Abstract
Differentiation of specialized cell types from stem and progenitor cells is tightly regulated at several levels, both during development and during somatic tissue homeostasis. Many long non-coding RNAs have been recognized as an additional layer of regulation in the specification of cellular identities; these non-coding species can modulate gene-expression programmes in various biological contexts through diverse mechanisms at the transcriptional, translational or messenger RNA stability levels. Here, we summarize findings that implicate long non-coding RNAs in the control of mammalian cell differentiation. We focus on several representative differentiation systems and discuss how specific long non-coding RNAs contribute to the regulation of mammalian development.
Keywords: lncRNA, cell differentiation, pluripotency
See the Glossary for abbreviations used in this article.
Glossary.
- AIR
antisense Igf2r RNA
- ANCR
anti-differentiation ncRNA
- c-Myc
myelocytomatosis oncogene
- Dlx5/6
distal-less homeobox 5/6
- EGO
eosinophil granule ontogeny
- Evf1/2
embryonic ventral forebrain 1/2
- Gad1
glutamate decarboxylase 1
- GABA
gamma-aminobutyric acid
- GFP
green fluorescent protein
- Hox
homeobox
- HOTAIR
HOX antisense intergenic RNA
- HOTAIRM1
HOX antisense intergenic RNA myeloid 1
- HOTTIP
HOXA transcript at the distal tip
- IRES
internal ribosome entry site
- ITPR1
inositol 1,4,5-trisphosphate receptor 1
- linc-MD1
linc muscle differentiation 1
- lincRNA-RoR
lincRNA regulator of reprogramming
- lincRNA-EPS
lincRNA erythroid pro-survival
- LTR
long terminal repeat
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- MECP2
methyl CpG-binding protein 2
- MLL
myeloid/lymphoid or mixed-lineage leukaemia
- Nkx2.2as
natural killer cell-associated antigen 2 locus 2 antisense
- NRON
non-protein coding RNA, repressor of NFAT
- OCT4
octamer-binding transcription factor 4
- PINC
pregnancy-induced ncRNA
- PRC2
polycomb repressive complex 2
- Pycard
pyrin domain and caspase activation and recruitment domain containing
- RNCR2
retinal non-coding RNA 2
- RNP
Ribonucleoprotein
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SOX2
SRY-box-containing gene 2
- TALE
transcription activator-like effector
- Tie-1AS
tyrosine kinase with immunoglobulin-like and EGF-like domains 1, antisense
- Tsix
X-inactive-specific transcript, antisense
- Vax2os1
ventral anterior homeobox-containing gene 2 opposite strand transcript
- WDR5
WD repeat domain 5
- Xite
X-inactivation intergenic transcription elements
- Xist
X-inactive-specific transcript
Introduction
A surprise from mammalian transcriptome annotation over the past ten years has been the identification of thousands of RNA transcripts that do not seem to be derived from known genes [1,2,3,4]. Many of these accumulate to significant levels and resemble messenger RNA in being capped, polyadenylated and spliced. Yet these RNAs do not overlap the exons of known protein-encoding genes [1,4,5,6]. Those longer than 200 nucleotides, which seem to have little to no protein-encoding capacity, have been typically termed long non-coding RNAs (lncRNAs; reviewed in [7]).
Among all cellular RNA species, the large number of putative lncRNA transcripts is among the least understood. A few lncRNAs have been functionally characterized in mammalian systems, and several have been associated with important cellular processes such as X-chromosome inactivation, imprinting, maintenance of pluripotency, lineage commitment and apoptosis [8,9,10,11,12]. An emerging theme among known lncRNA functions is thus the regulation of cell fate and differentiation decisions, often in response to developmental or environmental cues.
Here, we review evidence supporting a role for lncRNAs in the regulation of mammalian cell differentiation, and discuss how emerging ideas and technical innovations can help guide future studies. First, we review technical approaches for the global discovery and characterization of lncRNAs in mammals. We then discuss selected studies that illustrate advances in our understanding of how lncRNAs modulate a diversity of cell differentiation processes (Table 1). Finally, we synthesize emerging principles of lncRNA function and evaluate how they provide a framework for the integration of lncRNAs in known regulatory networks of mammalian cell differentiation.
Table 1. Examples of functional lncRNAs involved in mammalian cell differentiation.
