SUMMARY
A fundamental challenge in mammalian biology has been elucidating mechanisms linking DNA methylation and histone post-translational modifications. Human UHRF1 (ubiquitin-like, PHD and RING finger containing 1) has multiple domains that bind chromatin and is implicated genetically in DNA methylation maintenance. However, molecular mechanisms underlying DNA methylation regulation by UHRF1 are poorly defined. Here we show that UHRF1 association with methylated histone H3 lysine 9 (H3K9) is required for DNA methylation maintenance. We further show that UHRF1 association with H3K9 methylation is insensitive to adjacent H3 serine 10 phosphorylation – a known mitotic ‘phospho/methyl switch.’ Importantly, we demonstrate that UHRF1 mitotic chromatin association is necessary for DNA methylation maintenance through regulation of DNMT1 stability. Collectively, our results define a novel link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation, establishing an unexpected mitotic role for UHRF1 in this process.
Keywords: UHRF1, DNA methylation, histones, post-translational modifications, epigenetics
INTRODUCTION
Eukaryotic DNA is packaged in the nucleus by histone proteins to form chromatin. The functional subunit of chromatin is the nucleosome, in which approximately 147 base pairs of DNA is wrapped around a histone octamer containing two copies each of the four core histone proteins (H3, H4, H2A, and H2B)1–3. The unstructured tails of histones are rich in post-translational modifications (PTMs) such as methylation, acetylation, and phosphorylation4,5. Histone PTMs can alter the physical structure of chromatin6 and have been proposed to function in the form of a ‘histone code’ to coordinate the recruitment of effector proteins that elicit selective effects on gene expression and other chromatin-templated biological processes7–9. DNA methylation adds an additional layer of epigenetic regulation on chromatin. The faithful inheritance of DNA methylation patterns is essential for normal mammalian development and long-term transcriptional silencing10. While studies suggest there is evolutionarily conserved crosstalk between DNA methylation and histone PTMs11–13, biological mechanisms linking these two chromatin modifications in mammals have not been established14.
The E3 ubiquitin ligase UHRF1 is genetically linked to the establishment and maintenance of DNA methylation15,16 in mammals. Deletion of UHRF1 in mice is embryonic lethal, and mouse embryonic stem cells (mESCs) derived from these mice show a dramatic loss in DNA methylation, impaired maintenance of higher order chromatin structure, and spurious transcription of otherwise silenced repetitive DNA elements15,16. The UHRF1 SET and RING-associated (SRA) domain binds hemi-methylated CpG dinucleotides17–19. These results, along with others15,16, suggest that UHRF1 binds hemimethylated DNA during the semi-conservative replication of DNA in S-phase and recruits DNA methyltransferase 1 (DNMT1) to copy the methylation pattern on the daughter strand. We recently showed that through a conserved aromatic cage in the first subdomain of its tandem Tudor domain (TTD)20, UHRF1 binds to, and colocalizes with, histone H3 tri-methylated at lysine 9 (H3K9me3) – a transcriptionally repressive chromatin mark, enriched at pericentric heterochromatin and telomeres21–24. While the evidence of cooperative interplay between DNA methylation and UHRF1 has remained vague, the coordinated binding of UHRF1 to repressive chromatin signatures suggests an attractive effector-mediated mechanism for the propagation of epigenetic information. We therefore sought to determine whether the binding of UHRF1 to methylated H3K9 provides a mechanism for the propagation of DNA methylation patterns in mammals.
ONLINE METHODS
Materials
Histone peptides were synthesized, purified, and analyzed as described19. Antibodies used in this study: anti-GST (Sigma G7781; 1:1,000), anti-HIS (Santa Cruz sc-8036; 1:200), anti-myc (Millipore 05-419; 1:2,500), anti-Flag (Sigma F1804; 1:5000), anti-streptavidin HRP (Cell Signaling 3999; 1:10,000), anti-UHRF1 (Abcam ab57083; 1:1,000), anti-HP1γ (Cell Signaling 2619; 1:1,000), anti-β-tubulin (Cell Signaling 2146; 1:1,000), anti-H3 (Active Motif 39163; 1:20,000), anti-H3K9me3 (Active Motif 39765; 1:5,000), anti-H3K9me2/S10p (Millipore 05-1354; 1:1,000), anti-H3K9me3/S10p (Millipore 04-809; 1:10,000), anti-H3S10p (Active Motif 39253; 1:5,000), anti-5mC (Diagenode Mab-081; 1:100), anti-cyclin A (Santa Cruz sc-751; 1:2000), anti-cyclin E (Santa Cruz sc-247; 1:1,000). The UHRF1 TTD (human cDNA encoding residues 126–280) was cloned into pET28a-LIC (GenBank accession EF442785) as an N-terminal HIS fusion, expressed in Escherichia coli BL21(DE3) using standard procedures, and purified with Talon resin (ClonTech) according to the manufacturer’s protocol. HP1α (mouse full-length cDNA), HP1β (mouse full-length cDNA), HP1γ chromodomain (mouse cDNA encoding residues 11–129), and MPP8 chromodomain (human cDNA encoding residues 50–118) were cloned into pGEX-KG (GE Life Sciences). GLP ankyrin repeats (human cDNA encoding residues 734–968) were cloned into pGEX-6P1 (GE Life Sciences). GST fusion proteins were expressed in Escherichia coli BL21(DE3) using standard procedures and purified with GST-bind resin (Novagen) according to the manufacturer’s protocol. Full length human UHRF1 was cloned into pCMV-Tag 2 (Agilent) as an N-terminal Flag fusion for mammalian expression. Full length human DNMT1 (a gift from Zhenghe Wang; Case Western) was cloned into pCMV-3Tag (Agilent) as an N-terminal myc fusion for mammalian expression. Point mutations were generated by QuickChange site-directed mutagenesis (Stratagene).
