Summary
The Ndc80 complex is a key site of kinetochore-microtubule attachment during cell division. The human complex engages microtubules with a globular “head” formed by tandem calponin-homology domains and an 80 amino-acid unstructured “tail” that contains sites of phospho-regulation by the Aurora B kinase. Using biochemical, cell biological, and electron microscopy analyses, we have dissected the tail’s roles in microtubule binding and mediating cooperative interactions between Ndc80 complexes. Two segments of the tail that contain Aurora B sites become ordered at interfaces; one with tubulin and the second with an adjacent Ndc80 head on the microtubule surface, forming interactions which are disrupted by phosphorylation. We propose a model in which Ndc80’s interaction with either growing or shrinking microtubule ends can be tuned by the phosphorylation state of its tail.
Introduction
The Ndc80 complex (Ndc80c), a member of the conserved KMN (Knl1 complex, Mis12 complex, Ndc80 complex) network1, has been identified as a primary site of kinetochore-microtubule attachment2–5 and a necessary factor for the proper operation of the spindle assembly checkpoint (SAC)6–8. The complex is a heterotetramer of Ndc80 (also known as HEC1 in humans9), Nuf2, Spc24, and Spc25, each of which features a globular head domain and a coiled-coil stalk that mediates dimerization into subcomplexes: Ndc80 with Nuf2 and Spc24 with Spc2510,11. The tetramerization interface is formed by the dimerized coiled-coils, resulting in an elongated, dumbbell architecture10–12. The Ndc80–Nuf2 coiled-coil features a break halfway along its length that imparts flexibility by acting as a molecular hinge12, and which may also act as a binding site for other factors13. The Spc24–25 head, which has a unique fold14, mediates kinetochore association by direct interaction with the Mis12 linker complex15,16. The Ndc80-Nuf2 head is formed by the tandem association of calponin-homology (CH) domains contributed by each protein17,18 and mediates microtubule binding1,17,18.
In addition to the Ndc80–Nuf2 head, microtubule association in vitro and in vivo depends on the disordered N-terminal tail of the Ndc80 protein19,20. The tail can be phosphorylated on multiple residues by Aurora B kinase1,5,21, which reduces the affinity of the Ndc80c for microtubules in vitro1,18 and destabilizes improper kinetochore-microtubule attachments in vivo5,19,20,22. Although an unstructured, Aurora B-regulated tail is a conserved feature of the Ndc80 protein in all eukaryotes, the region is highly divergent in sequence and length, making functionally relevant features difficult to identify by bioinformatic methods.
We therefore set out to experimentally characterize in detail the role of this key 80 amino acid region in human Ndc80 complex function. We previously reported a sub-nanometer resolution cryo-EM reconstruction of an engineered version of the human Ndc80c (Ndc80 bonsai18) bound to the microtubule23, revealing tail-dependent oligomerization of the complex on the microtubule surface and details of the interaction between the Ndc80 CH domain and tubulin’s globular domains. Here we extend our analysis of the tail, delineating in detail its interactions with tubulin and the Ndc80–Nuf2 globular head when bound to the microtubule. Using a combination of biochemical, structural, and cell biological methods we dissect the physiological role of specific Aurora B sites and provide support for a model in which dynamic Aurora B phosphorylation of the Ndc80c can support directed chromosome motions during mitosis24 in addition to promoting proper attachment.
Results
Dissection of Ndc80’s N-terminal tail
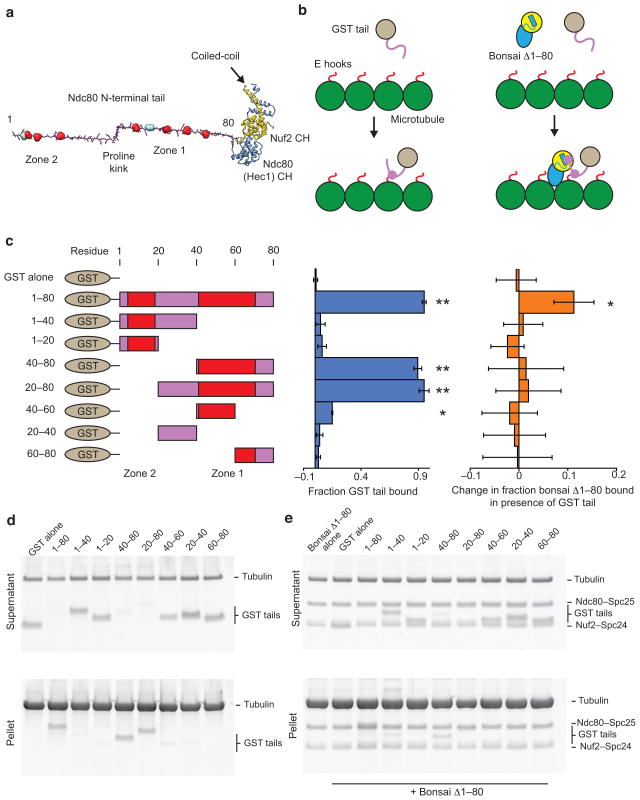
Examination of the primary sequence of the Ndc80 N-terminal tail (Fig. 1a) shows two distinct regions of Aurora B phosphorylation sites separated by a proline-rich linker. We refer to the cluster of sites found between amino acids 40–80 as “zone 1” and those found between amino acids 1–20 as “zone 2.” We previously found that the tail plays a role in the formation of phospho-regulated Ndc80c clusters along microtubule protofilaments in vitro23, suggesting that this segment mediates protein-protein interactions between Ndc80c molecules in addition to contacting tubulin. In order to probe these two activities of the tail, we removed it from the context of the entire complex and fused it to GST for primary structure-function analysis. All GST tail peptides were well-expressed and soluble (Supplementary Fig. 1). Guided by the positions of the two Aurora B phosphorylation regions, we created a number of constructs to test microtubule binding and tail–Ndc80c interactions (Fig. 1a,b).
Figure 1. The Ndc80 tail interacts with both the Ndc80c head and tubulin.