| Name | Expression | Loss-of-function phenotype | Assays | References |
|---|---|---|---|---|
| 26 ‘Lys 4–Lys 36’ lincRNAs | ES cells | Decreased expression of pluripotency markers; loss of ES cell morphology | shRNA knockdown | [11] |
| RNCR2, AK141205 | ES cells | Changes in expression of pluripotency markers; changes in expression of lineage markers; loss of ES cell morphology; altered proliferation | siRNA knockdown; overexpression | [10] |
| Xist | ES cells | Epigenetic silencing of inactive X-disrupted; embryonic lethal in females | Mouse knockout; directed mutagenesis; ectopic expression; overexpression; FISH | Reviewed in [51] |
| Tsix | ES cells | Epigenetic repression of Xist disrupted; embryonic lethal in both sexes | Mouse knockout; directed mutagenesis; ectopic expression | Reviewed in [51] |
| Xite | ES cells | Downregulation of Tsix | Mouse knockout; directed mutagenesis; ectopic expression | Reviewed in [51] |
| lincRNA-RoR | iPS cells | Impaired iPS cell generation | siRNA knockdown; overexpression | [75] |
| lncRNA_ES1–3 | ES cells | Downregulation of pluripotency markers; upregulation of lineage markers | siRNA knockdown | [98] |
| ANCR | Keratinocyte progenitors | Derepression of differentiation genes; differentiation within progenitor-specific epidermal layer | siRNA and shRNA knockdown | [76] |
| PINC | Mammary gland progenitors | Impaired cell cycle progression; reduced proliferation; increased apoptosis | siRNA knockdown; FISH | [77] |
| HOTAIR | Fibroblasts | Derepression of HoxD locus; decrease in cancer invasiveness | Mouse knockout; directed mutagenesis; siRNA knockdown; overexpression; FISH | [56,59,82] |
| HOTTIP | Fibroblasts | Decreased expression of HoxA cluster genes | siRNA and shRNA knockdown; ectopic expression; overexpression; FISH | [58] |
| Mistral | ES cells | Decreased expression of HoxA cluster genes | siRNA knockdown, FISH | [57] |
| EGO | Eosinophils | Downregulation of major basic protein and eosinophil-derived neurotoxin | siRNA knockdown | [86] |
| HOTAIRM1 | Myeloid progenitors | Downregulation of genes associated with myelopoiesis | siRNA and shRNA knockdown | [87] |
| lincRNA-EPS | Erythroblasts | Increased apoptosis; impaired enucleation | Directed mutagenesis; shRNA knockdown; ectopic expression | [50] |
| Tie-1AS | Vascular endothelial cells | Disrupts vascular tube integrity | Knockdown; ectopic expression | [88] |
| linc-MD1 | Myoblasts | Downregulation of genes asssociated with myogenesis | siRNA knockdown; directed mutagenesis; ectopic expression; overexpression | [65] |
| 1/2-sbsRNAs | Myoblasts | Increased stability of Staufen-1 target transcripts | siRNA knockdown | [91] |
| Nkx2.2as | Neural stem cells | N/A | Overexpression | [93] |
| Vax2os1 | Retinal progenitor cells | N/A | Directed mutagenesis; overexpression; FISH | [97] |
| Evf2 | Embryonic ventral forebrain | Reduction of GABAergic interneurons; synaptic inhibition | Mouse knockout; directed mutagenesis; FISH | [94,95] |
| RMST; lncRNA_N1–3 | Neuron progenitors | Impaired neuronal differentiation; downregulation of neuron markers; upregulation of glia markers | siRNA knockdown | [98] |
| RNCR2 | Retinal progenitor cells | Increased differentiation into non-retinal lineages | shRNA knockdown; directed mutagenesis; ectopic expression; overexpression; FISH | [96] |
1/2-sbsRNAs, half-STAU1-binding site RNAs; ANCR, anti-differentiation ncRNA; EGO, eosinophil granule ontogeny; ES, embryonic stem; Evf2, embryonic ventral forebrain 2; FISH, fluorescence in situ hybridization; GABA, gamma-aminobutyric acid; HOTAIR, HOX antisense intergenic RNA; HOTAIRM1, HOX antisense intergenic RNA myeloid 1; HOTTIP, HOXA transcript at the distal tip; HoxA/D, homeobox A/D; iPS, induced pluripotent stem; lincRNA, long intergenic non-coding RNA; lincRNA-EPS, lincRNA erythroid pro-survival; linc-MD1, linc muscle differentiation 1; lincRNA-RoR, lincRNA regulator of reprogramming; N/A, not applicable; Nkx2.2as, natural killer cell-associated antigen 2 locus 2 antisense; PINC, pregnancy-induced ncRNA; RMST, Rhabdomyosarcoma 2 associated transcript; RNCR2, retinal non-coding RNA 2; shRNA, short hairpin RNA; siRNA, small interfering RNA; Tie-1AS, tyrosine kinase with immunoglobulin-like and EGF-like domains 1, antisense; Tsix, X-inactive-specific transcript, antisense; Vax2os1, ventral anterior homeobox-containing gene 2 opposite strand transcript; Xist, X-inactive-specific transcript; Xite, X-inactive-specific transcript.
Discovery and characterization of long non-coding RNAs
About 30 years after the idea of mRNA was established [13], the first mammalian lncRNA was described in mice. In 1988, H19 was identified as an RNA induced during liver development that contained no large open reading frame (ORF), but rather small sporadic ORFs that were not evolutionarily conserved, could not be translated in vivo and did not produce detectable polypeptides [14,15]. In the ensuing 20 years, more lncRNA genes were functionally described, including XIST, AIR and NRON [8,9,16]. The advent of technical improvements in our ability to detect and catalogue the transcriptional output of entirely sequenced genomes propelled new efforts to detect and characterize lncRNAs at a global scale [3,4,6,17,18]. These efforts greatly increased the number of RNA transcripts ascribed to the lncRNA category, but doubts about their functional relevance also grew. Many investigators raised the important concern that many putative lncRNAs are just non-functional by-products of the transcription of neighbouring loci [19,20,21], whereas other long intergenic non-coding RNAs (lincRNAs) might actually encode small functional peptides [18,22,23]. These concerns in turn have led to new developments and approaches for the genome-wide discovery and characterization of bona fide lncRNAs.
