Abstract
Divergent natural selection promotes local adaptation and can lead to reproductive isolation of populations in contrasting environments; however, the genetic basis of local adaptation remains largely unresolved in natural populations. Local adaptation might result from antagonistic pleiotropy, where alternate alleles are favored in distinct habitats, and polymorphism is maintained by selection. Alternatively, under conditional neutrality some alleles may be favored in one environment but neutral at other locations. Antagonistic pleiotropy maintains genetic variation across the landscape; however, there is a systematic bias against discovery of antagonistic pleiotropy since the fitness benefits of local alleles need to be significant in at least two environments.
Here, we develop a generally applicable method to investigate polygenic local adaptation and identify loci that are the targets of selection. This approach evaluates allele frequency changes after selection at loci across the genome to distinguish antagonistic pleiotropy from conditional neutrality and deleterious variation. We investigate local adaptation at the QTL-level in field experiments, in which we expose 177 F6 recombinant inbred lines (RILs) and parental lines of Boechera stricta (Brassicaceae) to their parental environments over two seasons. We demonstrate polygenic selection for native alleles in both environments, with 2.8% of the genome exhibiting antagonistic pleiotropy, and 8% displaying conditional neutrality. Our study strongly supports antagonistic pleiotropy at one large-effect flowering phenology QTL (nFT): native homozygotes had significantly greater probabilities of flowering than foreign homozygotes in both parental environments. Such large-scale field studies are essential to elucidate the genetic basis of adaptation in natural populations.
Keywords: antagonistic pleiotropy, genetic tradeoff, common garden, conditional neutrality, field study, FLOWERING LOCUS T, flowering phenology, local adaptation
Introduction
Divergent natural selection can result in the evolution of locally adapted ecotypes, and can reproductively isolate populations inhabiting contrasting environments (Hedrick, 1986; Kawecki, Ebert, 2004; Schluter, Conte, 2009). Many studies have examined local adaptation to disparate selective regimes at the level of the whole organism or whole genome (e.g., Galloway, Fenster, 2000; Leinonen et al., 2010; Nagy, 1997; Schmitt, Gamble, 1990), and recent meta-analyses indicate that local adaptation is common (Hereford, 2009; Leimu, Fischer, 2008). However, the genetic factors that contribute to local adaptation and ecological speciation are poorly resolved in natural populations (Anderson et al., 2011b; Fry, 1996; Hall et al., 2010; Kawecki, 1997; Kawecki, Ebert, 2004; Mitchell-Olds et al., 2007; Schluter, Conte, 2009).
The genetic basis of local adaptation could be explained by: (1) antagonistic pleiotropy, wherein alleles at a single locus reverse rank fitness in alternative environments (i.e., single locus genetic tradeoffs), or (2) conditional neutrality, wherein an allele shows a fitness advantage in one environment, but is neutral in the contrasting environment (Anderson et al., 2011b; Fry, 1996; Hall et al., 2010; Kawecki, 1997; Kawecki, Ebert, 2004; Mitchell-Olds et al., 2007). These alternative hypotheses have only been investigated for a handful of species (e.g., Barrett et al., 2008; Fournier-Level et al., 2011; Gardner, Latta, 2006; Hall et al., 2010; Hawthorne, Via, 2001; Latta et al., 2010; Lowry et al., 2009; Lowry, Willis, 2010; Verhoeven et al., 2008; Verhoeven et al., 2004; Weinig et al., 2003), perhaps because testing between them requires growing large numbers of individuals from experimental mapping populations, such as recombinant inbred lines (RILs), in the ancestral environments in which the parental forms evolved (Anderson et al., 2011b).
Biologists have long been interested in the evolutionary processes that influence genetic variation within and among populations (Mitchell-Olds et al., 2007). Across the landscape, antagonistic pleiotropy can maintain genetic variation at ecologically-relevant loci since local alleles retain a fitness advantage over foreign alleles in each habitat (Anderson et al., 2011b; Hall et al., 2010; Mitchell-Olds et al., 2007). However, genetic tradeoffs have only very rarely been demonstrated at the level of the QTL (Hawthorne, Via, 2001; Weinig et al., 2003), candidate gene (Barrett et al., 2008), or chromosomal inversion (Lowry, Willis, 2010). Conditional neutrality is the more commonly described pattern in field studies (Gardner, Latta, 2006; Hall et al., 2010; Latta et al., 2007; Latta et al., 2010; Lowry et al., 2009; Verhoeven et al., 2008; Verhoeven et al., 2004; Weinig et al., 2003). If multiple loci determine fitness, alleles from different environments are conditionally beneficial at distinct loci, and gene flow among populations is limited, then conditional neutrality can contribute to transient local adaptation at the organismal level (Fournier-Level et al., 2011; Hall et al., 2010; Mitchell-Olds et al., 2007). However, high levels of gene flow could assemble recombinant genotypes with the conditionally advantageous allele at distinct loci across the genome, which would result in the evolution of generalist genotypes, not locally adapted ecotypes.
The current evidence supporting conditional neutrality might not have a biological basis, but instead might reflect systematic biases against detection of antagonistic pleiotropy, for two reasons. First, detection of genetic trade-offs at individual loci or QTL requires sufficient statistical power such that the fitness advantage of local alleles attain significance in two or more environments (Anderson et al., 2011b). Consequently, widespread evidence for conditional neutrality could be an artifact of small experiments, rather than a rarity of antagonistic pleiotropy in nature. Second, conditional neutrality might be prevalent in laboratory or artificial environments, whereas genetic trade-offs may be enriched in natural habitats. Heterogeneous natural selection can actively maintain polymorphism exhibiting antagonistic pleiotropy, whereas polymorphisms responsible for conditional neutrality can be eliminated by selection via fixation of the conditionally beneficial allele over time (Mitchell-Olds et al., 2007). Differentiating between antagonistic pleiotropy and conditional neutrality requires large field studies that explicitly consider the fitness effects of alleles in their native habitats (Anderson et al., 2011b).
