Abstract
Hemagglutinin (HA) of influenza A has been reported as the key protein in viral infection. Therefore, the density and the dynamic pattern of this protein in viral envelope will affect the virus to infect target cells. We used a lentiviral system to study the influenza A H1N1 viral infection. Herein we demonstrate that the influenza non-structural proteins (NS) significantly promote viral infection. By substituting NS gene segment from an H1N1 genome set of A/WSN/1933 with the NS segment isolated from another H1N1 substrain genome set, China246, we found that viral infection tropism was significantly altered. The reassortant H1N1 shows almost identical infectivity compared with its parental virus, A/WSN/1933, for the human epithelial cell line HOT, but shows only 1/100 infectivity of its parental virus when infecting the Madin-Darby canine kidney (MDCK) cell line. These results suggest that not only is NS important in the infectivity of human influenza virus, but that it may play a critical role in viral tropism, allowing the virus to mutate and spread to other species.
Keywords: Influenza A, H1N1, Lentivirus, influenza non-structural proteins, NS, Infection tropism
Introduction
Human viral influenza is highly contagious, with person-to-person spread by aerosol droplets that mainly infect the epithelial cells of the respiratory tract [1,2]. Identification of the mechanism by which influenza virus A infects human epithelial cells is important for planning strategies to prevent and treat influenza infection. Influenza viruses bind through a viral envelope protein, hemagglutinin, onto sialic acid sugars on the surfaces of epithelial cells. In addition to HA, there are other viral proteins found in viral envelope, including neuraminidase (NA), matrix protein 1 (M1), and non-structural protein 2 (NS2) [3,4,5,6,7]. NA cleaves terminal neuraminic acid residues from glycan structures on the surface of the infected cell [4,6,8]. This promotes the release of progeny viruses and the spread of the virus from the host cell to uninfected surrounding cells. NA may also be involved HA cleavage, which is required for viral entery [9]. Other proteins, such as non-structural protein 1 (NS1) and/or matrix protein 2 (M2), may also be involved in viral infection by regulating the presence of HA, NA, NS2, and/or M1 proteins on the viral envelope [10,11].
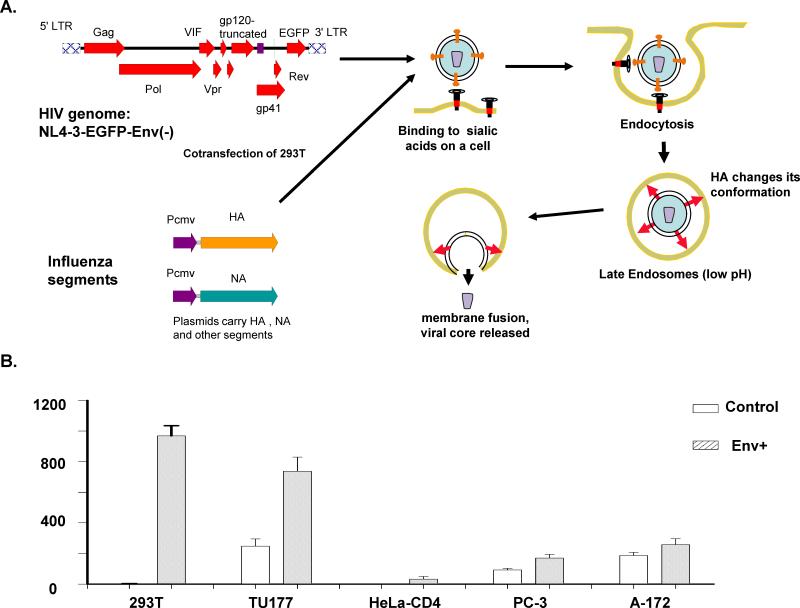
In this manuscript, we tested an HIV-based lentiviral system to study influenza viral proteins. In previous studies, the lentiviral system has been used to study the infection of vesicular stomatitis viral G-protein and Sindbis viral envelope proteins [12,13]. These non-HIV viral envelope proteins enable the HIV-based lentiviral genome to enter human cells similar to their parental viruses. We removed the genes encoding HIV envelope proteins from the viral genome and inserted the green fluorescent protein (GFP) gene into the genome (Fig. 1A). The expression of GFP provided a sensitive way to quantify the infection of virus. Using this system, we can selectively use different influenza proteins to construct viral envelopes and study the roles of these proteins in viral entry.