Finding and identifying lncRNAs
Advances in microarray technology and RNA sequencing revealed that most of the mouse and human genomes are transcribed in one cell type or another [2,3,4,6]. However, only a small portion of the transcripts could be recognized as protein-encoding or as previously known classes of ncRNAs, such as transfer RNAs, ribosomal RNAs, microRNAs and small nuclear RNAs, raising the possibility that some of the newly defined transcribed regions might encode new types of functional ncRNA [4,24,25,26]. This conjecture was supported by the clear evolutionary conservation of some putative lncRNAs [27,28], and by their often regulated expression patterns during development [18,29] and localization to specific subcellular structures [30,31,32,33,34,35]. However, as both their expression level and conservation seemed much lower than those of known coding genes [36,37], additional biological information was needed to distinguish between biologically relevant lncRNA candidates and transcriptional noise. A strategy devised by Guttman and colleagues to address this issue was to focus on intergenic regions marked by histone modifications indicative of stable RNA polymerase II (Pol II) transcription [2]. These regions were defined by a combination of two modifications—a short stretch of H3K4me3, marking Pol II initiation, followed by a longer stretch of H3K36me3, marking the region of Pol II elongation (Lys 4–Lys 36 domain). The strategy identified about 1,500 lincRNA loci expressed in four mouse cell types that were 5 kb or greater in length and did not overlap protein-encoding genes, microRNAs or endogenous small interfering RNAs. Extending the study to humans identified about 1,800 human lincRNAs [38]. However, there are important limitations to using this approach to discover lncRNAs. Loci actively transcribed by Pol II are not all marked by a Lys 4–Lys 36 domain; a study in mice found that approximately 25% of lincRNA or mRNA transcripts identified by RNA-seq alone are not marked [39], and in humans the number seems to be greater [40]. Conversely, the regions with a detectable Lys 4–Lys 36 domain do not all correspond to gene bodies; some correspond to transcribed enhancers [21,40]. Close examination of existing lncRNA catalogues indicates that approximately 10–15% actually overlap enhancers of protein-encoding genes [40]. Moreover, it is possible that some lncRNAs are transcribed by Pol III (discussed in [41]) and thus lack chromatin marks that are characteristic of Pol II transcription. These caveats indicate that both detection by sequencing and examination of the chromatin state need to be combined for the reliable discovery of stably transcribed lncRNA candidates.
Excluding functional coding capacity
The key feature of lncRNAs is that they do not have functional protein-encoding capacity. This is usually defined as the absence of a protein product from the tested transcript. The gold standard to discriminate whether a transcript is coding or non-coding is to determine whether a corresponding polypeptide can be detected from an ORF of the transcript. However, due to technical difficulties, such as the low abundance of putative target polypeptides and the absence of corresponding antibodies, the coding capacity of a newly identified transcript is usually determined indirectly by computational and biochemical approaches (reviewed in [18]). Computationally, evaluation of coding potential can be performed at a global scale by examining transcripts for presence and conservation of ORFs, by performing homology queries and by scrutinizing putative ORFs for biases in codon usage and in frequency of codon substitution through evolution.
It is worth noting that an ORF can occur in a transcript purely by chance, or alternatively be a vestige of an evolutionary former coding capacity [18,42]. For example, bona fide lncRNAs such as Xist and H19 do contain detectable small ORFs, but these are not evolutionarily conserved and fail to form a template for polypeptide synthesis in vivo [15,43]. Xist is believed to have originated in part from genes that at one time encoded proteins [44]. To distinguish functional from spurious ORFs, candidates can be tested for sequence features characteristic of functional coding regions. The bias in synonymous codon usage, for example, can be calculated for each putative ORF to determine whether it is significantly different from those of known protein-encoding genes. If so, it strongly suggests, but hardly proves, that the ORF might not be functional. The coding potential of a transcript can also be evaluated through comparative evolutionary analysis (reviewed in [45]). By using sequences from the target transcript and orthologues from other species, the preference for synonymous codon substitutions over missense or nonsense substitutions can be used as evidence for preservation of protein-encoding potential [46]. Non-coding transcripts are typically characterized by similar frequencies of synonymous and non-synonymous codon substitutions, whereas RNAs are significantly biased towards synonymous changes. However, conservation-based approaches might fail to detect newly evolved ORFs that lack orthologues. To address this, methods that do not require a multiple-sequence alignment should be considered (reviewed in [18]), as well as direct inspection of homology in protein family and domain databases—that is, BLASTX.
Collectively, computational approaches are powerful and cost-effective in testing the coding potential of large collections of lncRNA candidates. However, the candidates that pass these tests ultimately require experimental verification. The non-coding feature of a transcript implies that it is not associated with actively translocating ribosomes, a feature that can be experimentally tested by its presence on polysomes within cells [47]. This can be accessed through sucrose density gradients and ultracentrifugation to fractionate cell lysates. Those transcripts associated with ribosomes predominantly sediment with a higher velocity through the gradient whereas non-ribosome-associated transcripts remain at the top of the gradient (light fractions). However, care should be taken when interpreting results from polysome analysis. If a transcript sediments to the top of the sucrose gradient, it could be a non-coding transcript or alternatively a translationally repressed transcript. Conversely, if a transcript sediments to the bottom of the sucrose gradient, it only implies that the transcript is associated with a large particle, which could be ribosomes but could also be other large complexes. Specific disruption of translation, such as puromycin treatment, is required to discriminate between these two possibilities. An alternative approach is ribosome profiling coupled with high-throughput RNA sequencing, which can comprehensively analyse ribosome density and occupancy at high resolution [23,48]. This technique can specifically detect the translation status of known mRNAs. However, it cannot provide information about whether or not the bound ribosomes are actively translocating (making polypeptides). Thus, additional experiments are required to address this issue.
The coding or non-coding feature of a transcript can also be inferred from the localization of the transcript in the cell, as determined by RNA fluorescence in situ hybridization (FISH) or by fractionation of cell homogenates into nuclear and cytoplasmic fractions. Transcripts predominantly localized in the nucleus, such as Xist, are probably non-coding, because mRNA translation generally occurs in the cytoplasm. One caveat of this analysis, however, is that it only reveals the steady-state localization of the transcript. If the transcript is rapidly shuttling into and out of the nucleus or is specifically degraded in one compartment, for example, cytoplasm, the information obtained from steady-state RNA localization might be misleading [49].
Finally, from a functional perspective, in cases where the function of a transcript is known, its functional coding capacity can be directly determined by using frameshift mutations to disrupt any putative ORFs and by testing whether the function of the transcript is compromised [50]. If such function is independent of all the putative ORFs in the transcript, a strong claim can be made that functionally it is indeed non-coding. Altogether, under ideal conditions, computational large-scale approaches augmented by dedicated experiments are needed to determine convincingly whether or not a transcript truly functions as a non-coding RNA.