Cell culture and manipulation
HeLa cells (ATCC) were cultured in Minimal Essential Medium (Invitrogen) supplemented with 10% fetal bovine serum (PAA), maintained in a 37°C incubator with 5% CO2, and passaged every 2–3 days. E14 and NP95 −/− mouse ES cells (a gift from Haruhiko Koseki, RIKEN) were cultured on 0.1% gelatin (Sigma) in Glasgow’s Minimal Essential Medium (Invitrogen) supplemented with 15% ES-fetal bovine serum (PAA), 50 units/mL Leukemia Inhibitory Factor (Millipore), 2 mM L-glutamine (Invitrogen), 0.1 mM non-essential amino acids (Invitrogen), 55 µM beta-mercaptoethanol (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 1× penicillin-streptomycin solution (Invitrogen), maintained in a 37°C incubator with 5% CO2, and passaged every 2 days. HeLa cells were synchronized in mitosis with 0.05 µg mL−1 nocodazole for 16 hours. For double thymidine block, HeLa cells were synchronized by treatment with 2 mM thymidine (Sigma) for 16 hours, followed by release for 8 hours, and re-treatment with 2 mM thymidine for 16 hours. Transient transfections were performed using TurboFect (Fermentas) according to the manufacturer’s protocol. shRNAs obtained from The RNAi Consortium (TRC) are described in Supplementary Fig. 5. and were used following standard TRC Lentivirus production and infection protocols. The indicted concentrations of MG132 (Cayman Chemicals) or 0.05 µg mL−1 nocodazole (Sigma) in DMSO were added during the last 16 hours prior to harvest.
Histone peptide microarrays
Array fabrication and effector protein analysis was performed as described25,26. Heat maps were generated using Java TreeView.
In-solution peptide pulldowns
A 50 µL slurry of streptavidin magnetic beads (NEB) was equilibrated in binding buffer containing 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 0.1% NP-40 before being saturated with 1 nmole biotinylated peptide for 1 hour at 4°C with rotation. Unbound peptide was washed with binding buffer, and 100 pmoles of protein in binding buffer supplemented with 0.5% bovine serum albumin (BSA) (w/v) was incubated for 3 hours at 4°C with rotation. Unbound protein was washed with binding buffer, and bound protein and peptide were eluted from beads by boiling in 1× SDS loading buffer followed by western blot detection. Proteins were detected with anti-HIS (UHRF1) and anti-GST (MPP8) and peptides were detected with anti-streptavidin-HRP.
Fluorescence polarization
Peptides for fluorescence polarization (histone H3, residues 1–20) were synthesized as described25 with the addition of 5-carboxyfluorescin (5-FAM) at the N-terminus. Binding assays were performed in 40 µL volume in black flat-bottom 384-well plates (Costar). Protein was titrated with 50 nM peptide in buffer containing 20 mM Tris-HCl, pH 8.0, 250 mM NaCl, 1 mM DTT, and 0.05% NP-40. Following a 20 minute equilibration period at 25°C, plates were read on a POLARstar Omega (BMG Labtech) using a 480 nm excitation filter and 520/530 ± 10 nm emissions filters. Gain settings in the parallel (||) and perpendicular (⊥) channels were calibrated to a polarization measurement of 100 milli-polarization units (mP) for the fluorescent peptide in the absence of protein. Polarization (P) was determined from raw intensity values of the parallel and perpendicular channels using the equation P = || − ⊥ / || + 2(⊥) and converted to anisotropy (A) units using the equation A = 2P / 3 – P. Equilibruim dissociation constants (Kd) were determined by fitting anisotropy curves to a one-site binding model using GraphPad Prism 5.0.
NMR Spectroscopy and data analysis
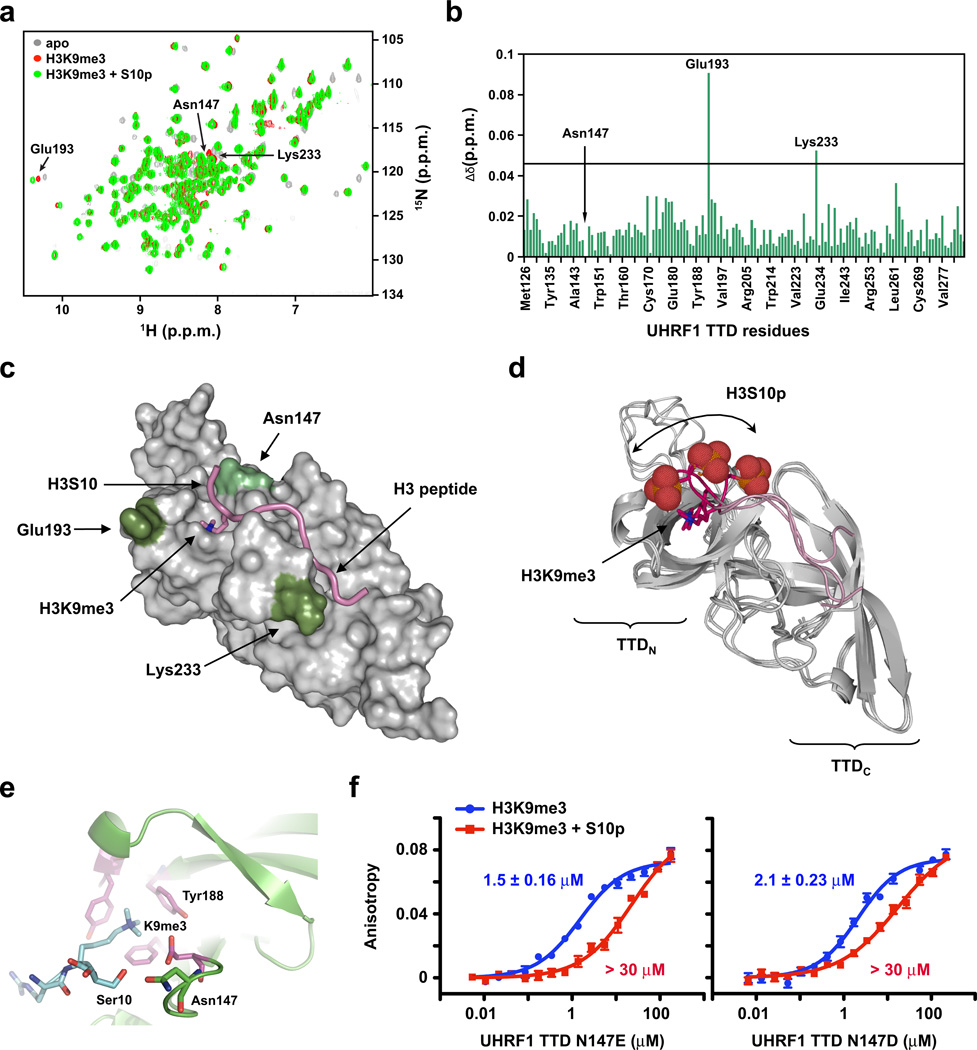
Chemical shift mapping on the UHRF1 TTD domain was performed by monitoring the 1H-15N HSQC spectra of the uniformly 15N-labeled TTD domain alone (0.45 mM) and with an excess of unlabeled H3 peptide (ARTKQTARK9me3/S10pT). Aliquots of unlabeled peptide were titrated into the labeled TTD domain in molar ratio 1:5 until no further changes in chemical shifts were detected in the 1H-15N HSQC spectrum. The HSQC spectra were recorded at 25°C in 20 mM NaPi, pH 7.0, 250 mM NaCl, 2 mM DTT, 1 mM benzamidine, 0.5 mM PMSF, and supplemented with 10% (v/v) D2O on a Bruker Avance 800-MHz spectrometer. Composite chemical shift perturbation values of TTD bound to the H3K9me3/S10p peptide (compared to those with peptide containing H3K9me3 and unphosphorylated S1020) were calculated using the equation Δcomp = [Δδ2HN + (ΔδN/6.5)2]1/2.