To investigate the genomic basis of local adaptation, we develop a novel permutation analysis of allele frequency change in response to natural selection. In conjunction with QTL mapping, this framework can be applied to other systems to identify loci that are likely targets of selection and to investigate the genetic basis of local adaptation. QTL by environment interactions can be detected in traditional QTL mapping, but these interactions could be due to significant effects of a QTL in one environment only (conditional neutrality), significantly positive effects of the local allele in both environments (antagonistic pleiotropy), or even significantly positive effects of the foreign allele in contrasting habitats. Thus, QTL by environment interactions require additional analysis to test between antagonistic pleiotropy and conditional neutrality. In contrast, our permutation analysis can readily distinguish loci displaying antagonistic pleiotropy from conditional neutrality at multiple loci distributed across the genome.
We further illustrate antagonistic pleiotropy using an ecologically-relevant life-history QTL. In a previous study, we mapped a large effect QTL (nFT) that influenced age and stage of first flowering in multiple laboratory treatments, as well as the probability of flowering at one field site in the mustard Boechera stricta(Anderson et al., 2011a). The probability of flowering is a key fitness component, as individuals that fail to flower have no reproductive fitness. Boechera stricta is an ideal system in which to investigate the genetic basis of local adaption as high rates of self-pollination (FIS = 0.89; Song et al., 2006) and strong differentiation between populations (Song et al., 2006; Song et al., 2009) suggest that gene flow is unlikely to inhibit adaptation to the local environment at our field sites. The objectives of our current study were to: (1) assess polygenic local adaptation by quantifying changes in native allele frequency across the genome in two contrasting environments; (2) examine antagonistic pleiotropy vs. conditional neutrality at markers throughout the genome using an analytical method that can be implemented with other systems and datasets; and (3) investigate the genetic basis of local adaptation at the nFT QTL in relevant field environments. Importantly, we realize these objectives in the native environments of the parental lines, and our experimental design affords sufficient statistical power to evaluate local adaptation at the level of the QTL.
Methods
Study species and field sites
Boechera stricta (Brassicaceae) is a short-lived perennial relative of the model organism Arabidopsis thaliana. This primarily selfing species is native to the western United States, spans an elevation gradient from 1500 to 3700 m, and occurs in a wide variety of habitats that differ in abiotic and biotic conditions (Anderson et al., 2011a; Song et al., 2006; Song et al., 2009). Our field sites in Montana and Colorado are relatively undisturbed by human activities (e.g., Brunelle et al., 2005) and differ in a variety of environmental conditions (e.g., elevation, latitude, precipitation, temperature, see Schranz et al., 2007), which could result in divergent natural selection on ecologically-relevant traits, and ultimately the evolution of local adaptation. Boechera stricta shows substantial population divergence between Western (Montana) and Southern (Colorado) groups (Song et al., 2006; Song et al., 2009). We local adaptation in 177 F6 recombinant inbred lines (RILs) of B. stricta created from a cross between a Montana father and a Colorado mother and outplanted into both parental environments. Details on the generation of the RILs can be found elsewhere (Anderson et al., 2011a; Schranz et al., 2007).
Common garden experiments
We established common garden experiments in the parental environments of the RILs in Montana and Colorado (Anderson et al., 2011a). In September 2008, we planted 2160 RIL and parental line plants (n= 6 individuals/RIL/garden and 30 individuals/parental line/garden; N=170 RILs and two parental lines). We replicated the experiment in September 2009 when we planted 1522 RIL and parental line plants [N=751 individuals in Colorado, and 771 in Montana; N= 1-5 individuals/RIL/garden (average of 4.23) and 15 individuals/parental line/garden, except in Montana, where only 2 individuals from the Colorado line were planted; N=174 RILs and two parental lines]. In the 2009 and 2010 growing seasons, we quantified the number of flowers produced by all experimental individuals. No plants flowered in Colorado in the 2009 growing season, likely due to intense herbivory in that garden during that year (Anderson et al., 2011a).
For the present study, we focus on the probability of flowering in the first growing season. This fitness component is highly relevant to the flowering phenology QTL we detected in our previous work (Anderson et al., 2011a). Survivorship of experimental individuals rapidly declines after the first growing season; furthermore, <5% of plants that failed to flower during their first summer were able to flower during subsequent growing seasons (Anderson and Mitchell-Olds, unpublished data). Therefore, transition to reproduction during the first growing season is a strong predictor of the lifetime probability of flowering in experimental populations. Throughout this study, we have excluded individuals that did not survive the first winter (i.e., from planting in September until the first census in June of the following year). Those individuals provide information on an earlier fitness component (overwinter viability), but not on reproductive biology. Furthermore, sources of overwinter mortality included maladaptation to the local environment, as well as indiscriminate sources of mortality that obscure patterns of local adaptation (e.g., damage from overwinter gopher burrowing activities, and runoff from melting snow). Future studies will investigate overwinter viability and fecundity components of fitness (Anderson and Mitchell-Olds, in preparation).
