EMBO Reports (2012) advance online publication; doi:; DOI: 10.1038/embor.2012.139
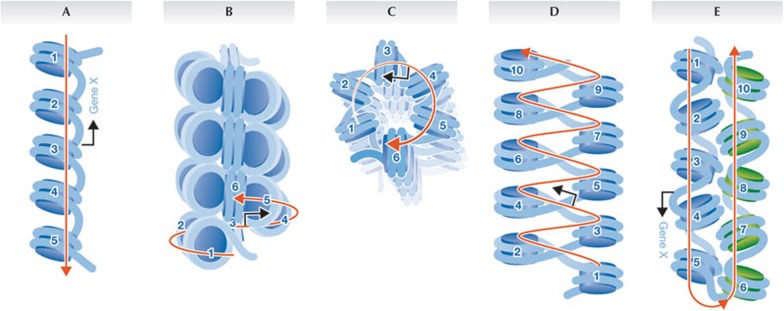
DNA in a human cell is approximately 2 m long but is compacted into micrometre-sized eukaryotic nuclei. To achieve this level of compaction, DNA is first wrapped up into nucleosomes, which are then thought to fold into fibres and loops. The basal unit of such folding is the 10 nm ‘beads on a string’ structure observed in all eukaryotic cells (Fig 1A; reviewed in [1]). The 10 nm fibre is subsequently folded into secondary and tertiary structures, the nature of which have been at the heart of a spirited debate for nearly 30 years [2].
Figure 1.
Models of chromatin organization. (A) 10 nm fibre. (B) Side-view of a 30 nm fibre or solenoid. (C) Top-down view of the solenoid. (D) Zig-zag model of the 30 nm fibre. (E) Interdigitation of two 10 nm fibres (blue against green) forming a boustrophedon. Numbered circles are nucleosomes in an array; red arrow follows the path of the DNA; blue arrow represents a gene promoter.
The first compelling description of the compacted interphase 30 nm chromatin fibre, found in all textbooks, originated from a seminal study by Finch and Klug, who used transmission electron microscopy and X-ray diffraction to investigate cell-extracted nucleofilaments. Now, nearly 40 years after Finch and Klug proposed their landmark ‘solenoid’ model, a series of papers including one published in this issue of EMBO reports [3] have questioned the existence of 30 nm chromatin fibres.
These data […] contradict decades of previous work that argued for the presence of 30 nm and thicker chromatin fibres
In their original study, Finch and Klug found that extracted chromatin appeared as a ‘solenoidal’ 30 nm wide helical fibre, with a radial distribution of adjacent nucleosomes and bent linker DNAs coiling continuously around a central axis (Fig 1B,C; [4]). This simple one-start helix required histone H1 for its stability, and in the absence of H1 collapsed back into the primary 10 nm fibre. Later, cryo-electron microscopy and computational modelling analyses showed that natural variations in DNA linker length, and nucleosome-free regions in vivo, affect the regularity of the 30 nm fibre [5]. These data supported an alternative model of chromatin fibre organization, namely the accordion-like two-start helix containing an interdigitated arrangement of adjacent nucleosomes, with straight linker DNA criss-crossing the interstitial space (Fig 1D). This model correctly represented how a tetra-nucleosomal chromatin fibre folds in vitro [2,6]. In situ data also revealed the existence of 30 nm chromatin fibres in starfish sperm, chicken erythrocyte and mouse photoreceptor cells through several optical approaches—for example, transmission electron microscopy, cryo-electron microscopy, energy spectroscopic imaging and immunogold labelling (reviewed in [2]). Furthermore, three-dimensional traces of chromatin fibres from electron microscopy images of serially sectioned G1 nuclei provided evidence for fibres with diameters of not only 30 nm, but also 60–80 nm and 100–130 nm, which seemed to have formed by progressive coiling of the 30 nm fibre [7]. In addition, analyses in live cells, where isolation or fixation artefacts cannot arise, revealed that tagged segments of chromatin domains decondensed on transcriptional activation into necklace-like structures, the overall length of which was consistent with an 80–100 nm fibre [8].