Probing lncRNA function
It has been known for decades that lncRNAs can modulate important biological processes. For example, the well-characterized lncRNA XIST is important in X-chromosome inactivation in female mammals [51]. For most lncRNAs identified through large-scale screens, however, their biological functions remain to be explored. Several interesting observations suggest that this might be a worthwhile endeavour. First, the proportion of non-coding DNA seems to increase with developmental complexity [52] and most of it is transcribed [2,3,6,24]. This has led to the proposal that regulation by non-coding RNAs, among them lncRNAs, might have been important in giving rise to the diversity of cell differentiation programmes underlying development in multicellular organisms [53,54]. Second, as expression of mammalian lincRNAs shows greater tissue specificity than that of coding genes [40], it seems conceivable that some might contribute to tissue-specific regulation. Third, dysregulation of lncRNAs has been observed under many pathological conditions (reviewed in [55]), including cancers, heart diseases and Alzheimer disease, suggesting that abnormal expression of some of these transcripts might contribute to the development of pathophysiological cellular states.
Importantly, studies have shown that lncRNAs are able to regulate gene expression through diverse mechanisms (Fig 1). For example, lncRNAs can function as molecular scaffolds to recruit repressive (such as PRC2) and activating (such as the Trithorax group) chromatin modifiers, thereby repressing or activating target gene expression, respectively [56,57,58,59]. In addition, lncRNAs can also modulate post-transcriptional events during gene expression, such as splicing [60], mRNA translation [61] and mRNA degradation [62]. Furthermore, some lncRNAs can inhibit microRNA function, thereby indirectly enhancing protein expression from certain mRNAs that otherwise would be downregulated by the microRNA [63,64,65]. The detailed molecular mechanisms by which lncRNAs regulate gene expression have been summarized in comprehensive reviews [66,67,68]. Such regulatory capacities enable lncRNAs to modulate several biological processes.
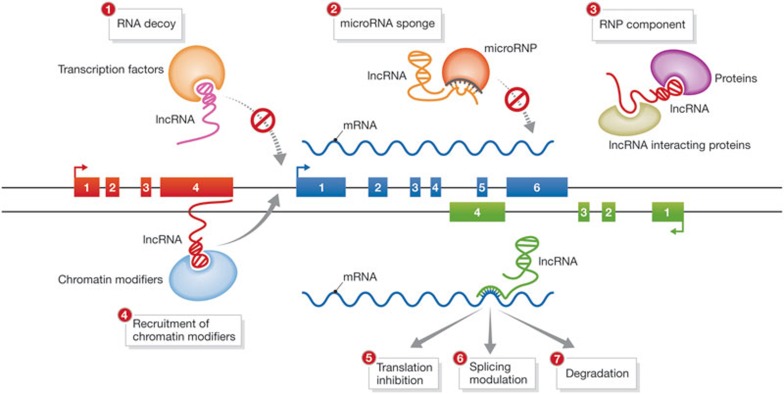
Figure 1.
Mechanisms of lncRNA function. Studies have described a range of mechanisms by which lncRNAs regulate their targets; many seem to depend on specific features of primary sequence, secondary structure and genomic positioning of lncRNA effector transcripts. (1) Several lncRNAs act as RNA decoys, titrating transcription factors away from their DNA targets by directly binding to them as target mimics [12,98]. (2) Others work at the post-transcriptional level as microRNA target site decoys, titrating microRNA effector complexes away from their mRNA targets [63,64,65]. lncRNAs, the microRNA target sites of which lack the structural sequence features needed for transcript degradation, have the overall effect of ‘sponging’ their microRNA regulators. (3) Many lncRNAs seem to bind to specific combinations of regulatory proteins, potentially acting as scaffold elements within ribonucleoprotein complexes [59,98]. (4) Recruitment of chromatin-modifying complexes to their DNA targets in cis has also emerged as a well-characterized function for several mammalian lncRNAs [57,58]. Recruitment in trans is not depicted [56]. A few lncRNAs seem to modulate direct processing of their mRNA targets, including translation (5), splicing (6) and degradation (7) [60,61,62]. lncRNA, long non-coding RNA; mRNA, messenger RNA, RNP, Ribonucleoprotein.
In the past few years, loss-of-function and gain-of-function studies have revealed that many lncRNAs are involved in a wide variety of biological processes in almost all eukaryotes, such as yeast, plants and animals [7,42,55,69]. These processes include, but are not limited to, cell fate determination, modulation of cell differentiation, cell cycle control and tumorigenesis. In the next section, we discuss selected examples of lncRNAs (Table 1) that illustrate advances in this fast-evolving field; we do not intend to provide a comprehensive review of all such studies. In particular, we focus on the regulation of mammalian cell differentiation by lncRNAs.
LncRNAs in ES cell maintenance and differentiation
Embryonic stem (ES) cells are unique in their ability to generate all terminally differentiated cells derived from all three primary germ layers—ectoderm, endoderm and mesoderm. Maintaining this pluripotent state requires precise and delicate transcriptional regulation mediated by key transcription factors, such as Oct4, Sox2 and Nanog [70]. In addition to these protein components, lncRNAs are also involved in maintaining the ‘stemness’ of ES cells (Fig 2A). Guttman et al systematically performed loss-of-function studies on 147 lincRNAs expressed in mouse ES cells by using lentiviral-based shRNAs [11]. For 90% of the lincRNAs, knockdown resulted in significant changes in ES cell gene expression. Interestingly, 26 lincRNAs were found to be involved in the maintenance of ES cell pluripotency, whereas 30 lincRNAs acted to repress specific gene expression programmes associated with differentiation. Importantly, expression of most of these lincRNAs is regulated by ES-cell-specific transcription factors, and many lincRNAs seem to bind to diverse chromatin regulatory proteins, potentially giving rise to specific nuclear RNA–protein complexes. Similarly, Lipovich's group identified four conserved lincRNAs that are regulated by the ES-cell-specific transcriptional factors Oct4 and Nanog [10]. Importantly, inhibition or misexpression of two of these lincRNAs impaired the ‘stemness’ state of ES cells. Collectively, these results implicate lincRNAs in the regulatory networks that maintain ES cell identity. One caveat in interpreting these data, however, is the uncertainty as to whether all of the studied transcripts are truly non-coding. Although the coding potential of these transcripts was computationally evaluated, experiments are required to verify this important point.