A total of 1853 individuals survived their first winter and final sample sizes are as follows: Montana garden planted 2008, N =776 individuals, 170 RILs and two parental lines; Montana garden planted 2009, N = 380 individuals, 166 RILs and two parental lines; Colorado garden planted 2009, N = 697 individuals, 171 RILs and two parental lines. Mortality due to gopher activity was particularly intense in the Montana garden, 2009 planting cohort. The results that we present here are qualitatively similar to results that we find in a much larger dataset, which excludes plants indiscriminately killed by gophers, but includes other individuals that died over the course of the winter (nFT × environment interaction: F1,1960=24.6, p<0.0001, Table S1).
Polygenic local adaptation
We investigated polygenic local adaptation in our common garden environments by examining selection for native alleles at loci distributed across the genome. In a previous study, we genotyped all RILs at 164 polymorphic molecular markers (62 microsatellite loci and 102 Single Nucleotide Polymorphisms, Anderson et al., 2011a). In this study, we calculated changes in allele frequency for the native allele in each common garden at all 164 markers by comparison of the population of plants alive at the beginning of the growing season (before selection; June 2009 and 2010), and the population of plants that successfully flowered during the first growing season in each garden (after selection; August 2009 and 2010). Local adaptation would be indicated by significantly positive changes in local allele frequency in each garden, suggesting that the local allele is favored at multiple markers throughout the genome. We tested for directional selection favoring local alleles, which would indicate polygenic adaptation, by comparing the actual allele frequency changes with permuted genome-wide frequency changes for all markers (1000 permutations).
In a complementary analysis, we investigated fitness differences of the parental lines only to assess local adaptation at the organismal level. This logistic regression (Proc Glimmix, SAS ver 9.2, SAS Institute, Cary, NC) modeled the probability of flowering as a function of environment, family and their interaction, with block crossed with site as a random effect. Here, family was considered a fixed effect.
Antagonistic Pleiotropy vs. Conditional Neutrality
To test between conditional neutrality and antagonistic pleiotropy, we developed the program CNAP (Conditional Neutrality Antagonistic Pleiotropy), which provides a null distribution of changes in allele frequency at all markers in each environment under the assumption that genotype is unrelated to reproductive status. This approach can easily be generalized to different fitness components (e.g., survivorship, fruit or seed set, etc.) or episodes of selection. This Python version 2.7.1 script is available from the corresponding author. For each of 2,000 iterations for each site, individuals were randomly permuted into one of two fitness categories (reproductive or non-reproductive) and the change in allele frequency was calculated at each locus, with positive values when the native allele was favored, and negative values when the foreign allele was favored. The permuted null distribution represents the maximum change in allele frequency of any locus in each of 2,000 randomizations. This permutation approach is equivalent to Doerge and Churchill (1996), and explicitly accounts for non-independence among linked loci.
For significance tests in CNAP, markers that fell in the extreme 2.5% tails of this distribution in either environment (Montana or Colorado) were then subject to a locus-specific permutation in the contrasting environment to determine whether the locus showed antagonistic pleiotropy or conditional neutrality. This locus-specific permutation randomized reproductive status among individuals in the second environment, and asked whether the observed change in allele frequency at a given marker is within the extreme 2.5% tails of the permuted null distribution. In summary, two approaches to permutation are used. First, in a given environment we ask whether the observed allele frequency change at a given locus falls within the most extreme 5% of changes for every marker in the genome. If this occurs, we consider this same locus in the other environment, and ask whether the observed allele frequency change falls within the 5% most extreme permuted values at this locus. Thus, this approach uses an α = 0.05 significance threshold specifically for this locus in the second environment, conditional on genome-wide significance of this locus in the first environment. This sequential testing approach may identify cross-environment QTL patterns which are not apparent from completely independent QTL analyses in two environments.
How does this approach compare to a standard QTL mapping experiment analyzed in two environments? Significance tests in QTL mapping also are based on Doerge and Churchill's (1996) permutation approach. However, by itself, QTL mapping of fitness components cannot distinguish between antagonistic pleiotropy and conditional neutrality, both of which would produce significant QTL × environment interactions. In contrast, our CNAP approach explicitly tests these alternative hypotheses by determining whether selection favors local alleles in contrasting environments at loci distributed across the genome.
To calculate the proportion of the genome displaying antagonistic pleiotropy vs. conditional neutrality, we determined the length of the chromosome region tagged by each marker, bordered by the midpoints of each intermarker interval (Anderson et al., 2011a). We summed these distances for loci with significantly positive local allele frequency changes in both sites (antagonistic pleiotropy), and for loci with significantly positive local allele frequency changes in only one environment (conditional neutrality), and divided by total genome length (872 cM).
nFT QTL × Environment interactions
Previous work highlighted the importance of the nFT QTL in the transition to flowering in both field and laboratory conditions (Anderson et al., 2011a). In this study, we evaluate the effect of genotype at nFT on the probability of flowering, which is a key fitness component as individuals that fail to flower have no reproductive fitness. Both conditional neutrality and antagonistic pleiotropy would result in a significant nFT QTL by environment interaction; therefore, we explicitly compared fitness values of Montana homozygotes and Colorado homozygotes in each experimental garden. These results only support antagonistic pleiotropy if local homozygotes significantly outperform foreign homozygotes in both environments.