A model of TTD bound to the (ARTKQTARK9me3/S10pT) histone H3 peptide was generated by using the NMR solution ensemble of TTD in a complex with the unphosphorylated peptide (PDB: 2L3R20) as the starting ensemble. We then re-refined the structure using CNS (version 1.3) in explicit water, but with the Ser10 sidechain phosphorylated, and included all experimental intraprotein and protein-peptide constraints. A total of three representative models that show various orientations of H3S10p are shown in Fig. 2a.
Figure 2. A structural basis for UHRF1 TTD insensitivity to H3S10 phosphorylation.
(a) 15N-1H heteronuclear single quantum coherence (HSQC) spectra of UHRF1 TTD alone (grey) or upon addition of H3K9me3 peptides in the absence (red) or presence (green) of H3S10p. (b) Composite chemical shift changes (Δδ) versus residue number for the UHRF1 TTD domain after binding H3K9me3 in the absence or presence of H3S10p. Prolines and residues that could not be assigned are not indicated on the histogram. Residues (Asn147) whose peaks disappeared upon addition of the peptide were given a value of zero. Residues (Glu193, Lys233) that were strongly affected are listed on the histogram. (c) Surface representation of the UHRF1 TTD bound to an H3K9me3 peptide in the presence of S10p. Colored are residues that have marked chemical shift perturbation values (dark green) or were missing (light green) as compared to those with peptide containing H3K9me3 alone (PDB: 2L3R). (d) Three NMR models of the UHRF1 TTD (ribbon representation) bound to an H3K9me3 peptide (pink backbone) (PDB: 2L3R) re-refined to also contain H3S10p (red space-fill). (e) Crystal structures of the UHRF1 TTD (PDB: 3DB3) bound to an H3K9me3-containing peptide (blue). Aromatic cage residues are depicted in magenta and the residues interacting with H3S10 are depicted as green sticks. (f) Fluorescence polarization binding assays of H3K9me3 peptides with the indicated protein domains in the absence (blue) or presence (red) of H3S10p. Error is represented as ± s.d. for three independent experiments. The y-axis is on the same scale for both plots.
Chromatin fractionation
Chromatin and associated proteins were isolated from asynchronously growing HeLa cells essentially as described38 with the following modifications: Cells lysis and washing steps were performed in cold buffer containing 10 mM PIPES, pH 7.0, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 1× EDTA-free protease inhibitor (Roche), 1× phosphatase inhibitor cocktail (Sigma), and 0.1% Triton X-100. Chromatin fractions were treated with benzonase (Sigma) prior to western blot analysis. H3 and β-tubulin antibody controls monitored the purity of fractionation.
DNA methylation analysis
For immunofluorescence analysis of 5mC content, HeLa cells growing in 8-well chamber slides (Lab-Tek) were fixed with ice-cold methanol at −20°C for 10 minutes before being treated with 2N HCl for 30 minutes at 37°C to denature the DNA. Following neutralization with 0.1 M Borate, pH 8.5, cells were blocked for 30 minutes in phosphate-buffered saline (PBS) containing 1% (w/v) BSA. Labeling was performed with anti-5mC for 1 hour at 25°C in PBS containing 1% BSA. Cells were washed with PBS and incubated with an anti-AlexaFluor 647 mouse IgG (Invitrogen; 1:1000) in PBS for 1 hour at 25°C protected from light. Cells were washed with PBS and mounted using SlowFade AntiFade (Invitrogen) containing DAPI. Images were acquired using an FV1000 Multiphoton inverted confocal microscope (Olympus) at 30× magnification following excitation with 405 nm and 633 nm single photon excitation lasers. Images were analyzed using the Measure RGB plugin on NIH ImageJ version 1.44o. Methylation percentage was derived from quantification of 5mC fluorescence (red channel) normalized to DNA fluorescence (blue channel) from at least four optical fields.
For bisulfite sequencing, genomic DNA was isolated from HeLa cells using a DNeasy Blood and Tissue Kit according to the manufacturer’s protocol. 500 ng of genomic DNA was treated with sodium bisulfite and desulphonated using an EZ DNA Methylation-Gold Kit (Zymo). Bisulfite DNA was used as a template to amplify the IGS-rDNA locus (Supplementary Fig. 6) with EpiMark DNA polymerase (NEB) using sense primer GAGGGGTATTTTTAGATTTTTTTT and antisense primer TCTCACTCACTCTACAACCTAAACC. PCR conditions consisted of 38 cycles, denaturing at 98°C for 30 seconds, annealing for 1 minute at 58°C, and extending at 68°C for 30 seconds. Following a final extension at 68°C for 5 minutes, PCR products were purified, directly ligated into a pDrive cloning vector (Qiagen) according to the manufacturer’s protocol, and miniprepped DNA was sequenced (Eurofins). Sequencing results were analyzed with Bisulfite Sequencing DNA Methylation Analysis39 (BISMA) using default stringency parameters. 5mC percentage was calculated from individual clones.