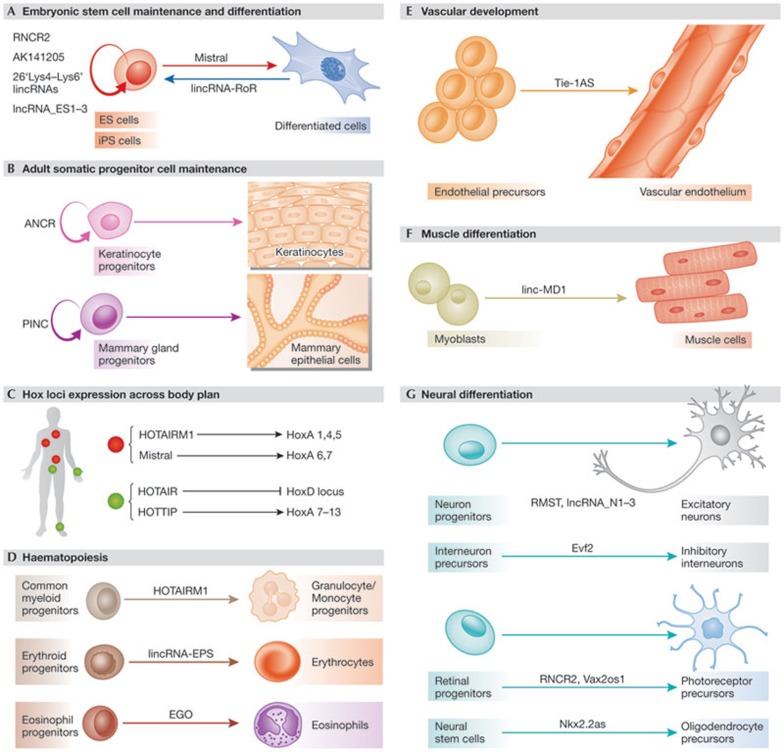
Figure 2.
Regulation of mammalian cell differentiation by lncRNAs. Examples are shown of lncRNAs implicated in modulating the differentiation of specialized cells from their progenitors. (A) Many lncRNAs are required for maintenance of the pluripotent state of ES cells, thereby antagonizing differentiation into specialized lineages [10,11,98]. Others favour differentiation [57], and yet others contribute to dedifferentiation of specialized cells into iPS cells [75]. (B) Other lncRNAs are important for the maintenance of adult epidermal lineage progenitor cells [76,77]. (C) Several lncRNAs transcribed from Hox clusters regulate the transcription of Hox genes in cis [57,58,87] or in trans [56], contributing to the distinct epigenetic profiles of Hox loci across cells from distinct anatomical positions. [82,83,84,87]. (D) lncRNAs have also been associated with the development of cells from haematopoietic [50,86,87] and vascular endothelial lineages (E; [88]). (F) Differentiation of muscle cells is also regulated by lncRNAs [65]. (G) Many lncRNAs are differentially expressed and specifically localized across neural tissues during development and disease. Many of them modulate differentiation of progenitors into excitatory, inhibitory or retinal photoreceptor neurons, whereas others favour oligodendrocyte differentiation [93,94,95,96,97,98]. ANCR, anti-differentiation ncRNA; EGO, eosinophil granule ontogeny; ES, embryonic stem; Evf2, embryonic ventral forebrain 2; HOTAIR, HOX antisense intergenic RNA; HOTAIRM1, HOX antisense intergenic RNA myeloid 1; HOTTIP, HOXA transcript at the distal tip; Hox, homeobox; iPS, induced pluripotent stem; linc-MD1, linc muscle differentiation 1; lincRNA-EPS, lincRNA erythroid pro-survival; lincRNA-ROR, lincRNA regulator of reprogramming; lncRNA, long non-coding RNA; Nkx2.2as, natural killer cell-associated antigen 2 locus 2 antisense; RMST, Rhabdomyosarcoma 2 associated transcript; RNCR2, retinal non-coding RNA 2; PINC, pregnancy-induced ncRNA; Tie-1AS, tyrosine kinase with immunoglobulin-like and EGF-like domains 1, antisense; Vax2os1, ventral anterior homeobox-containing gene 2 opposite strand transcript.
LncRNAs also regulate differentiation of ES cells (Fig 2A). One well-characterized example is the role of Xist in X-chromosome inactivation in mammalian females [51]. To equalize the dosage of X-chromosome-encoded genes between female and male cells, expression from one of two copies of the X chromosome must be silenced; this is achieved by formation of heterochromatin early during ES cell differentiation. A non-coding RNA transcript, Xist, is important in this developmental process. Xist is exclusively expressed from a region of the inactive X chromosome called the X-inactivation centre. Despite undergoing splicing, Xist remains in the nucleus to coat the inactive X chromosome, and to recruit—through its structured RNA domain termed Repeat A—the chromatin regulator PRC2 to this chromosome [71]. PRC2 facilitates the formation of heterochromatin through histone modifications, specifically H3K27. Thus, Xist-mediated recruitment of PRC2 contributes significantly to X-chromosome inactivation. Interestingly, the generation and the activity of Xist are regulated by several other lnc transcripts, such as Tsix and Xite [51]. Clearly, regulatory networks of lncRNAs are important in this differentiation process, even when the transcripts themselves show limited conservation in primary sequence between species [72].