(a) Structural model of the Ndc80c’s two-part microtubule binding module. The tail is modeled as an extended polypeptide colored in magenta. Aurora B sites are shown in space-filling representation: red sites were investigated in this study, while teal sites were not (see text). Kinks correspond to the positions of prolines. Ndc80’s CH domain is colored in blue and Nuf2’s in yellow. (b) Cartoons outlining the experiments presented in panels c–e. GST is grey, Ndc80 tail, magenta, tubulin, green, tubulin C-terminus (E hook), red. The Ndc80 head is colored and displayed in the same orientation as in A. (c) Left, legend of the constructs tested. Zones of Aurora B sites are indicated in red. Middle, quantification of d. Error bars represent s.d., n = 3. Double asterisk, P < 0.0015, single asterisk, P = 0.008, t-test vs. GST alone control. Right, quantification of e. Error bars represent s.d., n = 4. Single asterisk, P = 0.012, t-test vs. bonsai Δ1–80 alone control. For all co-sedimentation assays n represents technical replicates. (d) SDS-PAGE of microtubule co-sedimentation assays with GST tail constructs. Tubulin, 3 μM, GST tails, 1 μM. (e) SDS-PAGE of microtubule co-sedimentation assays with bonsai Δ1–80 in the presence of GST tail constructs. Tubulin, 3 μM, bonsai Δ1–80, 0.5 μM, GST tails, 1 μM.
Identification of the microtubule-binding region
We first performed microtubule co-sedimentation assays as described previously18,23. When compared to the negative control of GST alone, which we found does not interact with microtubules (Fig. 1c,d), we found that the full-length tail (GST 1–80) showed significant interaction with microtubules independently of the CH domains of Ndc80 and Nuf2, as had been reported previously for HEC1 residues 1–80 in the absence of GST20. This activity is conferred by residues 40–80, as both GST 40–80 and GST 20–80 bound microtubules as well as the full-length tail in our assay, while GST 1–40, GST 1–20 and GST 20–40 showed negligible microtubule binding activity. GST 40–60 displayed weaker but still significant microtubule binding activity, while GST 60–80 did not, suggesting that residues from both of these regions are necessary for full binding.
We therefore made finer truncations of the 40–80 region in three amino-acid steps and identified residues 47–68 as the minimal tubulin-binding region of the tail (Supplementary Fig. 2A–D), corresponding very closely to zone 1 of Aurora B sites. This is the most conserved region of the tail, with residues 49–61 showing modest but detectable conservation in all eukaryotes except nematodes and insects and the Aurora B site at Ser55 displaying the highest level of conservation (Supplementary Fig. 2e). Furthermore, most species feature prolines adjacent to this region (Fig. 1a, Supplementary Fig. 2e), suggesting that the architecture of the tail may be conserved although the sequence is not.
Phospho-regulated interaction between Ndc80 complexes
We next investigated the ability of the tail to interact with the Ndc80c in trans. Using GST pull-down assays we were unable to detect an interaction between the tail and the Ndc80c (Supplementary Fig. 3), in agreement with the observation that the Ndc80c does not form clusters in the absence of MTs23. We therefore turned to an indirect assay: we mixed microtubules with an Ndc80 complex lacking the tail (bonsai Δ1–80), which is deficient in microtubule binding18, in the presence and absence of our GST tail constructs, reasoning that constructs containing both microtubule-binding and Ndc80c-binding elements would increase the apparent affinity of the bonsai Δ1–80 for microtubules. Bonsai Δ1–80 showed a significant increase in binding only in the presence of GST 1–80 (Fig. 1c,e), suggesting the N-terminal half of the tail, particularly zone 2, containing the second cluster of Aurora B sites, is important for tail–Ndc80c interaction in trans in the context of the microtubule.
These results suggested an approximate functional segregation of the tail into two regions: a tubulin binding region and an Ndc80c binding region, each of which contains a cluster of Aurora B sites.
We next investigated if the two zones of Aurora B sites differentially regulated tail–tubulin binding and tail–Ndc80c binding, and by extension Ndc80c–microtubule binding and cluster formation, respectively. Nine Aurora B phosphorylation sites (S4, S5, S8, S15, S44, T49, S55, S62, S69) have been reported in the tail19 (Fig. 1a, teal and red residues), but only 7 of the sites (S5, S8, S15, S44, S55, S62, S69) were unambiguously verified in an in vitro Aurora B phosphorylation assay using the wild-type bonsai as a substrate18 (Fig. 1a, red residues). In a recent study investigating HEC1 phosphorylation by Aurora B in vivo Ser4 and Thr49 were not probed24. We therefore conservatively focused on the in vitro verified sites, although it is likely that Ser4 and Thr49 play a role in vivo. Importantly, both fall inside the two Aurora B zones we identified.
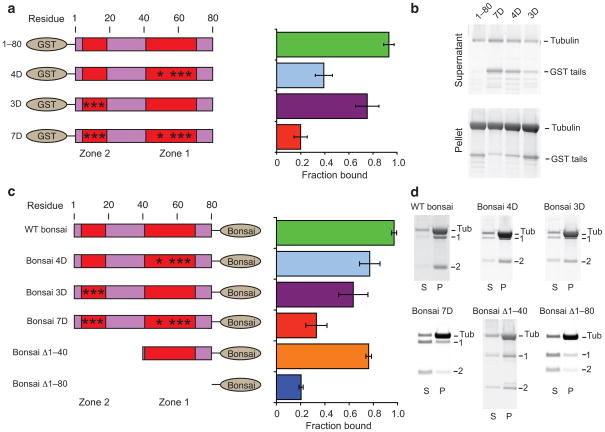
We made phosphomimetic serine to aspartic acid mutations in both zones individually and in combination. All phosphomimetic GST tail peptides showed significantly reduced microtubule binding relative to the wild-type control GST 1–80 (Fig. 2a,b). The tail phosphomimetic in zone 2 showed a modest (20%) reduction microtubule binding, while tails phosphomimetic in zone 1 (GST 4D) and both zone 1 and zone 2 (GST 7D) showed severe reductions (60% and 80%, respectively). These results demonstrate that phosphorylation of zone 1 has a stronger negative impact on microtubule binding than phosphorlyation of zone 2, consistent with our finding that residues in zone 1 form a direct interaction with tubulin and supporting a functional segregation between the two Aurora B zones.
