–MHC I interaction23, 27. Because the shape complementarity is high at site 4 (Sc = 0.81), any mutations that affect the geometrical fit between the α-helix of E3-19K and β2m is expected to disrupt the interconnected network of interactions at that site (Fig. 3). We have evidence in support of this view from studies in which we showed that for a given MHC class I molecule, Ad37 E3-19K of species D displays the weakest binding affinities relative to those of species B, C, and E23. E3-19K proteins of species D are not only most divergent at site 4 (Supplementary Fig. 1) but also carry non-conserved substitutions at each of the three contact residues; M89V, Q92L, and Y93H. Taken together, this analysis suggests that the binding mode of Ad2 E3-19K onto HLA-A2 is likely to represent a universal mode of interaction for all E3-19K proteins. Finally, site 3 is a region of the Ad2 E3-19K–HLA-A2 interface where the consensus sequences of HLA heavy chains are most divergent (Fig. 6), and it also lacks the kind of key interaction involving conserved residues that are present at the other three sites. As such, interactions at site 3 are expected to be responsible for the variability in binding affinities that E3-19K proteins display for MHC I.
There are many particular features of the Ad2 E3-19K–HLA-A2 structure (Fig. 1A) that exemplify the fine-tuned nature of the E3-19K immunomodulatory function. First, E3-19K ensures “cross-reactivity” with MHC I by binding largely to conserved MHC residues. Second, E3-19K has evolved to circumvent peptide specificity by binding away from the groove. Third, and importantly, E3-19K displays high-affinity interaction with MHC I22, 23. Fourth, Ad2 E3-19K has evolved an elongated structure that permits contacts over the entire length of HLA-A2, including the heavy chain and β2m. In this regards, we showed that the first N-terminal 93 residues of Ad2 E3-19K are sufficient to maintain high-affinity interaction with HLA-A*1101 (Ref. 29), in perfect agreement with our structure. Finally, E3-19K binds to the N-terminus, rather than the C-terminus, of the groove. The C-terminus of the groove is thought to represent the binding region for the class I assembly proteins onto immature MHC I36. Thus, in targeting the N-terminus of the groove, E3-19K likely ensures that it can maintain an association with both immature (peptide-free) and mature (peptide-filled) forms of MHC I. In support of this, we showed that Ad2 E3-19K binds to both peptide-free and peptide-filled HLA-A*1101 (Ref. 31).
The conformationally-sensitive monoclonal antibody (mAb) 64-3-7, which is specific for “open” MHC I37, was shown to bind to a region of MHC I comprising Arg48 and Pro50 (site 1 in Fig. 3). Because 64-3-7 can distinguish between “open” and “closed” MHC I, it was suggested that the region of the groove where 64-3-7 binds to is folded differently in these two forms of MHC I37. In the context of our structure, this is entirely consistent with our findings that Ad2 E3-19K displays differential binding affinities for peptide-free (“open”) and peptide-filled (“closed”) HLA-A*1101 (Ref. 31). It is also consistent with a previous report showing that E3-19K cannot be co-immunoprecipitated with 64-3-7 (Ref. 38). Taken together, this suggests that the stretch of MHC residues between Arg48 to Glu58 is likely to undergo major peptide-induced conformational changes, reinforcing the notion that peptide-induced conformational changes occur at the N-terminus of the groove37, 39.
We showed previously that E3-19K proteins bind to HLA-A and -B molecules but shows no avidity for HLA-C molecules22, 23. Other immunomodulatory proteins of large DNA viruses display the same locus-specificity40, 41, 42. An analysis of interaction between Ad2 E3-19K and HLA-A2 at site 3 (Fig. 3), in the context of the crystal structure of free HLA-Cw*0403 (PDB 1QQD), suggests that residues Glu183 and Gly207 in HLA-Cw*0403 cannot mediate interactions shown at site 3. We therefore suggest that these two residues play a role in the inability of E3-19K to bind HLA-C. Notably, we have new data showing that Ad2 E3-19K and Ad4 E3-19K are unable to associate with HLA-E (our unpublished results). HLA-E carries Glu183 and Gly207 and in the context of the structure of free HLA-E*0103 (PDB 3BZE), both of these residues also cannot mediate interactions shown at site 3. This lends further support to the idea that MHC residues Glu183 and Gly207 modulate the inability of E3-19K to associate with HLA-C and -E.