Fig. 1.
Use of a lentiviral vector to quantitatively study the roles of influenza envelope proteins. A) We hypothesize that an influenza viral envelope protein-modified lentiviral vector uses a similar way as influenza virus to infect human cells. A plasmid carrying the env gene-truncated HIV genome was used to co-transfect 293T cells with plasmids carrying influenza HA, NA, NS, and M segments. The virus produced was used to infect various cells. B) Infection of various human cell lines by a lentiviral vector wrapped with an influenza envelope. Plasmids carrying segments of HA, NA, M, and NS were used to co-transfect the packaging cells (293T) with plasmid NL4-3-EGFP-Env(-), which provides the viral core. The control virus was prepared using co-transfected plasmids NL4-3-EGFP-Env(-) and pCDNA3.1, which is a plasmid with no transgene.
Materials and Methods
Cell lines and maintenance
Epithelial cell lines 293T, HeLa-CD4, TU177, PC-3, CASK1, GH329, CaCo2, and VK2/E6E7 have been previously described [14,15,16]. Both the MDCK and Vero cell lines were purchased from the American Type Culture Collection (ATCC). Human oral epithelial cell line (HOT) was derived from normal human epithelial cells, as described previously [17]. All of these cell lines were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS).
Influenza A genome segments
One set of influenza A genome segments was from the A/WSN/1933 viral strain (NCBI Taxonomy ID: 382835). The sequences of these segments were inserted into the pHW12 plasmid backbone [18]. A NS viral DNA clone from China influenza A H1N1 strain #246 was obtained from the Chinese Academy of Medical Sciences.
EGFP gene-modified HIV generated by co-transfection with influenza envelope proteins
Virus was derived from HIVNL4-3, with part of the nef gene sequence replaced by the EGFP gene [15] and the viral Env proteins, gp120 and gp41 were deleted. We performed co-transfection to generate HIV preparations with influenza envelope proteins in their envelope as shown in Fig. 1A. The viral concentrations were determined by assessing the content of HIV p24 Gag protein in viral preparations. Generally, a viral aliquot containing one pg of p24 represents one to five virions with intact viral RNA genomes. The collected viruses were stored at -80° C.
Influenza viral A particle preparation
We performed co-transfection of 293T cells to prepare influenza A viral particles [18]. Real-time RT-PCR was performed to quantify HA ssRNA in the viral preparations to determine the amount of virus collected. Alternately, the viral preparations in limited dilutions were used to infect HOT cells. The dilution that showed approximately half probability to infect HOT cells was counted as one half-chance tissue culture infection dose (TCID50 ).
Infection of human cell lines
We plated 2×104 cells from each of the various epithelial cell lines into 24-well plates 24 hours prior to infection. A viral aliquot with 50 ng of HIV p24 Gag protein was added to the cell cultures for 0.5-ml total volume. The viral aliquot with 50 ng of HIV p24 Gag protein contained approximately 5×104 viral particles with an intact viral genome, a viral core structure, and an envelope that is similar to the corresponding influenza envelope. To avoid system errors, all infections were duplicated. We presumed that cell numbers doubled during the 24-hour period. Hence, the concentrations of virus were approximately 5×104 viral particles/4×104 cells per 0.5 ml for most cell lines. At 16 hours post-infection, the cell cultures were washed to remove unbound virus.
Quantification of EGFP-positive cells
Expression of EGFP was visualized and counted by fluorescent microscopy or FACS, as previously described [15].
Western blotting
Influenza envelope wrapped HIV preparations collected from transfected 293T cells were collected two to four days post the second transfection. The collected media with p24 Gag protein of 35 μg were ultracentrifuged at 16,000 rpm for one hour at 4°C. The pellets were resuspended with 50 μl of protein lysis buffer (0.5% NP40, 1.0% glycerol, 0.1% β-mercaptoethanol, 40 mM Tris, pH 6.8). The viral lysates were incubated at 100°C for 5 minutes before SDS-polyacrylamide gel electrophoresis (PAGE). Aliquots of 15 μl of protein lysate were applied to SDS-PAGE and followed by gel transfer to a nylon membrane (Poly Screen PVDF; Fisher Scientific, Pittsburgh, PA). A monoclonal antibody specific to HA2 (Santa Cruz, CA) was used according to the manufacturer's instructions to bind and detect HA2 protein in the PVDF membrane.