Figure 2. Aurora B zones regulate Ndc80c–Ndc80c and Ndc80c–tubulin interactions.
(a) Left, legend of the constructs tested in A and B, colored as in Fig. 1. Asterisks represent phosphomimetic S to D mutations. Right, Quantification of b. Error bars represent s.d., n = 3. All mutant constructs showed significantly reduced binding (P < 0.015, t-test) relative to GST 1–80. (b) SDS-PAGE of microtubule co-sedimentation assays with phosphomimetic GST tails. Tubulin, 1.5μM, GST tails, 1μM. (c) Left, legend of the constructs tested in c and d, colored as in a. Right, quantification of d. Error bars represent s.d., n = 3. All mutant constructs showed significantly reduced (P < 0.05, t-test) binding relative to WT bonsai. (d) SDS-PAGE of microtubule co-sedimentation assays with bonsai tail mutants. “Tub”, tubulin, “1”, Ndc80–Spc25, “2”, Nuf2–Spc24. Note that electrophoresis conditions varied between experiments and thus bands show different mobility. Tubulin, 2 μM, bonsai constructs, 0.5 μM.
Interplay between the N-terminal tail and CH domains
We next examined the same set of phosphomimetic constructs, but this time in the context of the Ndc80 bonsai complex (Fig. 2c,d). Again, all constructs showed a significant reduction in microtubule binding relative to the wild-type, although the effect of phosphomimetic mutations was more modest, presumably due to the contribution of the Ndc80–Nuf2 head domain to microtubule binding. Indeed, we found that the bonsai Δ1–80 complex retained a tail-independent residual microtubule binding activity, as has been previously reported17,18.
As was the case with the GST tail peptides, phosphomimetic mutations in both zone 1 and zone 2 in combination produced the greatest reduction in MT binding (70%, Fig. 2c,d). However, in the context of the Ndc80 bonsai phosphomimetic mutations in zone 2 (bonsai 3D) resulted in a slightly greater reduction in microtubule binding (35%) than mutations in zone 1 (bonsai 4D, 20%), the reverse of what we observed for the GST tail peptides. Since residues in zone 2 do not directly interact with tubulin, these results suggest that zone 2 mediates interactions between Ndc80c molecules that dominate apparent microtubule binding. Consistent with this observation, an Ndc80 complex lacking the first 40 amino acids of the tail (bonsai Δ1–40) also showed significantly reduced binding compared to the wild-type (20%), even though these amino acids do not interact with tubulin. This construct nevertheless bound microtubules more robustly than the bonsai 3D construct, suggesting that phosphorylation of zone 2 negatively regulates the interaction between Ndc80c molecules.
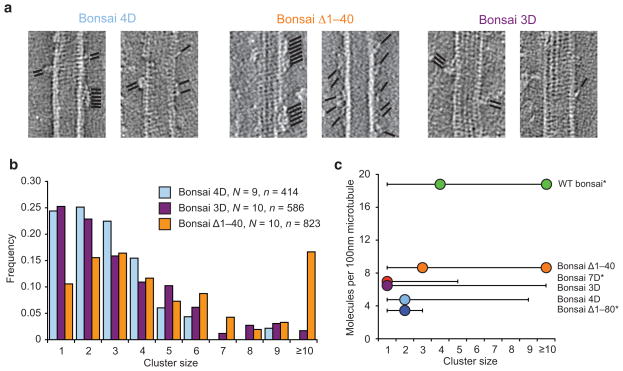
Although our co-sedimentation assays report on the relative contributions of elements of the Ndc80 complex to microtubule binding, they do not address the underlying mechanism. To directly investigate the effects of mutations on cooperative microtubule binding, we turned to electron tomography to visualize and quantify Ndc80c clusters along protofilaments23 (Fig. 3). We found that both the bonsai 4D and bonsai 3D mutants were significantly impaired in clustering (Fig. 3b), with a most probable cluster size of 2 and 1, respectively, distributions that are statistically indistinguishable from each other but distinct from wild-type bonsai23 (Fig. 3c, Supplementary Table 1). Both were capable of forming significantly larger clusters than the bonsai 7D mutant23, or a complex completely lacking the tail23 (bonsai Δ1–80). These results suggest that the region of the tail that directly interacts with tubulin may also contribute to Ndc80c–Ndc80c binding in the context of a cluster. The bonsai Δ1–40 mutant was still capable of clustering robustly, showing a distribution of cluster sizes similar to the wild-type bonsai (Fig. 3b), consistent with our observation of higher microtubule-binding affinity for this mutant. This result further supports our model that phosphorlyation of zone 2 results in repulsion between Ndc80c molecules, even though this region is not strictly required for clustering.
Figure 3. Zone 2 of Aurora B sites negatively regulates Ndc80c clustering.
(a) Representative slices from tomographic reconstructions of microtubules decorated with sub-stoichiometric amounts of the indicated constructs. The positions of Ndc80c molecules are indicated by black lines; all reconstructions featured both clusters and single molecules. Tubulin, 2.5 μM, bonsai constructs, 3.3 μM. (b) Cluster quantification of A. (c) Summary of cluster distributions for all bonsai constructs tested. Dots indicate mode(s), lines indicate range. Asterisks indicate datasets from ref. 23. See Supplementary Table 1 for pair-wise statistical comparisons between these distributions.