We conducted a logistic regression with data from both sites and both years. We excluded from this analysis the very few families (n=6) that were heterozygous at the nFT QTL. To control for genomic background, we also calculated the proportion of the genome that originated from the Colorado parent (%CO alleles) for each RIL. As above, our calculations accounted for intermarker distance along each linkage group. We excluded the nFT marker and four markers within 15 cM of the nFT marker (R6.B06, BstES0030, Bst011023, C01). We assessed whether fitness varied as a function of environment, nFT QTL genotype, %CO alleles, cohort, and two-way interactions between environment and nFT genotype, environment and %CO and environment and cohort in a logistic regression that included block and family cross classified with site as random effects (Proc Glimmix, SAS ver. 9.3). The cohort term accounted for differences in planting year. Since no plants flowered in CO during 2009, we were unable to assess three way interactions among cohort and other predictors.
Results
In our Montana garden, 58.8% and 21.8% of experimental individuals flowered in 2009 and 2010, respectively. Plants at the Colorado garden failed to flower during the 2009 growing season due to extensive herbivory and were not included in the analysis; however, 22% of experimental plants flowered in Colorado in 2010.
Polygenic local adaptation
Using the permutation program CNAP (Conditional Neutrality Antagonistic Pleiotropy) we identified significant increases in local allele frequency in both environments (mean allele frequency change = 0.016 in Colorado, and 0.048 in Montana; p<0.001 in both sites). This analysis supports the local adaptation hypothesis because the native alleles were favored at markers across the genome in both of our garden sites.
Similarly, the parental lines showed evidence congruent with local adaptation. In the Colorado garden, 5 of 10 individuals from the Colorado parent successfully flowered (50% success) compared with 3 of 13 Montana parental individuals (only 23.1%). In the Montana garden, 6 of 27 Montana parental individuals flowered (22.2%) compared with 1 of 6 Colorado parental individuals (16.7%). However, genotype by environment interaction for flowering of the parental lines was not significant (logistic regression, F1,27=1.54, p=0.22), likely because our statistical power was reduced by low survivorship of the foreign Colorado parents at our Montana garden site.
Antagonistic Pleiotropy vs. Conditional Neutrality
Based on 164 molecular markers (microsatellites and single nucleotide polymorphisms) genotyped in a previous study (Anderson et al., 2011a), 2.8% of the genome displays significant antagonistic pleiotropy, while 8.1% shows conditional neutrality for the probability of flowering. In the CNAP analysis almost all of the markers identified as significant in tests of antagonistic pleiotropy were linked to the known nFT QTL. We detected conditional neutrality for 18 markers, on linkage groups LG2 (1 locus), LG3 (12 loci) and LG7 (5 loci). The loci on LG 7 are in the genomic region of a QTL that underlies glucosinolate production and insect resistance (Schranz et al., 2009). One marker (C09, 129.9 cM on LG3) co-localizes with a minor effect QTL that influences flowering probability in the field, and another marker (Bst001594, 43.8 cM, LG3) co-localizes with a QTL controlling plant size at flowering in the field (Anderson et al., 2011a). The remaining markers that display conditional neutrality do no co-localize with previously mapped QTL.
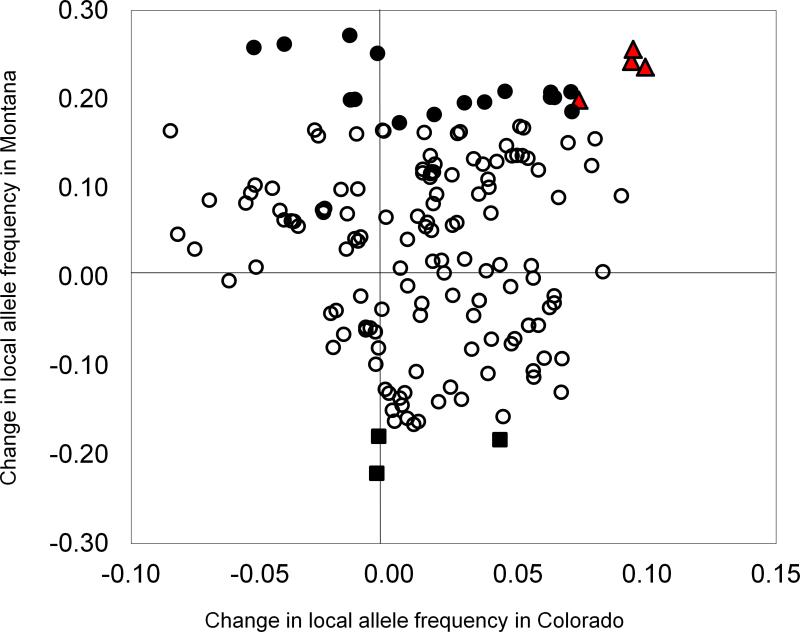
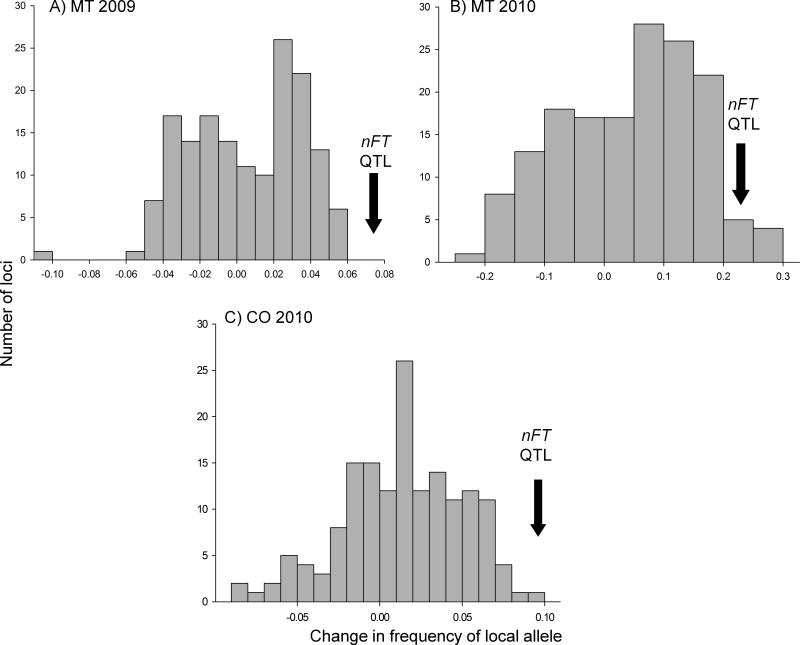
A bivariate plot (Fig. 1, SI Table 1) of changes in the frequency of the local alleles in Colorado and Montana reveals the specific loci under selection in each environment. Concordant with our other analysis, the nFT QTL exhibited a significant signature of antagonistic pleiotropy (p < 0.001 in MT; p < 0.04 in CO): an increase in local allele frequency in each environment (Figs. 1 and 2). Selection also favored the native nFT allele in our Montana 2009 database, which was not included in the CNAP analysis (Table 1, Fig. 2). In all datasets, the nFT QTL occurred in the extreme tails of the distributions of allele frequency changes (Fig. 2).