Results
Selective infection of human cells by influenza A viral envelope protein-wrapped HIV
We created specific HIV particles that were generated by co-tranfection with influenza A viral proteins HA, NA, M, and NS (Fig. 1A). The constructed viruses were used to infect five different human cell lines: 293T (embryonic kidney), TU177 (an oral cell line from head-neck cancer), HeLa-CD4 (a cervical cancer cell line), PC-3 (a prostate cell line), and A-172 (glial cell line), all of which are epithelial cells, except A-172. Compared with the control virus that did not contain any influenza viral proteins on its envelope, influenza A protein-modified HIV demonstrated significant increases in infectivity for the 293T and TU177 cell lines (Fig. 1B), suggesting that these two cell lines express influenza A-specific receptors. Human 293T cells showed very low background for the control virus, as seen in our previous studies [14,15]. Therefore, we used this cell line to study influenza proteins.
NS proteins in the viral envelope significantly increase viral infection
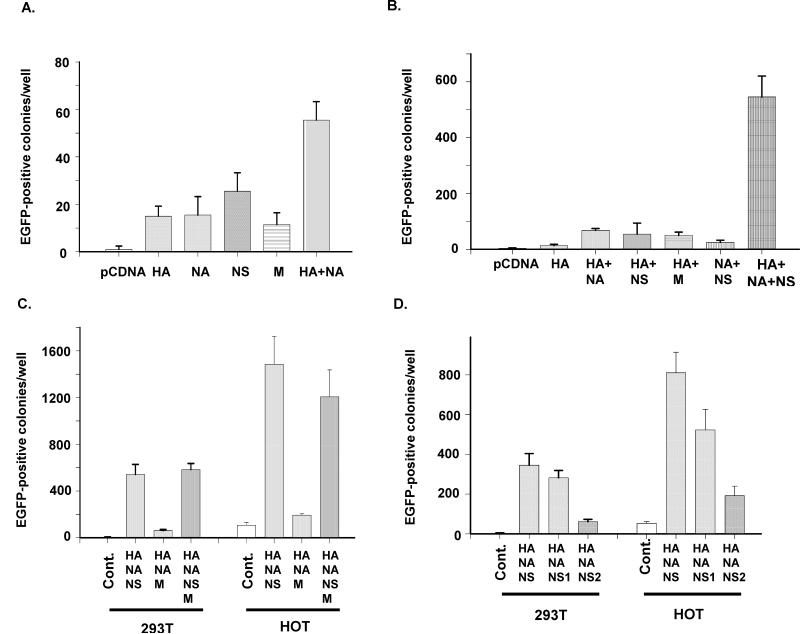
Subsequently, we studied the protein of HA, NA, NS, and M to identify their individual roles in viral infection. When both HA and NA proteins were added to the viral envelope, infectivity was increased (Fig. 2A), suggesting that NA is also required for viral infection. When NS proteins (NS) were added to viral preparations in addition to HA and NA, infectivity was significantly increased, suggesting that NS proteins both are required for viral infection (Fig. 2B). However, combinations of HA and NS or NA and NS did not show significant infectivity (Fig. 2B), suggesting that all three influenza proteins, HA, NA, and NS, are required for efficient infection. In Fig. 2C, we have also tested the combination of HA, NA, and NS using another human epithelial cell line HOT, the results of which are further described below.
Fig. 2.
The roles of the influenza viral envelope proteins in lentiviral-based infection. A) Minimal effects of any one of the viral envelope proteins. B) Effects of the combinations of two influenza viral envelope proteins in viral infection. C) Infection of 293T and HOT cells by HIV carrying HA, NA, and NS proteins, or HA, NA, and M. D) Comparison of NS1, NS2, and NS proteins. HIV carrying HA, NA, and either NS1 or NS2 prepared by co-transfection was used to infect either 293T or HOT cells.