RESULTS
UHRF1 binds H3K9 methylation regardless of H3S10p
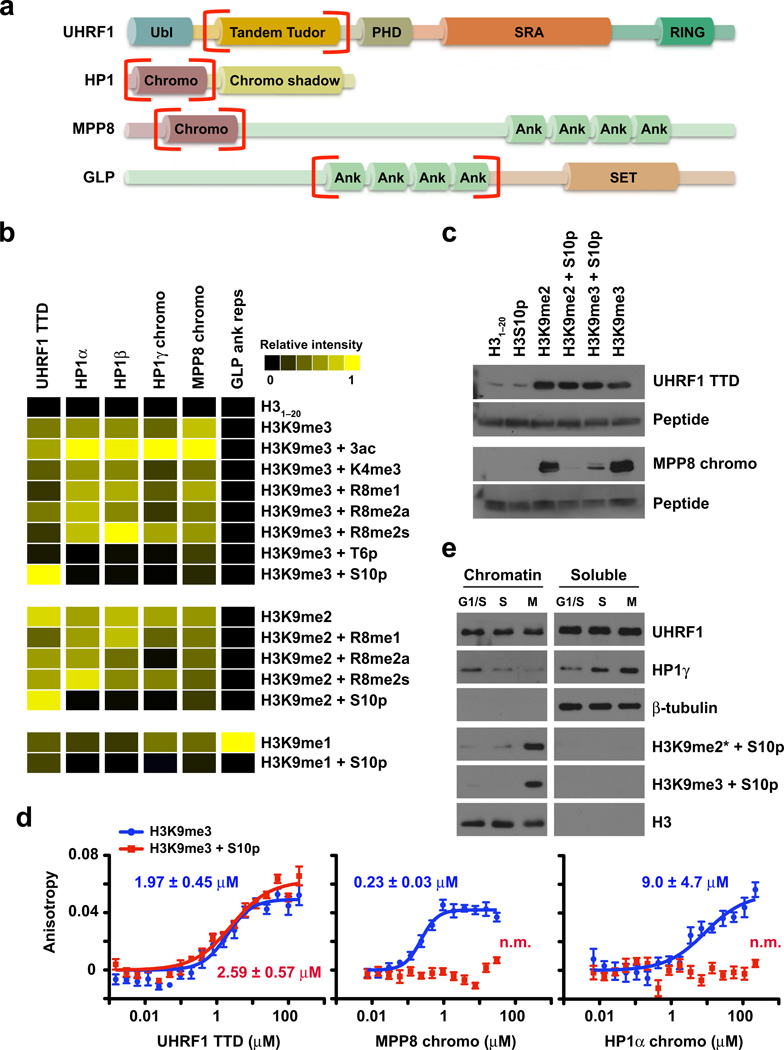
We began this study by using a recently developed histone peptide microarray platform to compare the binding properties of the UHRF1 TTD with other known H3K9 methyl effector protein domains, including the chromodomains of the three HP1 isoforms (α, β, γ), the MPP8 chromodomain, and the GLP ankyrin repeats (Fig. 1a). These peptide microarrays contain a library of 130 unmodified and modified histone peptides representing known single and combinatorial PTMs on the four core histones (H2A, H2B, H3, and H4), including lysine and arginine methylation, lysine acetylation, and serine and threonine phosphorylation (Supplementary Table 1). Arrays were spotted 24 times with each histone peptide as described25 and probed with the indicated HIS-tagged (UHRF1) or GST-tagged (HP1, MPP8, GLP) protein domains. Array analysis revealed that these effector proteins preferentially bound to H3K9 methylated peptides (Fig. 1b and Supplementary Fig. 1). With the exception of the GLP ankyrin repeats, which bound preferentially to H3K9me1, these effector proteins had a general preference for H3K9me2 and H3K9me3. Importantly, no binding was observed for H3K27 methylated peptides, which share a conserved ARKS binding motif with H3K9 (Supplementary Fig. 1c).
Figure 1. The UHRF1 tandem Tudor domain binds H3K9 methylation regardless of neighboring H3S10 phosphorylation.
(a) Domain map of H3K9 methyl effector proteins. Domains that bind methylated H3K9 are in red brackets. Ubl (ubiquitin-like); PHD (plant homeodomain); SRA (SET and RING-associated domain); RING (really interesting new gene); ank (ankyrin repeats); SET (Su(var)3–9 and Enhancer of zeste domain). (b) Peptide microarray analysis of the indicated H3K9 effector domains. Results of at least two arrays are presented as heat maps of normalized mean intensities on a scale from 0 (black; undetectable binding) to 1 (yellow; strong binding). (c) Western blot following in-solution peptide pulldowns with the indicated domains. (d) Fluorescence polarization binding assays of H3K9me3 peptides with the indicated protein domains in the absence (blue) or presence (red) of H3S10p. Error is represented as ± s.d. for three independent experiments. The y-axis is on the same scale for all plots. (e) Western blot analysis of HeLa cell lysates biochemically separated into chromatin and soluble fractions at 0 (G1/S), 5 (S), and 10 (M) hours following release from double thymidine block. See also Supplementary Fig. 2a.
Recent work from our lab and others demonstrates that neighboring PTMs often enhance or perturb the binding of effector proteins to their intended target25–28. Analyzing the influence of neighboring PTMs on the binding of these effector proteins to H3K9 methylated peptides revealed little influence of lysine acetylation, H3K4me3, or H3R8 methylation (mono-, symmetric, or asymmetric di-methylation), with the exception of the HP1γ chromodomain, whose binding to H3K9me2 and H3K9me3 was partially perturbed by H3R8me2a. In contrast, H3T6p perturbed the binding to H3K9me3 by all tested effector proteins (Fig. 1b). Interestingly, unlike other H3K9 methyl effectors tested, the UHRF1 TTD bound to H3K9me2 and H3K9me3 in the presence of H3S10p (Fig. 1b and Supplemental Fig. 1b), consistent with our previous observation using SPOT-array technology20. In-solution peptide pulldown assays verified that the UHRF1 TTD bound H3K9me2 and H3K9me3 regardless of the presence of H3S10p (Fig. 1c), unlike the MPP8 chromodomain (Fig. 1c) and HP1α (data not shown). Quantification of this interaction by fluorescence polarization showed the UHRF1 TTD bound to H3K9me3 peptides in the absence or presence of H3S10p with similar affinity (dissociation constants (Kd) of 2.0 µM and 2.6 µM, respectively) (Fig. 1d). In contrast, while the MPP8 chromodomain and HP1α bound to H3K9me3 peptides with Kd values of 0.23 µM and 9.0 µM, respectively, binding in the presence of H3S10p was not measurable (n.m.) (Fig. 1d).