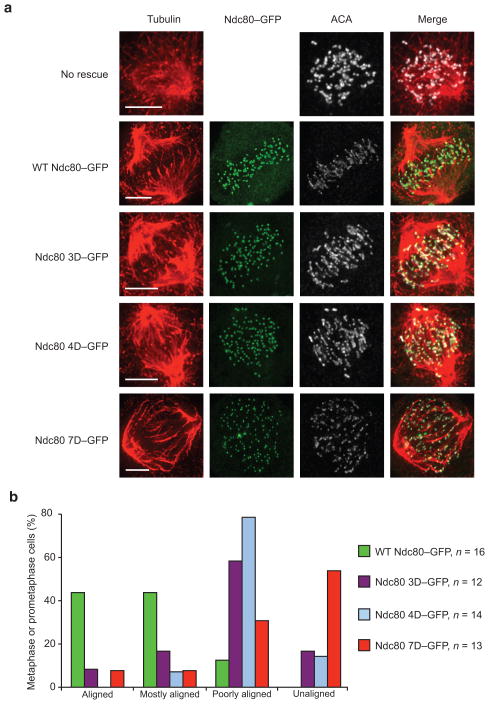
In order to investigate the role of the two Aurora B phosphorylation zones in vivo, we used an RNAi knockdown and replacement protocol20,25 in HeLa cells to assay kinetochore function during the first mitosis in cells that contained mutated Ndc80 proteins featuring phosphomimetic tails (Fig. 4). As has previously been reported, we found that wild-type Ndc80 (WT Ndc80–GFP) rescued the RNAi phenotype, as indicated by a well-organized metaphase plate, while Ndc80 with a N-terminal tail phosphomimetic in both zone 1 and zone 2 (Ndc80 7D–GFP) resulted in a disorganized plate, consisting mostly of unaligned chromosomes. Phosphomimetic mutants in both zone 1 (Ndc80 4D–GFP) and zone 2 (Ndc80 3D–GFP) individually produced intermediate phenotypes; most cells had chromosomes that were poorly aligned or unaligned in both cases. These results demonstrate that zone 1, which is involved in Ndc80c–tubulin interactions, and zone 2, which is involved in Ndc80c–Ndc80c interactions, are both critical for Ndc80c function in vivo.
Figure 4. Both Aurora B zones regulate kinetochore–microtubule interactions in vivo.
(a) Representative immunofluorescence images of prometaphase or metaphase HeLa cells depleted of endogenous Ndc80 and expressing GFP tagged rescue constructs stained for tubulin, GFP, and ACA. Note that non-rescued cells were not imaged for GFP. (b) Quantification of the chromosome alignment phenotypes for metaphase or prometaphase cells. “Aligned” indicates a tight metaphase plate, “Mostly aligned” indicates only 1–3 pairs of kinetochores off the metaphase plate, “Poorly aligned” indicates greater than 3 pairs of kinetochores off the metaphase plate, “Unaligned” indicates no visible metaphase plate.
Delineating the path of the N-terminal tail
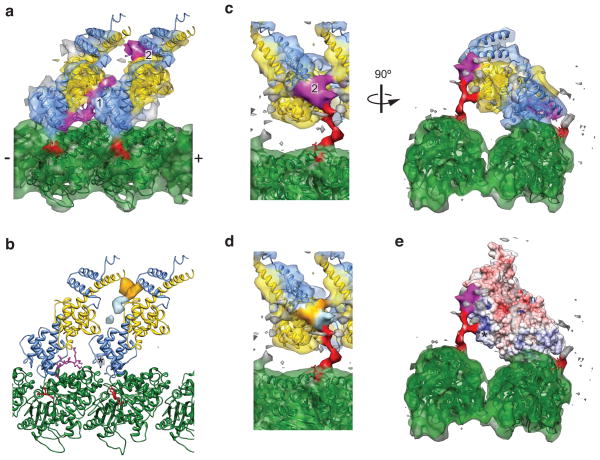
In order to structurally characterize tail–tubulin and tail–Ndc80c interactions, we turned to cryo-electron microscopy analysis (Fig. 5). Using a novel multi-model refinement strategy (Supplementary Fig. 4a) we were able to generate an improved reconstruction of wild-type Ndc80 bonsai bound to the microtubule (Fig. 5a,c, Supplementary Movie 1) with a resolution of 7.9 Å (FSC 0.143 criterion, Supplementary Fig. 4b). In this reconstruction all secondary structural features, including most loops, are resolved in both the bonsai globular head and tubulin. At a high threshold, two prominent densities are visible which are not accounted for by the docked crystal structure of bonsai Δ1–80 and tubulin, which we designate densities 1 and 2 (Fig. 5a). Density 1 extends directly from the amino terminus of the docked crystal structure of bonsai Δ1–80 and thus very likely corresponds to the residues immediately preceding amino acid 80. Based on its length and assuming an extended polypeptide chain, this density would correspond approximately to residues 67–80, containing the first Aurora B site found in zone 1.
Figure 5. Structural analysis of the Ndc80c–microtubule interface.
(a) Crystal structures of two bonsai Δ1–80 molecules (PDB 2VE7) and tubulin (PDB 1JFF) docked into the improved cryo-EM density map, colored as in Fig. 1a. Two densities not occupied by the crystal structures (magenta) were interpreted as corresponding to ordered regions of the N-terminal tail. (b) Positive difference density for reconstructions of wild-type bonsai minus either bonsai Δ1–40 (orange), or bonsai 4D (light blue), contoured at 3.5σ. The asterisk indicates a spurious density peak in the bonsai 4D difference map from the processing procedure (see Online Methods). (c) Same as a, but with the cryo-EM map displayed at a lower threshold, where the tubulin E hook (red) is visualized. (d) Same view as c, left panel, colored according to the locations of an ordered portion of residues 1–40 (orange), and the tubulin-binding portion of zone 1 (light blue). (e) Same view as c, right panel, displaying an electrostatic surface potential map of the bonsai Δ1–80 crystal structure, contoured at +/− 10 kT/e. Asterisk indicates the positive patch on the Nuf2 CH domain that interacts with the E hook of tubulin.