Figure 1.
Bivariate plot of local allele frequency changes in Colorado vs. Montana. Each data point represents a locus. Red triangles are loci displaying antagonistic pleiotropy (positive change in the local allele frequency in both environments), all of which are near the nFT QTL. Filled circles represent loci that exhibit patterns consistent with conditional neutrality (significant increase in local allele frequency in Montana, but not Colorado), and unfilled circles are neutral loci. Permutation analysis detected antagonistic pleiotropy at marker Bst027135, but local allele frequency change at this marker is only marginally significant in Colorado. In this figure, Bst027135 is coded as displaying conditional neutrality. Three loci showed negative allele frequency change in Montana, but were neutral in Colorado; those loci are represented by filled squares.
Figure 2.
Change in local allele frequency across the genome from the initial experimental population to the reproductive population. These figures exclude the nFT QTL and 4 nearby loci, resulting in n= 159 markers. The change in frequency of the local allele at the nFT QTL is indicated for each experimental garden and year.
Table 1.
Allele frequency change of the native allele at the nFT QTL and averaged across the genome (excluding the nFT marker, and 4 nearby loci, resulting in n=159 markers).
| Frequency of the native allele at the nFT QTL | Frequency of native alleles across the genome (n=159 markers) | |||||||
|---|---|---|---|---|---|---|---|---|
| Initial population | Reproductive population | Change in native allele frequency | Initial population | Reproductive population | Average change in native allele frequency | Minimum and maximum allele frequency changes | 95% allele frequency change | |
| Montana garden (2009) | 0.548 | 0.628 | 0.08 | 0.526 | 0.531 | 0.005 | -0.10 to 0.059 | 0.044 |
| Montana garden (2010) | 0.563 | 0.819 | 0.256 | 0.538 | 0.581 | 0.043 | -0.22 to 0.27 | 0.202 |
| Colorado garden (2010) | 0.461 | 0.556 | 0.095 | 0.479 | 0.493 | 0.014 | -0.085 to 0.09 | 0.066 |
The CNAP analysis also detected very marginally significant antagonistic pleiotropy (SI Table 1) for a genomic region (marker Bst027135 on linkage group 3 at 95.682 cM) that does not co-localize with any previously mapped QTL affecting our measured traits.
nFT QTL × E interactions
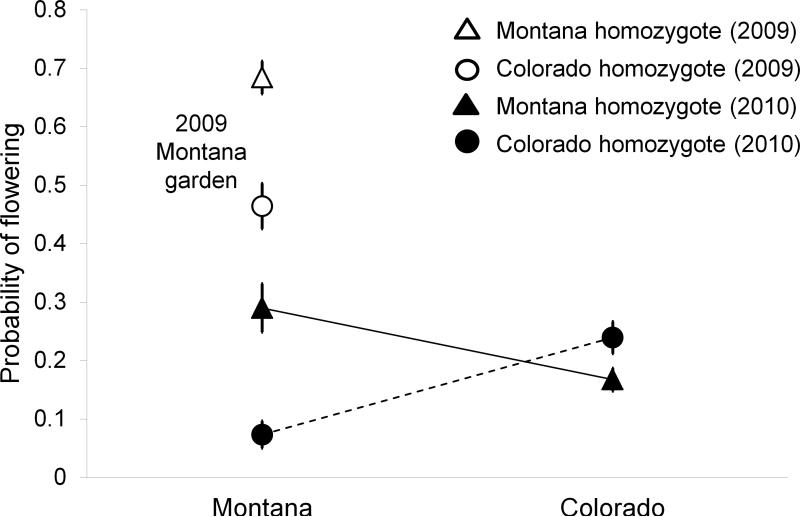
Logistic regression indicated a significant interaction between environment and the nFT QTL, which resulted from a tradeoff in fitness in the two environments, consistent with antagonistic pleiotropy (Table 2, Fig. 3). The local nFT allele had a higher probability of flowering than the foreign allele in both environments and both years (Fig. 2): in the Colorado garden, local CO homozygotes had 159% higher odds of flowering than foreign MT homozygotes (odds ratio= 2.59 for comparison of local vs. foreign genotype; 95% CL: 1.09 – 6.13, p-value after Tukey's adjustment for multiple tests=0.024); in the Montana garden, MT homozygotes had 294% greater odds of flowering than the foreign CO homozygotes (odds ratio = 3.94 for comparison of local vs. foreign genotype, 95% CI: 2.09 – 7.43, adjusted p-value <0.0001). These results are qualitatively similar to results of a model that included plants that did not survive the first winter, but excluded plants whose mortality could be unequivocally attributed to gopher activity (see Table S1).