Previous studies showed that Aurora kinase B-mediated H3S10p disrupts the binding of HP1 to H3K9 methylated chromatin in mitosis – a ‘phospho/methyl switch’ whose impairment leads to defects in chromosome alignment, segregation, spindle assembly, and cytokinesis29,30. Since our in vitro analysis revealed that the UHRF1 TTD bound H3K9 methylation regardless of H3S10p, we queried whether this ‘phospho/methyl switch’ affected the binding of UHRF1 to mitotic chromatin. Synchronized release of HeLa cells from double thymidine block showed that in mitosis all three states of H3K9 methylation (mono-, di-, and tri-methylation) occur in combination with H3S10p (Fig. 1e and Supplementary Fig. 2a,b). Biochemical separation of HeLa cells into chromatin-enriched and soluble fractions from time points along this synchronized released showed that unlike HP1γ, which was lost from chromatin fractions as cells progressed into mitosis and reciprocally was gained in the soluble pool, UHRF1 remained chromatin-bound through mitosis (Fig. 1e). Taken together, these results demonstrate that the UHRF1 TTD binds with specificity to H3K9me2 and H3K9me3 in vitro regardless of H3S10p, and they suggest that UHRF1 may evade the ‘phospho/methyl switch’, perhaps performing an unresolved mitotic function.
Structural basis for UHRF1 insensitivity to H3S10p
We reasoned that an understanding of the structural basis for the UHRF1 TTD insensitivity to H3S10p might allow us to uncouple UHRF1 from mitotic chromatin and explore its potential mitotic function. We showed previously that UHRF1 recognizes H3K9me3-containing histone peptides via its TTD with H3K9me3 sitting in the canonical aromatic cage20. To explain mechanistically the lack of sensitivity of UHRF1 towards the presence of H3S10p, we first obtained a crystal structure of the TTD in a complex with a short H3 peptide (residues 6–11, TARKme3SpT). While we were able to observe clear electron density for the H3K9me3 residue that fits in the aromatic cage, the electron density for the adjacent H3S10p residue was weak, suggesting multiple conformations of this side chain (data not shown).
We next examined the interaction of the UHRF1 TTD with a doubly modified H3 peptide (residues 1–11, ARTKQTARKme3SpT) using NMR spectroscopy and compared the results with our previously reported solution ensemble of the UHRF1 TTD bound to same peptide in the unphosphorylated form20 (Fig. 2a,b). Only three residues of the UHRF1 TTD (Asn147, Glu193, and Lys233) had substantial chemical shift differences when bound to an H3K9me3 peptide in the presence of S10p compared with the unphosphorylated peptide, indicating that both peptide-protein complexes have similar affinities and adopt similar conformations. Key among changes in chemical shift was the disappearance of the 15N-1H resonance of Asn147, whose side chain is closest to S10 in the unphosphorylated peptide-protein complex (Fig. 2c). This disappearance can be explained by intermediate conformational exchange on the NMR timescale near H3S10p, as suggested by our x-ray data, and implies that when bound to H3K9me3 in the presence of S10p, Asn147 adopts an altered conformation that may be important for this UHRF1 TDD interaction.
To better understand the potential conformations of H3S10p when bound to the TTD, we re-refined the NMR solution ensemble of the UHRF1 TTD in complex with H3K9me3 (PDB:2L3R)20, but with H3S10p. The ensemble of the H3K9me3 with S10p complex showed that the phosphoserine side chain can readily be accommodated by rotating outward into the solvent while maintaining all the experimental intra-protein and protein-peptide NOE (nuclear overhauser effect) values observed in the solution structure of the unphosphorylated peptide (Fig. 2d). The ensemble also offered an explanation for the change in 15N-1H resonance of Glu193, which may be influenced by a subset of the conformations of H3S10p.
A comparison of crystal structures of the UHRF1 TTD (PDB: 3DB320) (Fig. 2e), MPP8 chromodomain (PDB: 3QO231), and HP1α chromodomain (PDB: 1KNE32) in complex with H3K9me3 peptides identified a conserved, negatively charged glutamate residue in similar orientation and proximity (less than 3Å) to H3S10 as Asn147 of the UHRF1 TTD. Combined with our NMR results showing a chemical shift of Asn147 in the presence of an H3K9me3 and S10p-containing peptide, we rationalized that mutation of Asn147 to a glutamate or aspartate would place a repulsive negative charge near H3S10p and make the UHRF1 TTD sensitive to the ‘phospho/methyl switch.’ Indeed, UHRF1 N147E and UHRF1 N147D mutations reduced the binding affinity for the doubly-modified peptide by several orders of magnitude while minimally affecting binding to the H3K9me3 peptide (Fig. 2f). Importantly, HP1α E53N and MPP8 E91N mutations completely abolished binding of these domains to H3K9me3 (data not shown), suggesting chromodomain coordination of H3S10 through this conserved glutamate. Collectively, crystal and NMR structures of protein-peptide complexes coupled with mutagenesis studies indicate that the UHRF1 TTD can readily bind to both phosphorylated and unphosphorylated H3K9me3 peptides with almost identical conformations, and they suggest that H3S10p likely explores multiple solvent exposed conformations in the vicinity of Asn147 when bound to H3K9me3.