We next generated cryo-EM reconstructions of the bonsai 4D and bonsai Δ1–40 mutants (Supplementary Fig. 5), to structurally interrogate the positions of zone 1 and zone 2, respectively. Difference map analysis (Fig. 5b) shows distinct peaks of density present in the wild-type complex that are absent from each of the two mutants. The bonsai Δ1–40 difference density maps to the location of density 2 in the high-resolution wild-type reconstruction, suggesting that some of the residues in the 1–40 region, which contains zone 2 of Aurora B sites, make a major contribution to this density. The bonsai 4D difference density largely maps to the area between densities 1 and 2 and the base of density 2, consistent with a localization of zone 1 residues (40–67) to this area.
The tubulin E hook interacts with both the Ndc80 tail and Nuf2
An examination of the high-resolution wild-type reconstruction at lower threshold (Fig. 5c, Supplementary Movie 2) shows additional density extending from the C-terminal end of the docked crystal structure of tubulin; by parsimony we attribute this density to an ordered portion of the tubulin C-terminus, or “E hook”26. This density connects to both density 2, and to the CH domain of Nuf2 within an Ndc80c molecule whose toe (the region of the Ndc80 CH domain which contacts the globular portion of tubulin27) is bound to a laterally adjacent protofilament.
The density is branched, which has three potential explanations. The first is that our map represents a mixture of states in this region, because the E hooks of α and β-tubulin, which are indistinguishable in our reconstruction and thus averaged together, actually adopt different conformations. The second possibility is that one of the two branches corresponds to a portion of the N-terminal tail engaging the E hook (part of density 2). The third and most intriguing explanation is that the branch corresponds to polyglutamylation of the E hook28 a post-translational modification which is enriched in brain29 (the source of the tubulin used in our experiments), and the mitotic spindle30. Assuming an extended poly-peptide conformation, Glu254, the site of glutymalyation28, would be near the branch point. If Ndc80c binding to microtubules were sensitive to the glutamylation state of tubulin, it would allow for an additional mechanism to regulate the interaction between kinetochores and microtubules. This hypothesis could be tested by measuring the affinity of Ndc80c for de-glutamylated microtubules, an experiment currently rendered difficult by the lack of purified deglutamylating enzymes31.
The E hook contact site on the CH domain of Nuf2 corresponds to a positively charged patch (Fig. 5e) previously identified as being important for microtubule binding in vitro18. In vivo it was found that multiple charge-reversal substitutions in this patch were required to generate a deleterious phenotype32, consistent with an electrostatic interaction between Nuf2’s positive patch and the negatively charged E hook. The contact with density 2 corresponds to a direct interaction between the Ndc80 tail and the E hook that had been previously suggested based on biochemical studies 19,20. Based on the difference map analysis of the bonsai Δ1–40 and our biochemical data with the GST peptides, we propose that the base of density 2 corresponds to residues in zone 1 that directly interact with the E hook, while the remainder of density 2 is composed of residues in the 1–40 region that become ordered upon Ndc80c clustering, potentially including zone 2 (Fig. 5d). In the context of Ndc80c clusters, the tubulin-binding region of the tail is precisely positioned by the tail–Ndc80c interaction, suggesting that microtubule binding and cluster formation are intimately linked. We also find that under conditions of limiting microtubule binding sites, the tail is unable to effectively compete with the wild-type bonsai complex despite being in excess (2:1 GST tail:Ndc80 bonsai) (Supplementary Fig. 6). Collectively our results support a 3-way interaction between the tail, the Ndc80–Nuf2 globular head, and the microtubule that is significantly stronger than the binary tail–microtubule interaction. This model is also in agreement with our finding that the presence of the full-length tail increases binding of the bonsai Δ1–80 complex (Fig. 1c,e).
While these results provide a rich description of Ndc80c’s microtubule-binding inside a cluster, the mechanism by which the complex engages the microtubule prior to cluster formation or at the edge of a cluster remains unknown, and is an important area for future research.
Discussion
The existence of two different zones of Aurora B phosphorylation sites, one of which regulates both Ndc80c–tubulin and Ndc80c–Ndc80c interactions (zone 1), the other of which negatively regulates Ndc80c–Ndc80c interactions (zone 2), suggests that there are multiple mechanisms by which Aurora B can regulate the Ndc80c–microtubule interface. In our reconstruction of the wild-type Ndc80 bonsai bound to the microtubule, residues in zone 2 are located on the periphery of the cluster and are more accessible, while residues in zone 1 are buried within the cluster. Thus, we speculate that residues in zone 2 may still be susceptible to phosphorylation by Aurora B once a cluster has formed, while residues in zone 1 may be protected from re-phosphorylation. This model is consistent with the recent report that all Aurora B sites in the tail are highly phosphorylated in early stages of cell division, after which sites in zone 2 remain phosphorylated at an intermediate level while sites in zone 1 are dephosphorylated24. Our finding that phosphorylation in zone 2 negatively regulates cluster formation suggests that the cell may dynamically regulate the degree of Ndc80c clustering throughout mitosis, even after microtubule attachment has occurred.