Table 2.
Effect of genotype at the nFT QTL, environment, and interactions on the probability of flowering. Significance of the random effects was determined by likelihood ratio tests of models with and without these effects. Significant effects are highlighted in bold.
| Probability of flowering |
||
|---|---|---|
| F1,1430 | p-value | |
| nFT QTL genotype | 1.43 | 0.23 |
| Environment (E) | F1,29=1.8 | 0.19 |
| nFT × E | 24.3 | <0.0001 |
| Genomic proportion of Colorado alleles (%CO) | 0.07 | 0.80 |
| %CO × E | 3.6 | 0.057 |
| cohort | 2.55 | 0.11 |
| cohort × nFT | 4.8 | 0.029 |
| cohort × %CO | 2.7 | 0.10 |
| Family × E | χ2= 64.5 | <0.0001 |
| Block × E | χ2= 96.2 | <0.0001 |
Figure 3.
nFT QTL by environment interactions. Native alleles have significantly greater probability of flowering than foreign alleles in both environments. Means and standard errors were calculated based on family-level variation in the proportion of individuals that flowered. Triangles represent Montana homozygotes and circles signify Colorado homozygotes at the nFT QTL; filled symbols indicate data from RILs planted into both sites in September 2009 and monitored in 2010 (connected between sites by lines). RILs planted in fall 2008 into the Colorado garden failed to flower in 2009; experimental plants in the Montana garden from that year are indicated by unfilled, unconnected symbols.
Discussion
Here, we investigated the genomic basis of local adaptation in 177 F6 recombinant inbred lines (RILs) of Boechera stricta (Brassicaceae) created from a cross between a Montana father and a Colorado mother and outplanted into both parental environments. Our permutation approach revealed that polygenic selection favors local alleles in contrasting environments, indicative of local adaptation. Additionally, both antagonistic pleiotropy and conditional neutrality contribute to local adaptation in Boechera stricta. Furthermore, this analysis detected antagonistic pleiotropy at the nFT QTL, a QTL previously mapped for flowering phenology and the probability of flowering (Anderson et al., 2011a). Therefore, this permutation analysis is likely to identify loci that are the targets of selection in other systems. Previous investigations of antagonistic pleiotropy vs. conditional neutrality have focused on a relatively small number of loci. In contrast, this new analytical tool will allow researchers to elucidate the genetic basis of local adaptation across the genome and can be applied to existing databases and any relevant fitness component.
Owing to the pronounced effect of a previously mapped life history QTL on flowering phenology (Anderson et al., 2011a), we specifically investigated local adaptation at the nFT QTL. Our study clearly demonstrated fitness tradeoffs at the nFT QTL as native alleles resoundingly enhanced the probability of flowering in both field environments (Fig. 3). This study is one of the very few to investigate the genetic basis of local adaptation in the natural environments where polymorphism evolved (Barrett et al., 2008; Hall et al., 2010; Hawthorne, Via, 2001; Lowry, Willis, 2010; Weinig et al., 2003), and our results are highly consistent with antagonistic pleiotropy. As a consequence of selection for the native allele at the nFT QTL, our experiment revealed selective change in allele frequency in both environments within a generation after one episode of selection (Fig. 2). Here, we demonstrated that the nFT flowering phenology QTL has consistent effects on the probability of flowering under natural field conditions, corresponding to the known biological role of the Arabidopsis thaliana FLOWERING LOCUS T (FT) gene (Anderson et al., 2011a; Ehrenreich et al., 2009).
Previously, we showed that the nFT QTL influenced up to 27% of the phenotypic variation in the timing of flowering and up to 45% of the variation in size at flowering in laboratory environments that differed in ambient temperature, photoperiod, and vernalization (length of winter) (Anderson et al., 2011a). This QTL is centered on the genomic region that contains the A. thaliana FT gene, a key floral integrator in the Arabidopsis flowering time gene network (Anderson et al., 2011a; Ehrenreich et al., 2009). In Arabidopsis, FT receives signals from the vernalization, photoperiod, autonomous and gibberellin pathways, and encodes a protein that contributes to the initiation of flowering (Ehrenreich et al., 2009; Turck et al., 2008; Wellmer, Riechmann, 2010). A wealth of recent studies indicates that orthologs of FT affect flowering time in multiple plant species (Blackman et al., 2010; Böhlenius et al., 2006; Hayama et al., 2007; Lin et al., 2007; Skøt et al., 2011). Clearly, FT represents an important candidate gene at the nFT QTL in Boechera stricta.
Conditional neutrality can result in local adaptation at the organismal level, especially when limited gene flow prevents the assembly of recombinant genotypes carrying conditionally beneficial alleles at different loci (Fournier-Level et al., 2011; Hall et al., 2010; Verhoeven et al., 2008). Perhaps not surprisingly, conditional neutrality has been documented primarily in selfing species, such as Hordeum spontaneum (wild barley, Verhoeven et al., 2008; Verhoeven et al., 2004), Avena barbata (an annual grass, Gardner, Latta, 2006; Latta et al., 2007; Latta et al., 2010), and the model species Arabidopsis thaliana (Fournier-Level et al., 2011; Weinig et al., 2003), but also in the mixed mating system of Mimulus guttatus (yellow monkeyflower, Hall et al., 2010; Lowry et al., 2009). In contrast, although Boechera stricta is highly inbred, our results are consistent with both antagonistic pleiotropy (primarily in the region of the nFT QTL) and conditional neutrality (18 loci, on three distinct linkage groups). Our large sample sizes in contrasting natural environments facilitated discovery of fitness tradeoffs at the level of the QTL for a fitness component (probability of flowering) relevant to the flowering phenology nFT QTL.