The roles of NS1 and NS2 proteins in viral infection
The NS segment encodes two proteins, NS1 and NS2. To study the roles of these two NS proteins in viral infection, we generated two plasmids that expressed either NS1 or NS2. Using a co-transfection method, we generated HIV from cells that expressed HA, NA, and either NS1 or NS2. We found that NS2 did not significantly affect viral infection of 293T, whereas NS1 did. However, compared with the virus generated from 293T cells co-transfected by the plasmid that expresses both NS1 and NS2, the infectivity of the viral vector with NS1 plasmid-transfected cells was lower (Fig. 2D), suggesting that NS2 may also play positive roles in viral infection.
An oral epithelial cell line demonstrates high sensitivity to the influenza viral envelope
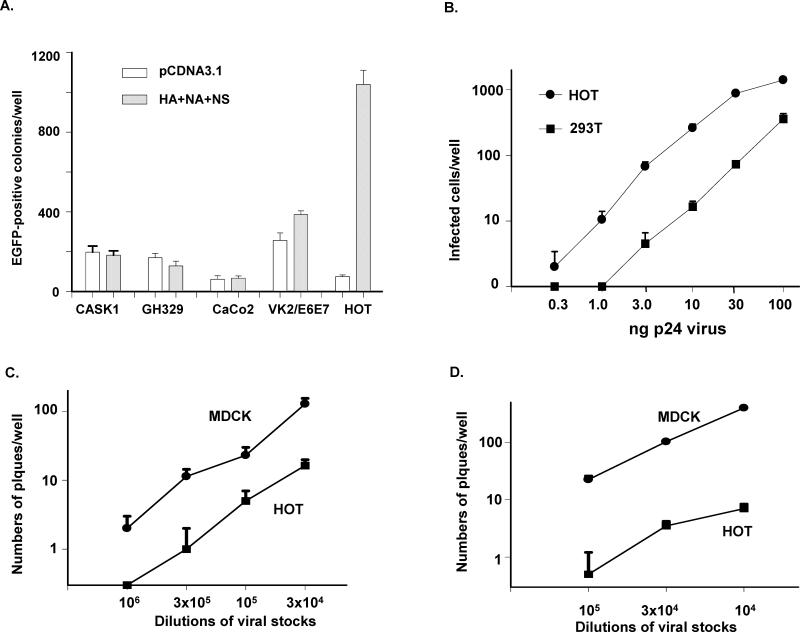
Although the low background of the 293T cell line is very helpful for identifying key proteins that are involved in viral infection, influenza A virus does not replicate well in this cell line. Therefore, it was necessary to find another human cell line for this study. We used HIV with HA, NA, and NS to infect additional epithelial cell lines, and found that the human epithelial cell line derived from oral epithelial cells, HOT [17], was highly sensitive (Fig. 3A). Using limiting dilution of the influenza envelope-wrapped HIV to infect both HOT and 293T cells, we found that the sensitivity of HOT cell line was approximately 5-to 30-fold greater than that of the 293T cell line (Fig. 3B). Thus, we infected HOT with HIV cotransfected with HA, HA/NA, HA/NA/ NS, or HA/NA/NS1 to quantify the infectivity of HOT for various influenza protein combinations. The results were very similar to those with viral infection of 293T cells, except that the infection scales were significantly higher (Fig. 2C & D), suggesting that HOT cells express higher levels of receptors for influenza A virus infection than 293T cells. It was of interest to determine whether HOT also supports replication of wild-type influenza A. We infected both HOT and the MDCK cell line with A/WSN/1933 influenza A virus (NCBI Taxonomy ID: 382835). Both cell lines were sensitive to the wild-type influenza virus that was collected from infected MDCK cells (Fig. 3C), suggesting that HOT is similar to the MDCK cell line in supporting viral infection and replication. The culture medium collected from the infected HOT cell cultures was also highly infectious (Fig. 3D), suggesting that influenza A virus can replicate and generate infectious virions in HOT cells.
Fig. 3.