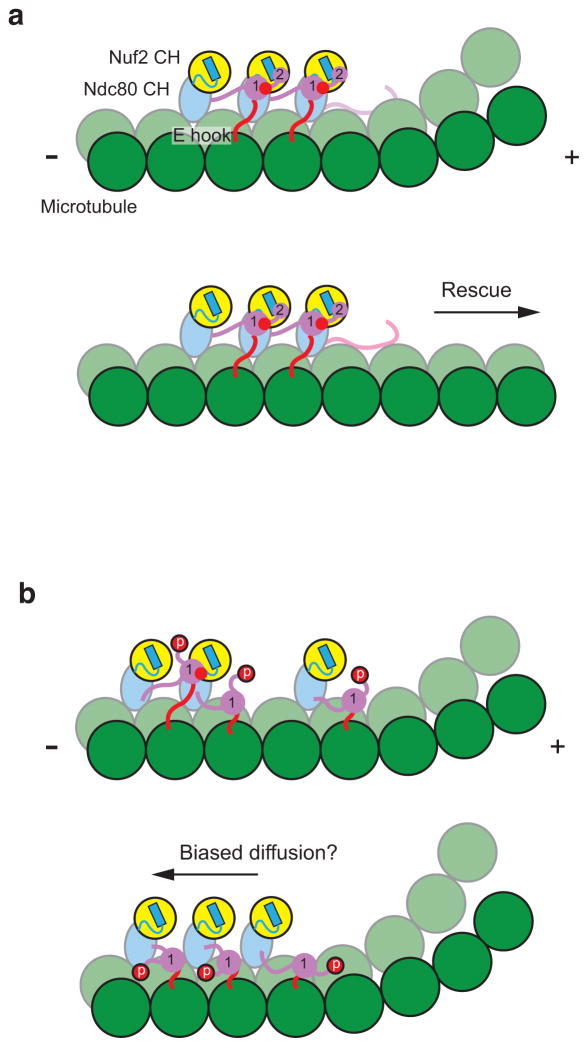
In our model the Ndc80 complex begins prometaphase with zones 1 and 2 maximally phosphorylated at an unattached kinetochore. We propose that once proper attachment is achieved the tail is de-phosphorylated and Ndc80c clusters are formed (Fig. 6a). Given the recent revised estimate that there are ~20 copies of the Ndc80 complex per kinetochore microtubule33, we believe that the high local concentration of the complex on the microtubule surface makes clustering likely in vivo. Zone 1 would become buried upon cluster formation and inaccessible for re-phosphorylation, while zone 2 would remain available as an Aurora B substrate, thereby acting as a cluster clutch. During metaphase, aligned chromosomes undergo oscillatory motions across the metaphase plate with a period of tens of seconds34,35. It was recently found that cells expressing a version of the Ndc80 complex which could not be phosphorylated by Aurora B did not support chromosome oscillations24; we thus speculate that zone 2 may be dynamically phosphorylated and dephosphorylated during oscillations.
Figure 6. Models of Ndc80c interacting with dynamic microtubule ends.
(a) Cartoon of fully de-phosphorylated Ndc80c molecules incorporated into a tight cluster on the surface of a dynamic microtubule, colored as in Fig. 1. The interactions formed by zones 1 and 2 of the Ndc80 tail are numbered, and tubulin E hook–Nuf2 CH interactions are depicted as red circles. For simplicity only interactions within the Ndc80c cluster are shown. In this configuration, the cluster can promote both tubulin longitudinal contacts along protofilaments, through its toe–tubulin interactions, and lateral contacts between protofilaments, by interacting with tubulin E hooks from a neighboring protofilament. This network of interactions has a cumulative effect on microtubule stability. (b) Cartoon of Ndc80c phosphorylated in zone 2 but not zone 1. We envision that in this state the complex will remain attached to the MT surface but in the context of smaller and looser clusters. In some cases a weak interaction between Ndc80 complexes may still be maintained via zone 1 (top panel, left), while in other cases all cooperative interactions may be absent (bottom panel). This intermediate phosphorylation state may enhance the mobility of the complex, enabling kinetochore tracking on depolymerizing microtubule ends by a biased diffusion mechanism.
If clusters were strongly disrupted, it is possible that zone 1 would also become re-accessible for phosphorylation by Aurora B, although this may not happen on a physiologically relevant time scale. Deluca et al. found that phospho-specific antibodies against Aurora B sites in the Ndc80 tail still reacted with the kinetochores of chromosomes that had not yet achieved biorientation upon nocodazole treatment to depolymerize microtubules24. Unexpectedly, however, antibodies against zone 1 did not react with the kinetochores of chromosomes that had achieved bi-orientation, even though kinetochore microtubules were no longer present and Aurora B activity was not down-regulated, while antibodies against zone 2 showed some reactivity. It is tempting to speculate that as a stable (bi-oriented) kinetochore–microtubule attachment matures, additional factors sequester zone 1 of the Ndc80 tail, either through direct interaction with this region or by stabilizing Ndc80c clusters independently from microtubules. Interestingly, it appears that zone 2 still remains somewhat accessible. The detailed kinetics of microtubule-bound Ndc80c phosphorylation by Aurora B is an important subject for future studies in vivo and in vitro, as is the potential role of the Ndc80 tail in mediating interactions between kinetochore complexes.
Our high-resolution cryo-EM reconstruction corresponds to the fully de-phosphorylated state (Fig. 6a). We previously found that the dephosporylated form of Ndc80c strongly enhanced microtubule stability against depolymerization by cold, promoting straight microtubule tubulin polymers23. Our improved cryo-EM reconstruction provides a richer mechanistic explanation for effect: in addition to promoting a straight tubulin conformation by wedging its toe into both intradimer and interdimer longitudinal interfaces, where protofilaments bend during microtubule depolymerization, and by oligomerizing along protofilaments, with the consequent stabilization of longitudinal contacts, the complex also engages a neighboring protofilament via the C-terminus of tubulin, stabilizing lateral contacts. Thus, the dephosphorylated complex is optimized, via a three-component mechanism, to promote microtubule stability and polymerization.
We envision zone 2 could be re-phosphorylated, loosening or disrupting Ndc80c clusters and making the complex more mobile on the microtubule surface and thus capable of tracking depolymerizing ends via a biased diffusion mechanism (Fig. 6b)36. The presence of other microtubule-binding outer kinetochore components, such as the Ska complex37–39, which cooperates with the KMN network40,41, kinesins, and plus-tip tracking proteins, are also likely to alter the architecture of the kinetochore–microtubule attachment site to promote microtubule growth under certain conditions and enable processive tracking along depolymerizing microtubule ends in others.
Online Methods
Expression cloning
All constructs were generated using standard molecular biology methods. The bonsai 4D and bonsai 3D constructs were generated as previously described for the bonsai 7D construct23. GST tail constructs were generated by Ligation Independent Cloning42 in the 2BT vector, which 5′ to 3′ codes for a poly-histidine tag, Glutathione S-Transferase (GST), and a Toboccao Etch Virus (TEV) protease cleavage sequence, followed by the expression cassette.