The CNAP procedure we outline here improves our ability to detect antagonistic pleiotropy; however, ascertainment bias could still influence the results we present. If native alleles at a given locus have only a small (~1%) fitness advantage relative to foreign alleles in both environments, then our CNAP permutation approach would not likely identify the locus as a target of selection in either environment. Nevertheless, even small fitness advantages of this sort could result in adaptive population divergence. The sequential testing approach of CNAP could facilitate detection of antagonistic pleiotropy at loci that would otherwise be overlooked in completely independent analyses in two environments. To understand the relative role of local adaptation in adaptive population differentiation, we must be able to estimate the relative contributions of antagonistic pleiotropy and conditional neutrality; however, this task remains difficult due to limited statistical power and multiple tests.
Future directions
There are very few well-documented examples consistent with antagonistic pleiotropy at the QTL-level in the natural environments in which polymorphisms originally evolved (but see Hall et al., 2010; Hawthorne, Via, 2001), and we know of no study that definitely demonstrates antagonistic pleiotropy at a polymorphic causal gene. Allelic variation at a locus can be the result of divergent local selection, population history, genetic drift, and/or gene flow among populations. To understand whether divergent selection maintains genetic variation at key loci, we need additional large-scale field studies of species that vary in life history (annual vs. perennial) and mating system (selfing vs. outcrossing) (Anderson et al., 2011b). These studies should measure fitness components in relatively-undisturbed native habitats to test whether selection favors local alleles. Complementary population genomic analyses can elucidate the historical origin of polymorphism at key loci, and can test for global signatures of balancing selection across the range of a species (Stinchcombe, Hoekstra, 2008).
Studies that examine allelic variation at causal genes, not just QTLs, will improve our understanding of the genetic basis of local adaptation. Hundreds of genes can occur within the confidence interval of a QTL, and antagonistic pleiotropy at the level of a fitness QTL could be the product of conditional neutrality of two or more tightly linked genes (i.e., pseudo-antagonistic pleiotropy). Indeed, the nFT QTL could represent a case of pseudo-antagonistic pleiotropy; thus, it is critical to identify the gene or genes underlying variation in fitness and ecologically-relevant QTLs. When the genetic basis of fitness is resolved, we can address advanced evolutionary questions, such as: Are conditionally beneficial alleles sweeping to fixation? Is antagonistic pleiotropy more prevalent when interhabitat gene flow is high, and divergent selection favors local alleles in contrasting environments? Does conditional neutrality predominate when interhabitat gene flow is low, e.g., in selfing species, or in outcrossing species when populations are geographically isolated?
Conclusions
For many loci across the genome, our permutation analysis revealed selection for the native allele in both environments, which is highly suggestive of polygenic local adaptation. Both antagonistic pleiotropy and conditional neutrality contributed to local adaptation. Our study is among the first to demonstrate antagonistic pleiotropy at the QTL-level in plants grown in native field environments. At a large flowering phenology QTL, the native allele is favored in each of our garden sites. Our CNAP permutation framework is a generally applicable method for illuminating the genetic basis of local adaptation across various episodes of selection, produced results that were highly consistent with previous analyses (Anderson et al., 2011a), and can be used with existing and future datasets in other systems in conjunction with traditional QTL mapping.
Ultimately, to evaluate the genetic basis of local adaptation, researchers must: (1) quantify fitness components of experimental individuals in at least two contrasting relatively undisturbed environments, within the native range of the species; (2) identify the gene(s) that influence fitness; (3) understand the mechanistic control of phenotype and fitness; and (4) gather population genetic data on allelic variation and historical origin of the polymorphism. To date, no study has met all four of these criteria. Investigations of ecologically relevant QTL (e.g., the nFT QTL) and candidate genes play a vital role in evaluating antagonistic pleiotropy, pseudo-antagonistic pleiotropy, and conditional neutrality. Testing among these hypotheses will illuminate the forces that maintain variation in genes underlying adaptive evolution.
Supplementary Material
Acknowledgements
We thank A. Manzaneda, B.-H. Song, C.-L. Huang, S. Mitchell-Olds, K. Dales, R. Doll, E. Raskin, and M. Wagner for assistance with field work, K. Springer and J. Rivera for greenhouse help, and C. Olson-Manning and T. Pendergast for thoughtful discussions. Jennie Reithel and Ian Billick at the Rocky Mountain Biological Laboratory facilitated the Colorado field study. We are indebted to Nancy Wicks for allowing us to conduct the Colorado common garden on her property. We thank Editor Rose Andrew and three anonymous reviewers for comments on a previous draft. Funding from the National Science Foundation (EF-0723447) and the National Institutes of Health (R01 GM086496) supported this research.