Because of the effects that E3-19K has on suppressing the expression of the MHC I homologs MICA and MICB on Ad-infected cells43, it is presumed that E3-19K binds to MICA and MICB and that E3-19K residues Thr14 and Met82 play a role in this interaction27. We have mapped the position of the side chains of Thr14 and Met82 in the structure of Ad2 E3-19K (Fig. 5). The implication of this analysis is that E3-19K likely uses a different surface to sequester MICA and MICB from that of its MHC I binding surface (Fig. 5). Furthermore, that mutation of Thr14 and Met82 in Ad2 E3-19K had essentially no effect on the MHC I binding function of E3-19K27 is entirely consistent with our structure (Figs. 1 and 5).
In conclusion, the structure of Ad2 E3-19K–HLA-A2 represents a convincing example of co-evolution between a viral immunomodulatory protein and its host MHC I ligand. Our studies provide structural explanations for how E3-19K achieves promiscuous and high-affinity binding onto HLA-A and -B, and insights into its inability to engage with HLA-C and HLA-E. Interestingly, a comparison of our structure with that of US2 immunomodulatory protein from human cytomegalovirus bound to HLA-A2 (Ref. 44) shows that both proteins have common contact residues on HLA-A2 at site 3 (Fig. 3). This has implications for understanding the molecular basis of co-infection of the same host cell by two different persistent viruses.
On-line METHODS
Cloning and protein expression
The complementary DNA (cDNA) encoding the ER-lumenal domain (residues 1–100) of Ad2 E3-19K (referred to as E3-19K) was generated by polymerase chain reaction. This was carried out using the plasmid pBS-Ad2E3-19K (gift from H.-G. Burgert) containing the cDNA of full-length Ad2 E3-19K as a template and the appropriate forward and reverse primers. The amplified cDNA was ligated into the PLM1 vector (gift from D.C. Wiley) using the BamH1 and EcoR1 restriction sites. Ad2 E3-19K, HLA-A2 heavy chain (residues 1–275), and β2m (residues 0–99) were expressed in the Escherichia coli strain BL21(DE3)pLysS (Stratagene, La Jolla, CA) as inclusion bodies. Inclusion bodies were isolated, purified from cell pellets, and solubilized as described previously46.
Assembly of HLA-A2–Tax and Ad2 E3-19K–HLA-A2
HLA-A2–Tax was reconstituted in vitro from the urea-solubilized inclusion bodies of HLA-A2 heavy chain (1 µM) and β2m (2 µM) in the presence of the Tax peptide (LLFGYPVYV) (10 µM) in an oxidative refolding buffer46. Stock solutions of purified HLA-A2–Tax (10–30 mg ml−1) in 20 mM Tris–HCl (pH 7.5), 150 mM NaCl were kept at −80°C. The Ad2 E3-19K–HLA-A2 complex was assembled using a modified version of this approach. The urea-solubilized inclusion bodies of Ad2 E3-19K were added at 4°C under rapid stirring to a final concentration of 1 µM to an oxidative refolding buffer consisting of 50 mM Tris–HCl (pH 8.0), 1 mM ethylenediaminetetraacetic acid , 400 mM L-arginine, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 1 µM phenylmethylsulfonyl fluoride, and containing submolar amounts (0.2 µM) of refolded, purified HLA-A2–Tax. The presence of refolded HLA-A2–Tax in the buffer of Ad2 E3-19K was absolutely essential for formation of the Ad2 E3-19K–HLA-A2 complex. The refolding mixture was incubated at 4°C. After 24 hours, the mixture was concentrated in an Amicon stirred cell followed by purification on a Superdex 200 HR 10-30 column in 20 mM Tris–HCl (pH 7.5), 100 mM NaCl. The purified Ad2 E3-19K–HLA-A2 complex generated from this rescue refolding strategy was characterized by sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE) gel and native PAGE gel. Stock solutions of purified Ad2 E3-19K–HLA-A2 (20-40 mg ml−1) in 20 mM Tris–HCl (pH 7.5), 100 mM NaCl were kept at −80°C. The rescue refolding strategy described here is applicable to other complexes involving recalcitrant proteins such as Ad2 E3-19K.