UHRF1 binds H3K9 methylation through mitosis to maintain 5mC
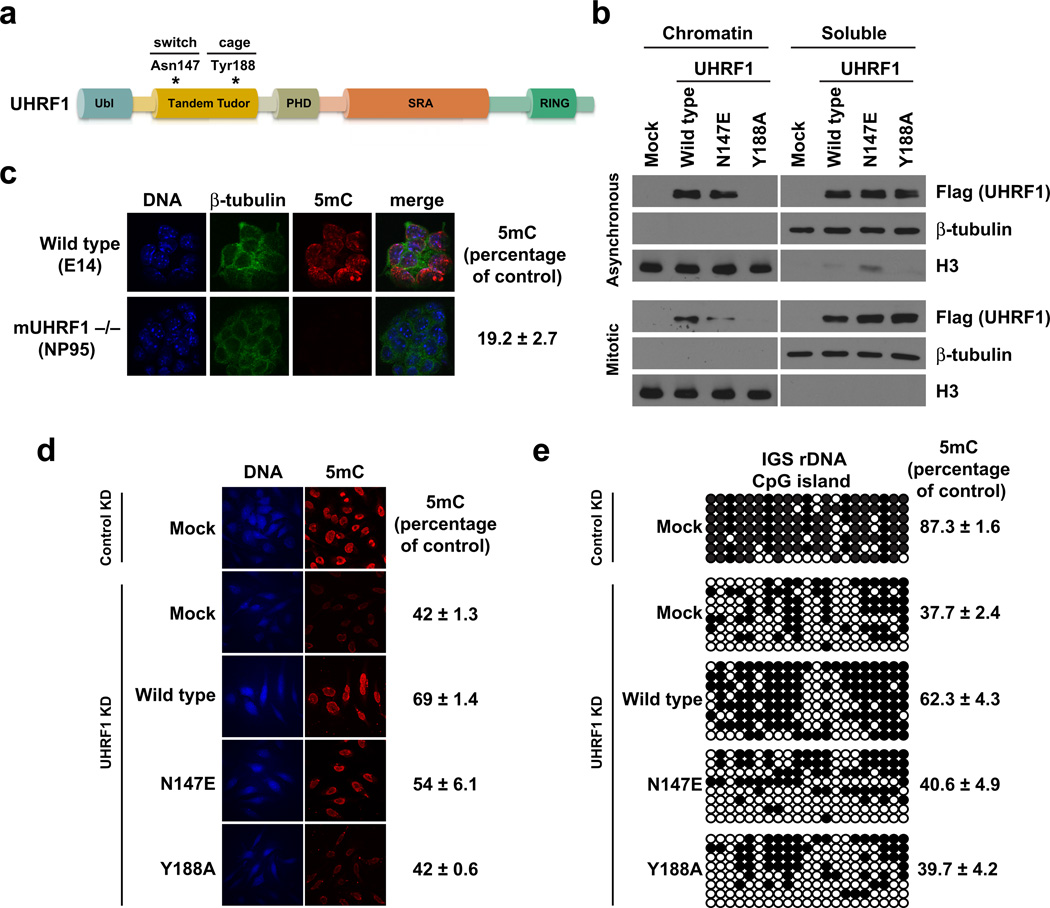
Next, we sought to determine whether the UHRF1 N147E mutation that made the UHRF1 TTD sensitive to the ‘phospho/methyl switch’ in vitro would affect the ability of UHRF1 to bind mitotic chromatin. In parallel, we examined the chromatin association of a UHRF1 aromatic cage mutant UHRF1 Y188A, incapable of binding H3K9me3 in vitro20. UHRF1 N147E “switch” and UHRF1 Y188A “cage” mutants (Fig. 3a) were transiently expressed in HeLa cells for 48 hours and resulting asynchronous and nocodazole-arrested cells were biochemically separated into chromatin-enriched and soluble fractions. UHRF1 Y188A was unable to bind bulk chromatin (Fig 3b), even with its intact SRA (binds CpG dinucleotides17–19) and PHD domains (binds to the unmodified H3 N-terminus33), thus indicating that binding of UHRF1 to H3K9 methylation is critical for its association with chromatin. Importantly, while UHRF1 N147E bound to asynchronous chromatin, its association with nocodazole-arrested mitotic chromatin was perturbed compared with wild type UHRF1 (Fig. 3b). Taken together, these results demonstrate that UHRF1 N147E confers upon UHRF1 sensitivity to the ‘phospho/methyl switch’ comparable to other H3K9 effector proteins (Fig. 1) and shows that H3K9 methylation plays a critical role in the association of UHRF1 with chromatin. However, it remains in question whether H3K9 methylation and/or mitotic binding by UHRF1 contributes to its function in DNA methylation maintenance.
Figure 3. Chromatin targeting of UHRF1 in mitosis is required for the maintenance of DNA methylation.
(a) Domain map of UHRF1. Residues mutated in this figure are indicated. (b) Western blot analysis of asynchronous and mitotically arrested HeLa cell lysates biochemically separated into chromatin and soluble fractions. Mock indicates no DNA control. (c) Immunofluorescence of mouse UHRF1 (NP95) wild-type (E14) or knockout embryonic stem cells. Error is represented as ± s.e.m. (d) Immunofluorescence staining in control and UHRF1 knockdown HeLa cells following genetic complementation with wild-type and mutant UHRF1. Mock indicates no DNA control. Error is represented as ± s.e.m. (e) Genetic complementation in HeLa cells as in d followed by sodium bisulfite conversion and sequencing of a CpG island in the intergenic spacer of the ribosomal DNA locus (IGS rDNA; see Supplementary Fig. 6). Individual CpG sites are represented as black (methylated) or white (unmethylated) circles. Error is represented as ± s.e.m.
Past studies15,16,33 and results from our lab (Fig. 3c,d) have shown that mouse UHRF1 (NP95) knockout and UHRF1 knockdown results in a reduction in the global levels of DNA methylation. Specifically, studies in mice suggest that UHRF1 physically interacts with DNMT1 and may facilitate its recruitment to chromatin15,16. We therefore hypothesized that the interaction between UHRF1 and H3K9 methylation might be necessary to maintain DNA methylation in cells. To test this hypothesis, we first stably knocked down HeLa cell UHRF1 with shRNA (Supplementary Fig. 2c,d) and analyzed global DNA methylation by immunofluorescence, which showed a 58% reduction in 5-methylcytosine (5mC) staining (Fig. 3d). Transient expression of wild type UHRF1 for 48 hours restored 5mC levels to 69% of that measured in control cells. However, the UHRF1 Y188A aromatic cage mutant was unable to restore 5mC levels, demonstrating for the first time that the H3K9 methylation binding function of UHRF1 is required for DNA methylation maintenance. Furthermore, the UHRF1 N147E ‘phospho/methyl switch’ mutant was able to only partially restore global 5mC levels, indicating that mitotic binding of UHRF1 to H3K9 methylation also contributes to DNA methylation maintenance.