Use of the HA/NA/NS- HIV to identify influenza-susceptible human cell lines. A) Comparison of the viral infectivity of five different epithelial cell lines. Lentiviral GFP vectors with 100 ng of p24 count was used to infect cells in each well in 24-well plates. B) Comparison of viral infection of 293T versus HOT cell lines. Cell cultures were infected by different doses of the HA/NA/NS lentiviral-GFP vector. C) Infection of HOT and MDCK cells by influenza virus A/WSN/1933 prepared from infected MDCK cells. D) Infection of HOT and MDCK cells by influenza virus A/WSN/1933 prepared from HOT cells. Subconfluent cell cultures of HOT or MDCK cells in 35-mm plates were infected with the same amount of influenza A virus collected from transfected 293T cells. Two days post-infection, viruses collected from either HOT or MDCK cell culture media were diluted, and 1 μl of the diluted viral preparations was used to infect either HOT or MDCK cells in 24-well plates. To restrict viral movement, cells were covered with 1.5% agarose in cell culture medium on cell culture plates. Viral plaques were counted 60 hours post-infection.
NS proteins are involved in viral tropism
We further investigated the roles of NS proteins using segment substitutions. A plasmid carrying the NS segment isolated from another H1N1 substrain of influenza A, #246 from China, was used to generate influenza A viral envelope protein-wrapped HIV containing HIVNL4-3-EGFP-Env(-), as well as the A/WSN/1933 HA, NA plasmid clones. The generated lentiviral virus was used to infect both the HOT and MDCK cell lines. We found that the substitution of NS from A/WSN/1933 with NS from #246 strain demonstrated infectivity similar to the control vector, which uses A/WSN/1933 NS proteins to infect HOT cells (Fig. 4A). These results suggest that substitution of A/WSN/1933 NS with #246 NS does not significantly alter viral infection of HOT human epithelial cells. To investigate whether such substitutions affect infection of MDCK cells, We co-transfected NS246 and seven other viral segments from A/WSN/1933, including HA, NA, NP, M, PA, PB1, and PB2, into 293T cells to generate a reassortant influenza A virus (NS246). As a control, a virus containing all 8 viral segments from A/WSN/1933 (NSWSN1933) was also prepared in parallel. Since both the wild-type and the reassortant viruses efficiently infect HOT cells, we titrated the virus with HOT cells. When equal amounts of either the reassortant or the control virus were used to infect HOT or MDCK cells, we found that both viral strains infected HOT cells with similar efficiency (Fig. 4B). However, infection of MDCK cells by the reassortant virus demonstrated much less efficiency than the control virus, A/WSN/1933 (Fig. 4B). When infecting MDCK cells with different doses of virus, we found that approximately 100 TCID50-HOT of the reassortant influenza A was required to generate significant pathogenic effect (Fig. 4B). However, only 1.0 TCID50-HOT of the control virus were required for infection of MCDK cells. These results strongly suggest that NS proteins are involved in viral tropism.
Fig. 4.
NS proteins in viral tropism of human and dog cells. A) Infection of human oral epithelial cell line HOT by lentiviral vectors carrying HA, NA and either NSWSN1933 or NS246. Results were from FACS analysis. B) Infection of human HOT and MCDK cells by influenza viruses. Photos were taken day 4 post-infection. C) Detection of the HA2 protein by Western blots from lentiviral vectors. The same amounts of influenza protein-modified lentiviruses titrated by p24 assays were used for Western blots. The virus titers were also quantified by Western blotting of Gag p24 and p55 (Experiment 1). Two independent experiments were performed.
Discussion
We used an HIV-influenza system to study the roles of the influenza envelope and envelope-associated proteins in viral infection. When the original HIV envelope proteins, gp120 and gp41, are deleted or truncated from an HIV genome, the virus is no longer able to infect its original target cells, such as CD4+ T-lymphocytes [14,15]. The low background of this modified HIV provides a system in which to study the roles of individual influenza envelope-related proteins in viral infection.
HIV with envelope proteins generated from other non-HIV viruses enables the virus to infect cells that carry receptors that match the envelope proteins from non-HIV viruses. For example, when HIV-based lentiviral core is enclosed with Sindbis viral envelope proteins modified envelope, the virus changes its infection mechanism from pH-independent to pH-dependent [12], suggesting that the envelope proteins from non-HIV viruses play critical roles in the infection tropism of HIV-based lentiviral vectors.