Crystallization and data collection
Crystallization conditions of the Ad2 E3-19K–HLA-A2 complex were searched with the aid of a Tecan Freedom EVO 200 robot using sitting drops. Complex solution (1 µL, 10 mg ml−1) was dispensed into each well of a Corning 96-well plate, mixed with an identical volume of precipitant solution, and equilibrated against 100 uL of reservoir solution. The plates were incubated at 17°C. The initial crystallization condition was found using Crystal Screen Index (Hampton Research) as solution #22, 0.8 M succinic acid (pH 7.0). Optimization of the crystallization condition was carried out using the Additive Screen (Hampton Research) and the hanging-drop vapor-diffusion method. Crystals used for data collection were grown by mixing 2 µL of 10 mg ml−1 protein complex solution with 2 µL of 0.8 M succinic acid (pH 7.0) and 0.2 µL of 0.1 M calcium chloride dihydrate as additive. Crystals grew over 20 d to dimensions of ~400 × 200 × 100 µm. Prior to data collection, the crystals were soaked for about 1–2 minutes in mineral oil (Sigma) as the cryoprotectant and then flash-frozen in liquid nitrogen. X-ray diffraction data for the Ad2 E3-19K–HLA-A2 complex was collected to 1.95Å resolution with a MAR-225 CCD detector at SER-CAT beamline 22-BM-D of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). The space group was determined to be I212121 (a=113.19 Å, b=133.67 Å, c=196.72 Å). Data were processed and scaled with the HKL-2000 program package47.
Structure determination and refinement
The structure of Ad2 E3-19K–HLA-A2 complex was determined by molecular replacement in AMoRe48 using free HLA-A2–Tax (PDB 1HHK) as a search model. Two solutions were identified for both the rotation and translation functions, corresponding to two molecules of Ad2 E3-19K–HLA-A2 in the asymmetric unit. After molecular replacement, the electron density maps were calculated and initial refinements were examined manually and residues with a poor fit to the electron density map were omitted from the model. The electron density improved using DM program from the CCP4 package49. Subsequent model rebuilding was carried out in Coot50. The extended model was then refined using simulated annealing, energy minimization, and restrained individual B factor in CNS51. In the first refinement cycles, noncrystallographic symmetry (NCS) restraints were applied to the main chain atoms (loops excluded) using data to 3.0 Å, resulting in Rfree and Rwork values of 0.43 and 0.39, respectively. In the final refinement cycles, NCS restraints were released and a composite omit map was calculated to eliminate model bias. Water molecules were added using the CNS program51 and checked with the FO-FC electron density map (>3 σ) at 3.5 Å or less from hydrogen bond donors or acceptors. A total of 472 water molecules were added to the model. Throughout refinement, agreement between the model and the observed data was monitored by calculating Rfree based on 10% of the reflections. The final Rwork and Rfree values (with a bulk solvent correction) are 21.0% and 24.1%, respectively, for all reflections between 37.06–1.95 Å (Table 1). In the final model, the structural geometry was checked using PROCHECK52. All backbone φ–ψ torsion angles of the model were within allowed regions of the Ramachandran plot. Because of poor quality or missing electron density, the final model lacks residue 1 of chain G, and residues 1, 99, and 100 of chain H. Buried surface areas were calculated with the program AreaIMol in the CCP4 package, using a probe radius of 1.4 Å. All structural figures were prepared using PyMol53.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in whole or in part, by the US National Institute of Allergy and Infectious Diseases grants R01 AI045070 and R56 AI102468 (to MB). We thank Bernard Santarsiero for help with x-ray data collection. Technical support by Lixia Qian for cell culture and protein biochemistry is also acknowledged.
Footnotes
ACCESSION CODE
The atomic coordinates and structure factors of the Ad2 E3-19K–HLA-A2 complex have been deposited in the PDB with ID code 4E5X and will be released immediately upon publication.