We further investigated the link between DNA and histone methylation by querying the methylation status of 21 CpG dinucleotides from the 5’ end of the intergenic spacer of ribosomal DNA (IGS rDNA) (Supplementary Fig. 3). Methylation of this CpG island was shown to be regulated by UHRF115. In a non-targeted control shRNA HeLa cell line, this locus was 87.3% methylated, and UHRF1 shRNA diminished the level of DNA methylation to 37.7% (Fig. 3e). Similar to our global analysis by 5mC staining, transient expression of wild type UHRF1 WT restored DNA methylation at this locus to 62.3% of control. However, neither UHRF1 Y188A nor UHRF1 N147E restored DNA methylation at this locus (Fig. 3e). Collectively, these results demonstrate that the association of UHRF1 with H3K9 methylation in mitosis is critical for maintaining DNA methylation in HeLa cells.
UHRF1 mitotic chromatin binding stabilizes DNMT1
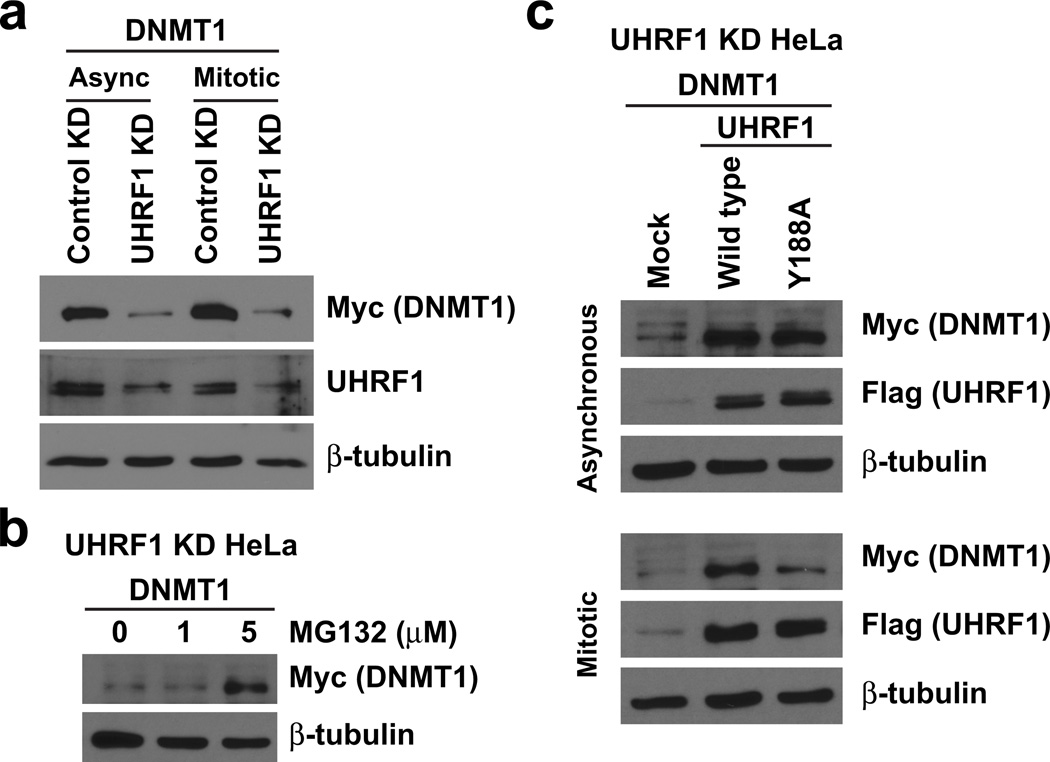
We next sought to determine the mechanism by which DNA methylation is regulated by the interaction of UHRF1 with H3K9 methylation. Previous studies suggested a lack of DNMT1 recruitment to chromatin in the absence of UHRF115. However, we and others34 find endogenous and tagged forms of DNMT1 to be only weakly associated with chromatin in wild-type cells; DNMT1 is primarily detected in the soluble nuclear component (data not shown). Importantly, we observed that stable knockdown of UHRF1 led to a dramatic decrease in DNMT1 protein levels in soluble HeLa lysates (Fig. 4a). Analysis of DNMT1 levels in the presence of a proteasome inhibitor (MG132) indicated that this effect was at the level of protein stability (Fig. 4b). To determine whether the ability of UHRF1 to regulate DNMT1 stability was mediated by its H3K9 methylation interaction, we re-introduced wild type UHRF1 WT and UHRF1 Y188A into these cells and re-examined the levels of DNMT1. Wild type UHRF1 stabilized DNMT1 in both asynchronous and mitotic cells (Fig. 4c). Conversely, UHRF1 Y188A stabilized DNMT1 in asynchronous cells but was unable to stabilize DNMT1 in mitosis – thus indicating that mitotic binding of UHRF1 with H3K9 methylation is required for maintaining DNMT1 stability. Similar results were observed for UHRF1 N147E (data not shown). UHRF1 H741A, a mutation perturbing UHRF1 E3-ubiquitin ligase function35,36, was unable to rescue DNMT1 stability alone or in combination with UHRF1 Y188A (Supplementary Fig. 4), indicating that mitotically driven DNMT1 stability occurs in a UHRF1 E3-ubiquitin ligase-independent manner.
Figure 4. Chromatin targeting of UHRF1 in mitosis is required for the stability of DNMT1.
(a–c) Western blot analysis of biochemically purified soluble protein from HeLa cells expressing control or UHRF1 shRNAs (see Supplementary Fig. 5) and myc-DNMT1. The indicated concentrations of (b) MG132 or (c) nocodazole in DMSO were added as described in Methods. Mock indicates transfection of myc-DNMT1 only.