In addition to modulating ES cell differentiation, lncRNAs are also involved in the generation of induced pluripotent stem (iPS) cells (Fig 2A). iPS cells can be derived from terminally differentiated somatic cells by ectopic expression of key pluripotency-associated transcription factors, such as Oct4, Nanog, Sox2 and c-Myc [73]. This cellular reprogramming is accompanied by an extensive global remodelling of the epigenome [74]. Loewer et al found that several lincRNAs contribute to this dedifferentiation process [75]. By comparing lincRNAs expressed in iPS cells with those expressed in ES cells, they identified 28 lincRNAs that are specifically enriched in iPS cells. These lincRNAs are regulated by pluripotency-associated master transcription factors (Oct4 and Nanog), suggesting a potential involvement in the generation of iPS cells. In particular, inhibition of one such lincRNA, lincRNA-RoR, impairs iPS cell formation. Conversely, overexpression of this lincRNA leads to an approximate 2.5-fold increase in cellular reprogramming, a modest yet significant effect. Collectively, these observations indicate that lncRNAs can activate or repress transcriptional programmes associated with ES cell pluripotency or differentiation, and that their impact on these processes can range from essential to subtle but detectable.
LncRNAs and the maintenance of somatic progenitor cells
Apart from acting in ES cells, lncRNAs are also involved in the maintenance of somatic progenitor cells (Fig 2B). Kretz et al demonstrated that a lncRNA is required for suppressing somatic progenitor cell differentiation [76]. By using high-throughput transcriptome sequencing, they identified lncRNAs expressed during the terminal differentiation of human primary keratinocytes. Among more than 1,000 dynamically expressed lncRNAs, they identified an 855 nt transcript (ANCR) with markedly reduced expression levels on keratinocyte differentiation. siRNA-mediated depletion of ANCR in keratinocyte progenitor cells resulted in rapid induction of differentiation genes; importantly, this gene induction occurred without any differentiation stimuli. In addition, epidermal knockdown of ANCR also leads to the expression of recognized differentiation markers.
Ginger et al also described a lncRNA, PINC, which is enriched in mammary gland cells with progenitor-like qualities [77]. PINC resides in both the cytoplasm and nucleus, and is induced by oestrogen and progesterone treatment. Inhibition of this lncRNA by siRNAs impairs cell cycle progression of mammary gland progenitor cells, limiting their proliferation and survival. These observations implicate PINC in the regulation of mammary gland development.
Although the detailed molecular mechanisms by which these two lncRNAs work remain to be explored, the evidence argues clearly that, as a new class of regulators, lncRNAs participate in maintaining the undifferentiated state of adult somatic progenitor cells.
Regulation of the Hox gene cluster by lncRNA
Hox genes are a group of related loci that are crucial for pattern formation and cell differentiation during early embryonic development in animals [78]. They encode transcription factors that regulate a variety of developmental loci by binding to their regulatory enhancer sequences through a protein domain known as the homeodomain. There are 39 Hox genes in mammals, and they are clustered into four chromosomal loci (HoxA to HoxD), which regulate different genetic programmes along the anterior–posterior axis of the body. Precise temporal and spatial expression of Hox genes and accurate maintenance of their expression patterns are essential for animal development and cell fate determination. Thus, Hox genes are subject to intensive regulation at both transcriptional and post-transcriptional levels [79,80]. In addition to transcription factors and microRNAs, genomic approaches revealed that Hox gene clusters also encode hundreds of lncRNAs [56]. Interestingly, some of these RNAs are important in modulating the expression of Hox genes (Fig 2C).
Rinn et al identified a 2.2 kb lncRNA named HOTAIR that can repress the HOXD locus in trans in mammalian cells [56]. HOTAIR resides in the HoxC cluster and is transcribed antisense to protein-encoding HoxC genes in cells with posterior and distal positional identities. HOTAIR knockdown results in upregulation of genes across the HoxD locus, the strongest effect being an approximate twofold increase in HOXD10 expression, again a modest but significant effect. Such transcription activation is accompanied by a decrease in the repressive histone modification mark H3K27me3, and reduced occupancy of PRC2 and of a second chromatin-modifying complex containing the lysine demethylase LSD1 [59]. Thus, the HOTAIR lncRNA might function as a scaffold for the recruitment of chromatin-modifying complexes that repress transcription of the HoxD locus. This function is consistent with greater evolutionary constraint on the inferred RNA structure than in the primary sequence among mammalian HOTAIR orthologues [81]. HOTAIR has also been implicated in disease, as overexpression occurs in a wide variety of cancers [82]. In breast and colorectal cancer, for example, HOTAIR seems to modulate tumour invasiveness by enhancing PRC2-mediated repression of genes that suppress metastasis [83,84]. Therefore, HOTAIR plays a crucial role during both development and cancer by helping to specify gene expression programmes.
In addition to repressing transcription, lncRNAs from Hox clusters can also facilitate transcriptional activation. Two lncRNAs from the HoxA cluster, HOTTIP and Mistral, have such a capacity [57,58]. HOTTIP resides in the 5′ tip of the HoxA locus. Although expressed at low levels, this 3,764 nt transcript can be specifically detected at distal–posterior anatomic sites of both mouse and chicken embryos. The positive correlation between HOTTIP expression and that of its neighbours at the HoxA locus suggests that HOTTIP might help regulate their transcription. Consistent with this, inhibition of HOTTIP by siRNA specifically reduces the expression levels of HoxA genes in a unidirectional trend inversely proportional to their distance from HOTTIP; the decrease ranges from 30–80%. This reduction is associated with the appearance of the repressive H3K27me3 chromatin mark and disappearance of the active chromatin mark H3K4me3, together with reduced occupancy by the WDR5–MLL1 complex, an epigenetic activator of the Trithorax group. Biochemical analysis has revealed that WDR5 can specifically interact with HOTTIP; this interaction results in activation of targets only when HOTTIP is physically positioned near them, as tested by direct tethering to the promoter region of a reporter gene. Hence it seems that productive interaction of this lincRNA with chromatin modifiers is restricted to its localization to target chromosomal domains. This is supported by detection of endogenous chromatin interactions between HOTTIP and target loci by chromosome conformation capture, and by the fact that its low copy number—less than one copy per cell measured by single-molecule RNA FISH—might limit significant activity in trans. Thus, HOTTIP can help maintain the active chromatin state of the HoxA locus in cis by recruiting the WDR5–MLL1 complex.