However, advances from cryo-electron microscopy, small-angle X-ray scattering (SAXS) and energy spectroscopic imaging approaches present cogent arguments for a eukaryotic nucleus composed of almost exclusively 10 nm fibres. In situ mitotic chromosome analyses by cryo-electron microscopy and SAXS have found the 10 nm chromatin fibre but no other structures [9,10], although interactions between adjacent fibres leading to a more compact structure were described in the same study. In addition, an energy spectroscopic imaging study of several cell types and constitutive heterochromatin did not reveal the 30 nm fibre [11]. Finally, data presented in this issue of EMBO reports, as well as a study published in Nucleus, further support the argument that the 30 nm fibre does not exist [3,12]. Energy spectroscopic imaging is a spectrographic approach that can distinguish phosphorus atoms and thereby enable a ‘trace’ of the path of the DNA in a chromatin fibre. By using this method, Fussner and colleagues have found that although they can detect approximately 24 nm fibres in starfish sperm—serving as a control for the most compacted chromatin structures—chromatin in cultured mouse embryonic fibroblasts and native mouse tissues has almost exclusively 10 nm fibre organization. More provocatively, Joti and colleagues have used cryo-electron microscopy and ultra-SAXS to show that previous SAXS data indicative of 30 nm fibres can potentially be attributed to contamination from ribosomes, which are arranged in a 30 nm periodicity when they coat mitotic and interphase chromatin preparations. Indeed, when the chromosomes and nuclei are extensively washed to remove ribosomes, only the 6 nm and 11 nm peaks remain in the SAXS profile, representing individual nucleosomes and the 10 nm chromatin fibre, respectively.
…the functional difference between 10 and 30 nm chromatin fibres is profound
These data fundamentally contradict decades of previous work which have argued for the presence of 30 nm and thicker chromatin fibres. Is there any way to reconcile the two opposing views? As suggested above, a potential answer might be that the conditions used for chromatin extraction studies favoured the assembly of the thicker 30 nm fibre that Fussner and co-workers found in starfish sperm—for example, magnesium, sodium chloride and chromatin concentrations in such studies are typically far lower than those found in vivo. Previous electron microscopy studies are also subject to the concern that fixation or staining procedures might generate thicker fibres due to cross-linking artefacts. It is also conceivable that studies using either light or transmission electron microscopy missed isolated 10 nm fibres because their staining intensity was too weak for detection relative to the higher signal density of 30 nm fibres. The issue is more perplexing when one considers some of the intact cell studies in which specimens were neither stained nor fixed. The simplest explanation might be that it is nearly impossible to distinguish between two discontiguous 10 nm fibres that are interdigitated in a polymer melt, folded in a boustrophedon fashion or otherwise annealed by proteins, and a true contiguous 30 nm fibre composed of an one-start or two-start helix (Fig 1E).
Does it matter whether chromatin organizes itself as a 10 or 30 nm fibre? Although it might seem an esoteric argument, the functional difference between 10 and 30 nm chromatin fibres is profound. The 30 nm fibre theory was satisfying precisely because it held that the genome-wide transcriptional quiescent state would be maintained by lack of access to DNA (Fig 1). RNA polymerases or transcriptional activators could not easily reach a gene buried within the solenoidal core sealed by the histone H1. By contrast, the emerging 10 nm fibre model is a free-for-all, with all potential target sites accessible. In this scenario, gene regulation becomes largely dependent on the local affinity of a chromatin site, and perhaps also on the speed at which that site can be reached depending on the local density of 10 nm fibres. This hypothesis is supported by ENCODE data which demonstrate that large swathes of the genome, previously thought to be quiescent, are accessible and transcriptionally active, thus supporting a paradigm shift in our view of stochastic protein–chromatin-binding in vivo.
Precise structure–function relationship questions can be addressed by using tools for single-molecule tracking of transcriptional activators, RNA polymerases and other factors in nuclear domains. Also crucial for future work is the application of energy spectroscopic imaging, cryo-electron microscopy and emerging technologies such as cryo-X-ray tomography to some of the classic systems in which 30 nm or thicker fibres were visualized. Only by this direct cross-validation of approaches can we ultimately resolve the chromatin structure conundrum that confronts us.
Footnotes
The authors declare that they have no conflict of interest.
References
- Olins DE et al. (2003) Nat Rev Mol Cell Biol 4: 809–814 [DOI] [PubMed] [Google Scholar]
- Grigoryev SA, Woodcock CL (2012) Exp Cell Res 318: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Fussner E et al. (2012) EMBO Rep (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch JT, Klug A (1976) Proc Natl Acad Sci USA 73: 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz RA et al. (1994) J Cell Biol 125: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T et al. (2005) Nature 436: 138–141 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bruce K (1994) J Cell Biol 127: 287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T et al. (1999) J Cell Biol 145: 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltsov M et al. (2008) Proc Natl Acad Sci USA 105: 19732–19737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino Y et al. (2012) EMBO J 31: 1644–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed et al. (2010) PLoS ONE 5: e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joti Y et al. (2012) Nucleus 3: [Epub ahead of print] doi:; DOI: 10.4161/nucl.21222 [DOI] [PMC free article] [PubMed] [Google Scholar]