Using this system, we have studied the contributions of individual influenza A envelope or envelope-related proteins in viral infection. Our findings indicate that NS proteins regulate both cell infection and tropism, suggesting that in addition to HA and NA, NS proteins are also important. These results suggest that NS sequences should be considered as an important factor to assess the infectivity of newly emergent viral strains.
Our results also demonstrate that cells such as HOT, which are sensitive to influenza HA, NA, and NS protein-wrapped HIV infection, are also sensitive to wild-type influenza A virus, suggesting that HA, NA, and NS protein-wrapped HIV can be used as a quantitative tool to predict viral infectivity. The lentiviral approach provides rapid analysis to assess viral infectivity of influenza strains. In fact, it is easier and safer to use RT-PCR to clone HA, NA and NS genes from viral samples obtained from patients than to obtain an amount of infectious viral quantity from patients for infectivity studies. Lentiviral vectors express their genes very efficiently in human cells and thus, such a system can be used to quantify viral infectivity.
Using our substitution of NS A/WSN/1933 for NS246, we found that viral tropism was altered by NS. While the modified virus maintained its infectivity for human oral epithelial cells (HOT), the infectivity of the NS246-modified virus showed only approximately 1/100 of the infectivity of its parental strain, A/WSN/1933, in infecting MDCK. These results suggest that NS is very important in viral tropism. It also suggests a possible mechanism by which influenza A may mutate its NS proteins to change its tropism so that viruses can spread from its original host species to a new species. We expect that if the reassortant influenza virus A with NS246 can be used as a prototype for mutating the NS sequence into NSF A/WSN/1933, allowing the virus to jump species from human to dog. The use of reassortant influenza strains to study viral tropism also provides a very useful means to predict the potential risk of currently existing influenza A substrains in disease propagation in the future.
The mechanism of NS proteins in viral infection is under investigation. Since viral infection is directly mediated by the binding of HA1 protein to its receptors and followed by the fusion of viral envelope to the cell membrane mediated by HA2, the most plausible explanation is that NS proteins mediate the presence of either HA1 or HA2, or both in viral envelope. In our lentiviral system, all the three groups of viral proteins, HA, NA and NSs are driven by the CMV promoter. Therefore, the mRNA expressions of these proteins are relatively independent. Therefore, this is likely that NS proteins post-transcriptionally regulate HA on viral envelope. We found that at least HA2 levels in lentivirus can be upregulated by NS proteins (Fig. 4C) supporting our hypothesis that NS proteins regulate influenza infection by mediating the level of HA on viral envelope. However, since the deletion of NS segment completely abolishes the production of influenza virus, we cannot test our hypothesis using real influenza virus.
Our results also demonstrated that NS proteins are involved in viral tropism. Substitution of NS segment of A/WSN/1933 with NS from another H1N1 strain, China#246, significantly decreases viral infectivity of MDCK cells while retains viral infectivity of HOT cells. Since MDCK cell line has insufficient interferon-induced antiviral mechanism [19], it is unlikely that the low infectivity of the substitution of NS from China#246 is related to such cell innate immunity. IFN signalling and subsequent activation of the antiviral state in MDCK cells show only a minor effect on influenza A and B virus replication [20], therefore, the effect of NS in virus propagation in MDCK or HOT cells cannot simply contribute to the interaction between NS1 and interferon induction. Our results here propose another explanation to elucidate the role of NS1 in viral infection. NS1 may affect the abundance and the distribution pattern of HA on viral envelope. Such change of HA on viral envelope may significantly affect viral infectivity. In summary, our results demonstrated that influenza A NS proteins are involved in both viral infection and tropism.
Highlights.