DISCUSSION
Taken together, these results demonstrate that mitotic binding of UHRF1 to H3K9 methylated chromatin is necessary for DNMT1 stability and the maintenance of DNA methylation (Fig. 5). To our knowledge, this finding is the first example showing a requirement for H3K9 methylation in the maintenance of DNA methylation in human cells. In addition to the TTD, UHRF1 possesses an adjacent PHD domain, recently shown to contribute to the association of UHRF1 with histones via its interactions with the unmodified N-terminus of H333, and an SRA domain shown to bind CpG dinucleotides17–19. Identification of this latter interaction suggested an attractive model whereby UHRF1 facilitates DNMT1-mediated maintenance methylation by binding to hemi-methylated DNA during semi-conservative DNA replication. Importantly, our result that UHRF1 Y188A cannot bind chromatin (Fig. 3b) demonstrates that the TTD interaction with H3K9 methylation is the primary means by which UHRF1 is maintained on chromatin and implies that the UHRF1 PHD and SRA domains, at least by themselves, cannot maintain UHRF1 chromatin association and DNA methylation in the absence of a functional TTD (Fig. 3).
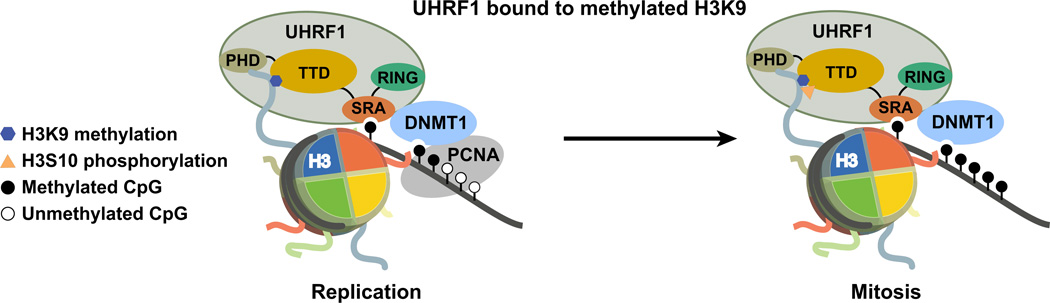
Figure 5. Proposed interaction between UHRF1, DNMT1, and H3K9 methylated histones in replicating and mitotic chromatin.
Shown is a cartoon representation of the interactions between UHRF1, chromatin, and DNMT1. We find that the UHRF1 TTD interaction with H3K9 methylation through mitosis is necessary to faithfully propagate DNA methylation patterns. Additionally, we demonstrate that the TTD is insensitive to H3S10p, thus evading the mitotic ‘phospho/methyl switch.’ Mitotic chromatin binding of UHRF1 is further shown to regulate the stability of DNMT1. These studies provide mechanistic insight into the interplay between H3K9 methylation and DNA methylation in human cells.
Our studies also identify the UHRF1 TTD as the first H3K9 methyl effector protein that is insensitive to the mitotic ‘phospho/methyl switch’29,30 and describe a function for binding to this dually marked histone tail in mitosis (i.e., DNMT1 stability and the maintenance of DNA methylation). We further reveal that the faithful propagation of DNA methylation patterns by DNMT1 requires events occurring outside of S-phase. DNMT3a and 3b have been shown recently to be unstable when not physically associated with chromatin34,37. We postulate that, similar to these de novo methyltransferases, DNMT1 stability and subsequent DNA methylation maintenance are dependent on chromatin association indirectly through its associations with UHRF1 and the replication machinery in S-phase, and solely by its interaction with UHRF1 in mitosis (Fig. 5).
Our DNA methylation analysis with UHRF1 N147E (sensitive to H3S10p and fails to localize to mitotic chromatin) revealed partial rescue of global DNA methylation in UHRF1 knockdown cells (Fig. 3d), but no restoration of DNA methylation at the late-replicating (heterochromatic) IGS rDNA CpG island (Fig. 3e). We therefore hypothesize that UHRF1 insensitivity to the ‘phospho/methyl switch’ is necessary to maintain DNA methylation at these late-replicating heterochromatic loci demarcated with H3K9me3. It is indeed plausible that Aurora B phosphorylates H3S10 in these regions before the DNA methylation pattern has been copied. While these intriguing ideas await further testing, our data nonetheless define an important role for UHRF1 as an intermediate link between H3K9 methylation and the faithful epigenetic inheritance of DNA methylation in human cells, which involves mitotic association and insensitivity to H3S10p as necessary properties of UHRF1 to maintain DNMT1 stability.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Strahl Lab for helpful discussions and H. Fried (UNC Chapel Hill) for critical reading of the manuscript. We also thank the North Carolina Biotechnology Center (NCBC) and the UNC School of Medicine for support in the establishment of the High Throughput Peptide Synthesis and Array Core Facility at UNC Chapel Hill. This work was supported in part by research grants from the National Institutes of Health (GM085394) to B.D.S. and (T32CA09156) to S.B.R., the NCBC (2010-IDG-I003) to B.D.S, and the Cancer Prevention and Research Institute of Texas (RP110471) to M.T.B.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.B.R. and B.D.S. designed experiments. S.B.R. performed and analyzed peptide array, biophysical, molecular biology, and cellular studies. K.K. performed peptide synthesis and contributed to data analysis. N.N., W.T., S.X, and C.H.A. designed, performed, and analyzed structural studies. A.I.B., and J.Y.M. contributed to cloning and protein production. D.B.L. produced lentivirus. S.M.F. contributed key technical assistance for array studies. S.B.R., N.N., C.H.A., and B.D.S. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Van Holde KE, Allen JR, Tatchell K, Weischet WO, Lohr D. DNA-histone interactions in nucleosomes. Biophysical journal. 1980;32:271–282. doi: 10.1016/S0006-3495(80)84956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 7.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics. 2012 doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 12.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 13.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Current biology : CB. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 14.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews. Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 15.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 16.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 17.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 18.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nady N, et al. Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein. The Journal of biological chemistry. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 22.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell stem cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothbart SB, Krajewski K, Strahl BD, Fuchs SM. Peptide Microarrays to Interrogate the "Histone Code". Methods in Enzymology. 2012;512:107–135. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Current biology. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bock I, et al. Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC biochemistry. 2011;12:48. doi: 10.1186/1471-2091-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garske AL, et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nature chemical biology. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 30.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 31.Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. Journal of Molecular Biology. 2011;408:807–814. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 33.Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Molecular cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Molecular and cellular biology. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Citterio E, et al. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Molecular and cellular biology. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins Y, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Molecular biology of the cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS genetics. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo AJ, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.