Another lncRNA from the HoxA locus, Mistral, can also recruit the WDR5–MLL1 complex to activate the expression of target Hox genes. Mistral is an unspliced and polyadenylated 798 nt transcript. It is located between HoxA6 and HoxA7 in the HoxA locus and is induced on differentiation of ES cells. The recruitment of the WDR5–MLL1 complex can result in chromosomal conformational changes in the HoxA locus, which contributes to the activation of HoxA6 and HoxA7 transcription during ES cell differentiation.
These three examples indicate that, as with proteins and microRNAs, lncRNAs are also important in repressing and activating target Hox genes. Therefore, lncRNA-mediated regulation can contribute to the precise temporal and spatial control of genes that specify the body plan in animals.
Modulation of haematopoiesis by lncRNAs
Haematopoiesis, the developmental process by which mature blood cells are generated from primary progenitors, is essential for mammals. All haematopoietic effector cells—erythroid cells, myelocytes and lymphocytes—are derived from haematopoietic stem cells through regulated lineage specification and differentiation. Haematopoietic multipotent and lineage-determined progenitor cells can be readily purified by using cell-surface markers and have been extensively studied, making the haematopoietic system one of the best paradigms for studying cell-lineage determination and differentiation in mammals [85]. In addition to well-characterized transcription factors and microRNAs, studies are revealing that lncRNAs are also involved in haematopoiesis, particularly in the development of cells of the myeloid lineages (Fig 2D).
Wagner et al observed that an intronic lncRNA, EGO, is involved in the development of eosinophils [86], one of the immune system components that has a role in parasitic immunity and allergic diseases such as asthma. EGO is a conserved transcript derived from an intron of the ITPR1 gene. It becomes upregulated during eosinophil differentiation, and biochemical analysis indicates that the transcript is non-coding, as it does not associate with ribosomes. siRNA-mediated knockdown of EGO impairs the expression of genes important for eosinophil development, such as major basic protein and eosinophil-derived neurotoxin. These results suggest that EGO can modulate the differentiation of cells in the eosinophil lineage.
LncRNAs are also implicated in the regulation of myelopoiesis, the formation of granulocytes and monocytes. Zhang et al identified a lincRNA (HOTAIRM1) encoded in the human HoxA cluster that is markedly upregulated during granulocytic differentiation of myeloid progenitor cells induced by retinoic acid [87]. Transcribed antisense to coding genes in the HoxA cluster, HOTAIRM1 is about 500 nt in length and does not associate with ribosomes. Knockdown of this lincRNA inhibits retinoic acid-induced HoxA1 and HoxA4 expression during myeloid differentiation and specifically impairs the expression of several markers of differentiated myeloid cells, such as CD11b and CD18. These observations implicate HOTAIRM1 in regulating myelopoiesis, possibly by regulating the transcription of genes at the HoxA locus.
Our group found that one lincRNA has an essential role in the maturation of red blood cells [50]. We performed transcriptome profiling on mouse erythroid cells at different developmental stages and observed that more than 400 lncRNAs are differentially expressed during this developmental process. Among these, we characterized a lincRNA, lincRNA-EPS, which is required for the maturation of mouse erythroid cells by inhibiting apoptosis. LincRNA-EPS is markedly induced during the terminal differentiation of mouse erythroid cells. Its inhibition results in apoptosis and severely compromises differentiation and downstream enucleation from erythroid cells. Conversely, ectopic expression of lincRNA-EPS can protect erythroid progenitor cells from apoptosis triggered by erythropoietin starvation. Thus, this erythroid-specific lincRNA shows potent anti-apoptotic activity. Mechanistic studies suggest that lincRNA-EPS regulates apoptosis by repressing expression of several pro-apoptotic proteins, most prominently the caspase-activating adaptor protein Pycard.
LncRNAs can also regulate the function of haematopoietic-related tissues (Fig 2E). Li et al characterized an antisense lncRNA, Tie-1AS, that has a role in regulating the function of vascular endothelial cells, which derive from the same progenitors as blood cells during embryogenesis [88]. Tie-1AS is an evolutionarily conserved, non-coding transcript antisense to the Tie-1 gene, which encodes a cell surface tyrosine kinase receptor for angiopoietin ligands. It seems to modulate the function of endothelial cells by regulating Tie-1 mRNA levels, presumably by forming Tie-1AS–Tie-1 mRNA hybrids. Transient transfection of Tie-AS disrupts vascular tube formation both in zebrafish in vivo and in human-cultured vascular endothelial cells. Consistent with this phenotype, the ratio of Tie-1 mRNA to Tie-1AS is altered in human vascular pathological samples. This study suggests that a certain level of Tie-AS might be required to maintain the normal state of vascular endothelial cells. However, loss-of-function experiments are needed to clarify further the physiological role of this antisense lncRNA.
Collectively, these studies indicate that lncRNAs contribute to the regulation of differentiation in several haematopoietic and related lineages.