We generated a lentiviral system to study influenza A virus infection
Virus infection can be quantified by GFP expression
The NS proteins significantly increase viral infection
Substitution of NS segment significantly changes viral tropism
NS proteins may increase the presence of HA on viral surface
Acknowledgments
We thank Dr. Debi P. Nayak for providing important comments, and Wendy Aft for editing the manuscript. This work was supported by grants from Department of Defense Grant W81XWH-04-1-0084 and National Institutes of Health (NIH) grant AI047722 to SP; the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khanna M, Kumar P, Choudhary K, Kumar B, Vijayan VK. Emerging influenza virus: a global threat. J Biosci. 2008;33:475–482. doi: 10.1007/s12038-008-0066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamboulian D, Bonvehi PE, Nacinovich FM, Cox N. Influenza. Infect Dis Clin North Am. 2000;14:141–166. doi: 10.1016/s0891-5520(05)70222-1. [DOI] [PubMed] [Google Scholar]
- 3.Etchison J, Doyle M, Penhoet E, Holland J. Synthesis and cleavage of influenza virus proteins. J Virol. 1971;7:155–167. doi: 10.1128/jvi.7.1.155-167.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 5.Mori I, Yokochi T, Kimura Y. Role of influenza A virus hemagglutinin in neurovirulence for mammalians. Med Microbiol Immunol. 2002;191:1–4. doi: 10.1007/s00430-002-0110-9. [DOI] [PubMed] [Google Scholar]
- 6.Ohuchi M, Asaoka N, Sakai T, Ohuchi R. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 2006;8:1287–1293. doi: 10.1016/j.micinf.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Schlesinger RW, Bradshaw GL, Barbone F, Reinacher M, Rott R, Husak P. Role of hemagglutinin cleavage and expression of M1 protein in replication of A/WS/33, A/PR/8/34, and WSN influenza viruses in mouse brain. J Virol. 1989;63:1695–1703. doi: 10.1128/jvi.63.4.1695-1703.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 9.Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci U S A. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grambas S, Hay AJ. Maturation of influenza A virus hemagglutinin--estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 12.Morizono K, Ringpis GE, Pariente N, Xie Y, Chen IS. Transient low pH treatment enhances infection of lentiviral vector pseudotypes with a targeting Sindbis envelope. Virology. 2006;355:71–81. doi: 10.1016/j.virol.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 14.Chow YH, Yu D, Zhang JY, Xie Y, Wei OL, Chiu C, Foroohar M, Yang OO, Park NH, Chen IS, Pang S. gp120-Independent infection of CD4(-) epithelial cells and CD4(+) T-cells by HIV-1. J Acquir Immune Defic Syndr. 2002;30:1–8. doi: 10.1097/00042560-200205010-00001. [DOI] [PubMed] [Google Scholar]
- 15.Pang S, Yu D, An DS, Baldwin GC, Xie Y, Poon B, Chow YH, Park NH, Chen IS. Human immunodeficiency virus Env-independent infection of human CD4(-) cells. J Virol. 2000;74:10994–11000. doi: 10.1128/jvi.74.23.10994-11000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng J, Xie Y, Campbell R, Song J, Wang RQ, Chiu R, Berenson J, Razi M, Massachi S, Yang OO, Chen IS, Pang S. gp120-independent HIV infection of cells derived from the female reproductive tract, brain, and colon. J Acquir Immune Defic Syndr. 2006;43:127–136. doi: 10.1097/01.qai.0000228149.17669.08. [DOI] [PubMed] [Google Scholar]
- 17.Zheng J, Yang OO, Xie Y, Campbell R, Chen IS, Pang S. Ethanol stimulation of HIV infection of oral epithelial cells. J Acquir Immune Defic Syndr. 2004;37:1445–1453. doi: 10.1097/01.qai.0000129572.13008.db. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann E, Neumann G, Hobom G, Webster RG, Kawaoka Y. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology. 2000;267:310–317. doi: 10.1006/viro.1999.0140. [DOI] [PubMed] [Google Scholar]
- 19.Seitz C, Frensing T, Hoper D, Kochs G, Reichl U. High yields of influenza A virus in Madin-Darby canine kidney cells are promoted by an insufficient interferon-induced antiviral state. J Gen Virol. 2010;91:1754–1763. doi: 10.1099/vir.0.020370-0. [DOI] [PubMed] [Google Scholar]
- 20.Frensing T, Seitz C, Heynisch B, Patzina C, Kochs G, Reichl U. Efficient influenza B virus propagation due to deficient interferon-induced antiviral activity in MDCK cells. Vaccine. 2011;29:7125–7129. doi: 10.1016/j.vaccine.2011.05.069. [DOI] [PubMed] [Google Scholar]