Footnotes
Data Accessibility:
Genotypic data: Available as an appendix (Table S1) associated with Anderson et al. (2011). http://onlinelibrary.wiley.com/doi/10.1111/j.1558-5646.2010.01175.x/suppinfo Fitness data deposited in the Dryad repository: doi:10.5061/dryad.c7ss600f
References
- Anderson JT, Lee C-R, Mitchell-Olds T. Life-history QTLs and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution. 2011a;65:771–787. doi: 10.1111/j.1558-5646.2010.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends in Genetics. 2011b;27:258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Rogers SM, Schluter D. Natural selection on a major armor gene in Threespine Stickleback. Science. 2008;322:255–257. doi: 10.1126/science.1159978. [DOI] [PubMed] [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AW, Michaels SD, Rieseberg LH. The role of recently derived FT paralogs in sunflower domestication. Current Biology. 2010;20:1–7. doi: 10.1016/j.cub.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Brunelle A, Whitlock C, Bartlein P, Kipfmueller K. Holocene fire and vegetation along environmental gradients in the Northern Rocky Mountains. Quaternary Science Reviews. 2005;24:2281–2300. [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Hanzawa Y, Chou L, et al. Candidate Gene Association Mapping of Arabidopsis Flowering Time. Genetics. 2009;183:325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Fry JD. The evolution of host specialization: Are trade-offs overrated? American Naturalist. 1996;148:S84–S107. [Google Scholar]
- Galloway LF, Fenster CB. Population differentiation in an annual legume: Local adaptation. Evolution. 2000;54:1173–1181. doi: 10.1111/j.0014-3820.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- Gardner KM, Latta RG. Identifying loci under selection across contrasting environments in Avena barbata using quantitative trait locus mapping. Molecular Ecology. 2006;15:1321–1333. doi: 10.1111/j.1365-294X.2005.02835.x. [DOI] [PubMed] [Google Scholar]
- Hall MC, Lowry DB, Willis JH. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Molecular Ecology. 2010;19:2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- Hayama R, Agashe B, Luley E, King R, Coupland G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. The Plant Cell. 2007;19:2988–3000. doi: 10.1105/tpc.107.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: A decade later. Annual Review of Ecology and Systematics. 1986;17:535–566. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. American Naturalist. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ. Sympatric speciation via habitat specialization driven by deleterious mutations. Evolution. 1997;51:1751–1763. doi: 10.1111/j.1558-5646.1997.tb05099.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Latta RG, Gardner KM, Johansen-Morris AD. Hybridization, recombination, and the genetic basis of fitness variation across environments in Avena barbata. Genetica. 2007;129:167–177. doi: 10.1007/s10709-006-9012-x. [DOI] [PubMed] [Google Scholar]
- Latta RG, Gardner KM, Staples DA. Quantitative trait locus mapping of genes under selection across multiple years and sites in Avena barbata: Epistasis, pleiotropy, and genotype-by-environment interactions. Genetics. 2010;185:375–385. doi: 10.1534/genetics.110.114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimu R, Fischer M. A meta-analysis of local adaptation in plants. Public Library of Science One. 2008;3:e4010. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen PH, Remington DL, Savolainen O. Local adaptation, phenotypic differentiation, and hybrid fitness in diverged natural populations of Arabidopsis lyrata. Evolution. 2010;65:90–107. doi: 10.1111/j.1558-5646.2010.01119.x. [DOI] [PubMed] [Google Scholar]
- Lin M-K, Belanger H, Lee Y-J, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hall MC, Salt DE, Willis J. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytologist. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. Public Library of Science Biology. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Nagy ES. Selection for native characters in hybrids between two locally adapted plant subspecies. Evolution. 1997;51:1469–1480. doi: 10.1111/j.1558-5646.1997.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Gamble SE. The Effect of Distance from the Parental Site on Offspring Performance and Inbreeding Depression in Impatiens capensis - a Test of the Local Adaptation Hypothesis. Evolution. 1990;44:2022–2030. doi: 10.1111/j.1558-5646.1990.tb04308.x. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Manzaneda AJ, Windsor AJ, Clauss MJ, Mitchell-Olds T. Ecological genomics of Boechera stricta: identification of a QTL controlling the allocation of methionine- vs branched-chain amino acid-derived glucosinolates and levels of insect herbivory. Heredity. 2009;102:465–474. doi: 10.1038/hdy.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Windsor AJ, Song BH, Lawton-Rauh A, Mitchell-Olds T. Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiology. 2007;144:286–298. doi: 10.1104/pp.107.096685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skøt L, Sanderson R, Thomas A, et al. Allelic variation in the perennial ryegrass Flowering Locus T gene is associated with changes in flowering time across a range of populations. Plant Physiology. 2011;155:1013–1022. doi: 10.1104/pp.110.169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BH, Clauss MJ, Pepper A, Mitchell-Olds T. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Molecular Ecology. 2006;15:357–369. doi: 10.1111/j.1365-294X.2005.02817.x. [DOI] [PubMed] [Google Scholar]
- Song BH, Windsor AJ, Schmid B, et al. Multilocus patterns of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics. 2009;181:1021–1033. doi: 10.1534/genetics.108.095364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Poorter H, Nevo E, Biere A. Habitat-specific natural selection at a flowering-time QTL is a main driver of local adaptation in two wild barley populations. Molecular Ecology. 2008;17:3416–3424. doi: 10.1111/j.1365-294X.2008.03847.x. [DOI] [PubMed] [Google Scholar]
- Verhoeven KJF, Vanhala TK, Biere A, Nevo E, van Damme JMM. The genetic basis of adaptive population differentiation: a quantitative trait locus analysis of fitness traits in two wild barley populations from contrasting habitats. Evolution. 2004;58:270–283. [PubMed] [Google Scholar]
- Weinig C, Dorn LA, Kane NC, et al. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics. 2003;165:321–329. doi: 10.1093/genetics/165.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends in Genetics. 2010;26:519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.