Protein purification
All constructs were expressed in BL21-CodonPlus-(DE3)-RIL cells. Ndc80 bonsai constructs were purified as described18. GST tail constructs were induced at OD 0.4–0.6 for 4 hours at 37°C with 400 μM Isopropyl β-D-1-thiogalactopyranoside (IPTG) in Luria Broth. Cells were harvested by centrifugation, washed once with phosphate buffered saline (PBS), and flash frozen in liquid nitrogen. Cell pellets were resuspended in 50mM Tris-Cl pH 8, 1 mM di-thiothreitol (DTT), 300 mM NaCl, 10 mM imidazole, augmented with protease inhibitor cocktail tablets (Roche) and lysed by sonic disruption. Lysates were clarified by centrifugation at 17640 RCF, then applied to His-Select Nickel Affinity resin (Sigma), followed by washing and elution with buffer supplemented with 20 mM and 250 mM imidazole, respectively. By using a limiting amount of affinity resin, a homogenous purification was achieved in a single step. Proteins were desalted into storage buffer (50mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM EDTA), and flash frozen in liquid nitrogen.
Microtubule co-sedimentation assays
Microtubules and binding proteins were prepared as described23 in Binding Buffer (80mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 5% sucrose), supplemented with 20 μM taxol as appropriate. Co-sedimentation assays were performed as described18,23: briefly, binding proteins were mixed with microtubules in Binding Buffer supplemented with 20 μM taxol in a 50 μl reaction volume. After a 20 minute incubation at 25°C, the reactions were layered on to a 50% glycerol cushion in 80mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 20 μM taxol, and centrifuged in a TLA 100 rotor (Beckman) at 312,530 RCF for 10 minutes at 25°C. Supernatant and pellet fractions were collected, precipitated with 90% ethanol, then analyzed by SDS-PAGE and densitometry. Statistical analyses were performed with Microsoft Excel.
Electron tomography
Negatively stained samples were prepared as described previously23 using 2.5 μM tubulin and a single application of 3.3 μM Ndc80 bonsai construct. Tilt series were collected from −65 to 65 degrees with a Tecnai F20 operating at 120kV using the Leginon software package43. Images were recorded on a Gatan Ultrascan 4K × 4K CCD camera at a nominal magnification of 49,000X and 1 micron underfocus. Images were filtered to 25 Å, prior to the first inversion of the Contrast Transfer Function, and binned by two prior to performing the reconstructions as described previously with the IMOD software package44.
RNAi rescue experiments
RNAi rescue experiments were performed as described previously25. HeLa cells grown in DMEM/10% FBS were plated at 95% confluency and transfected with pEGFP plasmids expressing siRNA-resistant WT Ndc80–GFP, Ndc80 3D–GFP, Ndc80 4D–GFP, Ndc80 7D–GFP, or empty plasmid, using Lipofectamine2000 (Life Technologies). After 6 hours, cells were trypsinized and plated with thymidine at 25% confluency onto lysine-treated coverslips. After 18 hours, the cells were released from thymidine and treated with 75 nM siRNA against endogenous Ndc80 (GAGTAGAACTAGAATGTGATT) (Qiagen) using Lipofectamine RNAiMAX (Life Technologies). After 12 hours, the cells were blocked in thymidine and treated with a second round of siRNA against Ndc80. The following morning, the cells were released from thymidine and incubated for 10 hours, at which point they were fixed in 2% paraformaldehyde and 0.5% TX-100. Fixed cells were processed for immunofluorescence using standard procedures using antibodies against Tubulin (Thermo Scientific, cat. no. MS-581-P1; 1:500), GFP (Abcam, cat. no. ab290; 1:5,000), and ACA (Antibodies Incorporated, cat. no. 15-234; 1:250). Images were collected using a DeltaVision microscope (Applied Precision) and 100X lens and processed using ImageJ (NIH).
Cryo-electron microscopy
The dataset used to generate the high-resolution reconstruction of the wild-type Ndc80 bonsai bound to the microtubule corresponds to that used to generate the previously reported reconstruction23.
The four datasets used for difference map calculation were generated as follows: cryo-EM grids were prepared as previously reported23,45. Briefly, proteins and taxol-stabilized microtubules were prepared in EM Buffer (80mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.05% Nonidet P-40) supplemented with 20 μM taxol. 2 μl of 2.5 μM microtubules were applied to C-Flat grids (Protochips) glow-discharged with an Edwards carbon coater and incubated for 30 seconds in the humidity chamber of a Vitrobot. 4 μl of 5 μM Ndc80-bonsai was applied and incubated for 1 minute. Excess buffer was removed, then another 4 μl of Ndc80 bonsai was applied and incubated for 1 minute. Excess buffer was removed, then the sample was blotted and plunge-frozen in ethane slush. To ensure maximum stability and concentration of Ndc80 bonsai, the protein was stored on ice and was rapidly warmed to room temperature in a water bath seconds before preparing grids. Data was collected semi-automatically using the Leginon software package46 on a Tecnai F20 electron microscope operating at 120 kV. Images were collected on a Gatan Ultrascan 4K × 4K CCD camera between 1.2 and 2.5 micron underfocus with a dose of 20 electrons/Å2 at a nominal magnification of 80,000X.
Image processing
The power spectra of images were manually inspected for quality: those featuring obvious drift were excluded. The defocus of the images was estimated with CTFFIND347. Microtubule segments were selected using BOXER48, extracted, phase-flipped with EMAN48 and normalized with XMIPP49. The data were then subjected to reference-free classification using IMAGIC50 and a topology-representing network algorithm51 as described23,52; segments which contributed to averages that did not feature well-ordered Ndc80 decoration were excluded from further processing.
Remaining segments were then processed using a multi-reference implementation of the IHRSR method53 in the EMAN254 and SPARX55 processing packages, using naked 13- and 14-protofilament microtubule reconstructions as starting references to avoid model bias. The refined 14-protofilament reconstructions, which generally incorporated more segments and were of higher resolution, were selected for further analysis. The 4 reconstructions for difference map calculation featured between 1,600–2,700 segments, corresponding to 67,200–113,400 asymmetric units.