AUTHOR CONTRIBUTIONS
LL, crystallographic and biochemical studies and manuscript preparation; YM, biochemical studies; MB, project supervisor and manuscript preparation and principal manuscript author.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Berk AJ. Adenoviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, editors. Fields Virology. 5th. Philadelphia: Lipincott Williams & Wilkins; 2007. pp. 2395–2436. [Google Scholar]
- 2.Wold WSM, Horwitz MS. Adenoviridae: The viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, editors. Fields Virology. 5th. Philadelphia: Lipincott Williams & Wilkins; 2007. pp. 2355–2394. [Google Scholar]
- 3.Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 4.Burgert H-G, Ruzsics Z, Obermeier S, Hilgendorf A, Windheim M, Elsing A. Subversion of host defense mechanisms by Adenoviruses. In: Koszinowski UH, Hengel H, editors. Viral Proteins Counteracting Host Defenses. Berlin: Springer-Verlag; 2002. pp. 273–318. [DOI] [PubMed] [Google Scholar]
- 5.Andersson M, Paabo S, Nilsson T, Peterson PA. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell. 1985;43:215–222. doi: 10.1016/0092-8674(85)90026-1. [DOI] [PubMed] [Google Scholar]
- 6.Andersson M, McMichael A, Peterson PA. Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 1987;138:3960–3966. [PubMed] [Google Scholar]
- 7.Burgert HG, Kvist S. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell. 1985;41:987–997. doi: 10.1016/s0092-8674(85)80079-9. [DOI] [PubMed] [Google Scholar]
- 8.Burgert H-G, Kvist S. The E3/19K protein of adenovirus type 2 binds to the domains of histocompatibility antigens required for CTL recognition. EMBO J. 1987;6:2019–2026. doi: 10.1002/j.1460-2075.1987.tb02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flomenberg P, Piaskowski V, Truitt RL, Casper JT. Human adenovirus-specific CD8+ T-cell responses are not inhibited by E3-19K in the presence of gamma interferon. J. Virol. 1996;70:6314–6322. doi: 10.1128/jvi.70.9.6314-6322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawle FC, Tollefson AE, Wold WSM, Gooding LR. Mouse anti-adenovirus cytoxic T lymphocytes. J. Immunol. 1989;138:3960–3966. [PubMed] [Google Scholar]
- 11.Ginsberg HS, et al. Role of early region 3 (E3) in pathogenesis of Adenovirus disease. Proc. Natl. Acad. Sci. USA. 1989;86:3823–3827. doi: 10.1073/pnas.86.10.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wold WSM, Gooding LR. Adenovirus regions E3 proteins that prevent cytolysis by cytotoxic T cells and tumor necrosis factor. Mol. Biol. Med. 1989;6:433–452. [PubMed] [Google Scholar]
- 13.Feuerbach D, et al. Identification of amino acids within the MHC molecule important for the interaction with the adenovirus protein E3/19K. J. Immunol. 1994;153:1626–1636. [PubMed] [Google Scholar]
- 14.Flomenberg P, Gutierrez E, Hogan KT. Identification of class I MHC regions which bind to the adenovirus E3-19K protein. Molec. Immunol. 1994;31:1277–1284. doi: 10.1016/0161-5890(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 15.Gabathuler R, Levy F, Kvist S. Requirement for the association of adenovirus type 2 E3-19K wild-type and mutant proteins with HLA antigens. J. Virol. 1990;64:3679–3685. doi: 10.1128/jvi.64.8.3679-3685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermiston TW, Tripp RA, Sparer RA, Gooding LR, Wold WSM. Deletion mutation analysis of the adenovirus type 2 E3-gp19K protein: identification of sequences within the endoplasmic reticulum lumenal domain that are required for class I antigen binding and protection from adenovirus-specific cytotoxic T lymphocytes. J. Virol. 1993;67:5289–5298. doi: 10.1128/jvi.67.9.5289-5298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox JH, Bennink JR, Yewdell JW. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J. Exp. Med. 1991;174:1629–1637. doi: 10.1084/jem.174.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paabo S, Bhat BM, Wold WSM, Peterson PA. A short sequence in the COOH-terminus makes an adenovirus membrane glycoprotein a resident of the endoplasmic reticulum. Cell. 1987;50:311–317. doi: 10.1016/0092-8674(87)90226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Severinsson L, Martens I, Peterson PA. Differential association between two human MHC class I antigens and an adenoviral glycoprotein. J. Immunol. 1986;137:1003–1009. [PubMed] [Google Scholar]
- 20.Deryckere F, Burgert H-G. Early region 3 of adenovirus type 19 (subgroup D) encodes an HLA-binding protein distinct from that of subgroups B and C. J. Virol. 1996;70:2832–2841. doi: 10.1128/jvi.70.5.2832-2841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korner H, Burgert H-G. Down-regulation of HLA antigens by the adenovirus type 2 E3/19K protein in a T-lymphoma cell line. J. Virol. 1994;68:1442–1448. doi: 10.1128/jvi.68.3.1442-1448.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Fu J, Bouvier M. Allele- and locus-specific recognition of class I MHC molecules by the immunomodulatory E3-19K protein from adenovirus. J. Immunol. 2007;178:4567–4575. doi: 10.4049/jimmunol.178.7.4567. [DOI] [PubMed] [Google Scholar]
- 23.Fu J, Li L, Bouvier M. Adenovirus E3-19K proteins of different serotypes and subgroups have similar, yet distinct, immunomodulatory functions towards major histocompatibility class I molecules. J. Biol. Chem. 2011;286:17631–17639. doi: 10.1074/jbc.M110.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermiston TW, Tripp RA, Sparer RA, Gooding LR, Wold WSM. Deletion mutation analysis of the adenovirus type 2 E3-gp19K protein: identification of sequences within the endoplasmic reticulum lumenal domain that are required for class I antigen binding and protection from adenovirus-specific cytotoxic T lymphocytes. J. Virol. 1993;67:5289–5298. doi: 10.1128/jvi.67.9.5289-5298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flomenberg P, Szmulewicz J, Gutierrez E, Lupatkin H. Role of the adenovirus E3-19K conserved region in binding major histocompatibility complex class I molecules. J. Virol. 1992;66:4778–4783. doi: 10.1128/jvi.66.8.4778-4783.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee D, Maizel JV. Homology of adenoviral E3 glycoprotein with HLA-DR heavy chain. Proc. Natl. Acad. Sci. USA. 1984;81:6039–6043. doi: 10.1073/pnas.81.19.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sester M, et al. Conserved amino acids within the adenovirus 2 E3/19K protein differentially affect downregulation of MHC class I and MICA/B proteins. J. Immunol. 2010;184:255–257. doi: 10.4049/jimmunol.0902343. [DOI] [PubMed] [Google Scholar]
- 28.Menz B, Sester M, Koebernick K, Schmid R, Burgert H-G. Structural analysis of the adenovirus type 2 E3-/19K protein using mutagenesis and a panel of conformation-sensitive monoclonal antibodies. Molec. Immunol. 2008;46:16–26. doi: 10.1016/j.molimm.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Fu J, Bouvier M. Determinants of the endoplasmic reticulum (ER) lumenal-domain of the Adenovirus serotype 2 E3-19K protein for association with and ER-retention of major histocompatibility complex class I molecules. Molec. Immunol. 2011;48:532–538. doi: 10.1016/j.molimm.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wold WSM, Cladaras C, Deutscher SL, Kapoor QS. The 19-kDa glycoprotein coded by region E3 of adenovirus. Purification, characterization, and structural analysis. J. Biol. Chem. 1985;260:2424–2431. [PubMed] [Google Scholar]
- 31.Liu H, Stafford WF, Bouvier M. The endoplasmic reticulum lumenal domain of the adenovirus type 2 E3-19K binds to peptide-filled and peptide-deficient HLA-A*1101 molecules. J. Virol. 2005;79:13317–13325. doi: 10.1128/JVI.79.21.13317-13325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sester M, Burgert H-G. Conserved cysteine residues within the E3/19K protein of adenovirus type 2 are essential for binding to major histocompatibility complex antigens. J. Virol. 1994;68:5423–5432. doi: 10.1128/jvi.68.9.5423-5432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acid Res. Trends Biochem Sci. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three-dimension. Acta Crystallogr. Sect. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 36.Bouvier M. Accessory proteins and the assembly of human class I MHC molecules. A molecular and structural perspective. Mol. Immunol. 2003;39:697–706. doi: 10.1016/s0161-5890(02)00261-4. [DOI] [PubMed] [Google Scholar]
- 37.Yu YYL, Myers NB, Hilbert CM, Harris MR, Balendiran GK, Hansen TH. Definition and transfer of a serological epitope specific for peptide-empty forms of MHC class I. Int. Immunol. 1999;11:1897–1905. doi: 10.1093/intimm/11.12.1897. [DOI] [PubMed] [Google Scholar]
- 38.Morris CR, et al. The amyloid precursor-like protein 2 and the adenoviral E3/19K protein both bind to a conformational site on H-2Kd and regulate H-2Kd expression. J. Biol. Chem. 2003;278:12618–12623. doi: 10.1074/jbc.M208203200. [DOI] [PubMed] [Google Scholar]
- 39.Mage MG, et al. The peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics. J. Immunol. 2012;189:1391–1399. doi: 10.4049/jimmunol.1200831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gewurz BE, Wang EW, Tortorella D, Schust DJ, Ploegh HL. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 2001;75:5197–5204. doi: 10.1128/JVI.75.11.5197-5204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishido S, Wang C, Lee B-S, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen GB, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 43.McSharry BP, et al. Adenovirus E3/19K promotes evasion of NK cell recognition by intracellular sequestration of the NKG2D ligands major histocompatibility complex class I chain-related proteins A and B. J. Virol. 2008;82:4585–4594. doi: 10.1128/JVI.02251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gewurz BE, et al. Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6794–6799. doi: 10.1073/pnas.121172898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 46.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. U S A. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology: Macromolecular Crystallography, Part A. Vol. 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 48.Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 49.Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Cryst. 1993;D49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 53.Delano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