Initial alignments for the high-resolution wild-type Ndc80 reconstruction were performed in this manner. The 14-protofilament reconstruction was further refined using FREALIGN56 v8.9, which implements helical processing. Since the multi-reference approach was used to sort the different symmetry groups, rather than reference-free classification, a larger proportion of the dataset could be aligned and incorporated into the reconstruction (5,268 segments, corresponding to approximately 220,000 asymmetric units). A negative B-factor of 1,000 Å2 was applied with a peak at 8.3 Å and high-resolution cutoff of 7.6 Å, which showed features that matched those of docked crystal structures well. All rigid-body docking of protein crystal structures (tubulin: 1JFF57, Ndc80 bonsai: 2VE718), and preparation of molecular graphics illustrations was performed with UCSF Chimera58.
Difference map analysis
In order to minimize artifacts caused by differences in data collection or image processing, all reconstructions used for difference map analysis were performed identically. When comparing reconstructions of the bonsai 4D and bonsai Δ1–40 reconstructions with the wild-type bonsai reconstruction it was apparent that the mutants were decorating the microtubule sub-stoichiometrically (Supplementary Fig. 5). This interfered with the direct calculation of amplitude scaled difference maps, since the signal from the microtubule outweighed that of the bound Ndc80 bonsai in the mutants at low resolution, but was equivalent in the wild-type, where stoichiometric binding is observed. Thus, we generated a reconstruction of a naked taxol-stabilized microtubule, which we pre-subtracted from the three Ndc80 bonsai reconstructions without amplitude scaling using the SPIDER software package59 after rigid-body docking of the reconstructions using the wild-type as the reference in UCSF Chimera.
We subsequently calculated amplitude-scaled difference maps with DIFFMAP (http://emlab.rose2.brandeis.edu/diffmap), which largely showed positive differences along the expected path of the Ndc80 tail and minimal artifacts when filtered to 12 Å resolution. A single minor peak was observed between microtubule protofilaments in the case of the wild-type bonsai minus bonsai 4D difference map (Fig. 3B). We also found that applying a soft mask to the edges of the segmented volume and padding by two before difference map calculation with DIFFMAP reduced edge artifacts. No amplitude scaling was performed prior to difference map calculation, which we also found to introduce artifacts.
Supplementary Material
Acknowledgments
We gratefully acknowledge G. Lander for assistance with image processing. GST tail expression constructs were generated by the QB3 Macrolab at UC Berkeley. We also thank T. Houweling, P. Grob and G. Kemalyan for computer and electron microscopy support. G. Alushin is partially supported by an NIH training grant. This work was funded by grants from US National Institutes of Health (P01GM051487 to E. Nogales and RO1GM081576 to P. Stukenberg). E. Nogales is a Howard Hughes Medical Institute Investigator.
Footnotes
Author contributions
G.M.A. and E.N. designed research. G.M.A. and V.M. purified proteins and performed microtubule-binding assays. G.M.A. carried out electron microscopy experiments and image processing. D.M. and J.T. performed cell biology experiments and generated new constructs. G.M.A. and E.N. wrote the paper. All authors contributed to data analysis and editing of the manuscript.
Accession codes
Reconstructed volumes have been deposited in the EMDataBank (EMDB) with the following ID codes: EMD-5489 (improved reconstruction of the Ndc80c–microtubule interface); EMD-5490 (WT bonsai), EMD-5491 (bonsai 4D), EMD-5492 (bonsai Δ1–40) and EMD-5493 (microtubule alone), the four reconstructions used for calculating difference maps.
References
- 1.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–60. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNUF2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–55. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCleland ML, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore–microtubule attachment. Curr Biol. 2004;14:131–7. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by HEC1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Lluesma S, Stucke VM, Nigg EA. Role of HEC1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–70. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca JG, et al. Nuf2 and HEC1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol. 2003;13:2103–9. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 8.McCleland ML, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–14. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–56. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciferri C, et al. Architecture of the human ndc80–hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–95. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 11.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–7. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HW, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maure JF, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–13. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei RR, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–9. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Petrovic A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–52. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–34. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore–microtubule attachment. Nat Struct Mol Biol. 2007;14:54–9. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 18.Ciferri C, et al. Implications for kinetochore–microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore–microtubule attachment relies on the disordered N-terminal tail domain of HEC1. Curr Biol. 2008;18:1778–84. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(HEC1) Curr Biol. 2008;18:1785–91. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheeseman IM, et al. Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–72. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka TU, et al. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–29. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 23.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–10. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in HEC1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–34. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 Complex Employs a Tripartite Attachment Point to Couple Microtubule Depolymerization to Chromosome Movement. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikkawa M, Okada Y, Hirokawa N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–52. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 27.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edde B, et al. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–5. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 29.Redeker V. Mass spectrometry analysis of C-terminal posttranslational modifications of tubulins. Methods Cell Biol. 2010;95:77–103. doi: 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
- 30.Bobinnec Y, et al. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39:223–32. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Rogowski K, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–78. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Sundin LJ, Guimaraes GJ, Deluca JG. The Ndc80 complex proteins Nuf2 and HEC1 make distinct contributions to kinetochore–microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–68. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–82. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–75. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amaro AC, et al. Molecular control of kinetochore–microtubule dynamics and chromosome oscillations. Nat Cell Biol. 2010;12:319–29. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–75. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaitanos TN, et al. Stable kinetochore–microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. Embo J. 2009;28:1442–52. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theis M, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. Embo J. 2009;28:1453–65. doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore–microtubule attachments by inhibiting Ska complex–KMN network interaction. J Cell Biol. 2012;196:563–71. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang G, et al. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012 doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- 42.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–74. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–8. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 45.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–61. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 48.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 49.Sorzano CO, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 50.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 51.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Ramey VH, Wang HW, Nogales E. Ab initio reconstruction of helical samples with heterogeneity, disorder and coexisting symmetries. J Struct Biol. 2009;167:97–105. doi: 10.1016/j.jsb.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 54.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–57. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 58.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–7. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.