LncRNAs in muscle differentiation
The differentiation of muscle cells is another extensively studied somatic developmental process in mammals. Many key transcription factors and microRNAs that control the expression of genes involved in muscle growth, morphogenesis and differentiation are well-characterized in both in vitro tissue culture and in vivo mouse models [89]. A report of cross-talk between lncRNA and microRNA function adds an interesting layer of regulation to this biological process (Fig 2F). Cesana et al characterized a muscle-specific lincRNA, linc-MD1, which can inhibit two important microRNAs, miR-133 and miR-135, during muscle differentiation [65]. Linc-MD1 becomes activated on myoblast differentiation, but unlike the lincRNAs that regulate chromatin modification, it resides in the cytoplasm, suggesting that it might regulate cytoplasmic events. Sequence analysis has revealed that linc-MD1 contains highly conserved binding sites for miR-133 and miR-135. Functional studies indicate that this non-coding transcript can ‘sponge’ miR-133 and miR-135 during muscle differentiation, thereby downregulating the microRNAs and upregulating their mRNA targets. Importantly, inhibition of linc-MD1 compromises muscle differentiation. This example is consistent with the proposed idea that endogenous RNAs can modulate each other indirectly by competing for the available pool of common microRNA regulators [90].
In addition to regulating microRNA activity, lncRNAs can also modulate mRNA decay during muscle differentiation. By using C2C12 myoblasts as an in vitro culture system, Gong et al observed that two mRNA decay pathways, named Staufen1-mediated mRNA decay (SMD) and nonsense-mediated mRNA decay (NMD), contribute to muscle differentiation by regulating the abundance of target mRNAs [91]. Cytoplasmic lncRNAs can trigger SMD by base-paring with the 3′ untranslated region of a subset of SMD target mRNAs. This lncRNA–mRNA interaction can recruit Staufen1, a key component of the SMD pathway, which results in degradation of target mRNAs [62]. Thus, it seems that certain cytoplasmic lncRNAs can contribute to muscle differentiation by modulating mRNA stability.
These two examples implicate lncRNAs directly in the regulation of muscle differentiation. Therefore, in addition to regulating chromatin modification in the nucleus, lncRNAs can also modulate cytoplasmic events, such as microRNA activity and mRNA stability.
LncRNAs in neural cell differentiation
The central nervous system of mammals is mainly composed of two types of cell: the neuron and the glia cell. These are generated from neural stem cells, which can be isolated from adult brain or derived from ES cells. Several lines of evidence indicate that lncRNAs modulate neural cell differentiation (Fig 2G).
First, many lncRNAs are differentially expressed in the central nervous system. Large-scale RNA FISH analysis revealed that many lncRNAs expressed in the brain are specifically detected in distinct neuroanatomical regions, cell types and subcellular compartments [17]. Such highly specific expression patterns suggest the possibility that some of these lncRNAs might be involved in various neurological functions. Consistent with this, transcriptome profiling during neuronal cell differentiation indicated that many lncRNAs are differentially expressed during neuronal–glial fate specification and during oligodendrocyte lineage maturation [92]. Moreover, altering the expression levels of several lncRNAs uncovered important roles in regulating neural cell differentiation.
For example, overexpression of the antisense lncRNA Nkx2.2as in neural stem cells promotes their differentiation along the oligodendrocyte lineage [93]. Although Nkx2.2as is a cytoplasmic transcript, its ectopic expression can modestly increase the mRNA level of Nkx2.2, a transcription factor involved in oligodendrocyte differentiation. Thus, it was hypothesized that Nkx2.2as enhances differentiation through modulation of Nkx2.2 [93], a possibility that should be further explored by loss-of-function analysis.
Inhibition of lncRNAs might also impair neural cell differentiation. For instance, Evf2 is a well-characterized lincRNA transcribed from the conserved intergenic region of the Dlx5 and Dlx6 loci [94]. Evf2 is an alternatively spliced isoform of Evf1, another lincRNA within this genomic locus. During the development of mouse ventral forebrain, Evf2 functions as a transcriptional co-activator that recruits DLX and MECP2 transcription factors to modulate the expression of the neighbouring Dlx5, Dlx6 and Gad1 genes [95]. Importantly, suppression of mouse Evf2 by poly(A) site insertions reduces the number of GABAergic interneurons and compromises synaptic inhibition in the early postnatal hippocampus and dentate gyrus [95]. Such marked phenotypes suggest that Evf2 has a crucial role during early development of the hippocampus.
Inhibition of certain lincRNAs also impairs mouse retinal cell differentiation [96]. Specifically, RNCR2 is a lincRNA selectively expressed in retinal progenitor cells. Knocking it down by shRNAs results in differentiation of progenitor cells towards non-retinal cell lineages, such as amacrine cells. This suggests that RNCR2 is involved in retinal cell fate specification. Interestingly, mislocalization of this nuclear-retained lincRNA to the cytoplasm, through fusion with an IRES-controlled GFP transgene, phenotypically copies the shRNA knockdown, indicating that the correct cellular localization of this lincRNA is important for its cellular function. Similarly, Meola et al reported that overexpression of Vax2os1, a lncRNA selectively expressed in the developing retina, can inhibit retinal progenitor cell proliferation [97] probably through impairment of cell cycle progression and increased apoptosis.
Inhibition of lncRNAs also impairs neuronal differentiation of human ES cells [98]. Transcriptome profiling revealed that many lncRNAs are differentially expressed during the differentiation of human ES cells towards neuronal progenitor cells and ultimately neurons. Among these lncRNAs, 35 are highly expressed in terminally differentiated neurons, suggesting potential roles in neuronal differentiation. siRNA-mediated knockdown of four of these neuron-enriched lncRNAs results in significant changes of gene expression patterns and impairment of neuronal differentiation. Mechanistically, three of these lncRNAs seem to act in the regulation of the chromatin state, as they are localized predominantly in the nucleus and bind to the PRC2 complex. Interestingly, they also seem to bind to the pluripotency-associated transcription factor Sox2, thus suggesting that lncRNAs can also act as scaffolds for combinations of chromatin modifiers and transcription factors. These examples indicate that, as a group of gene-expression regulators, lncRNAs can modulate neural cell differentiation through diverse mechanisms.
Perspectives and challenges for the future
Thanks to the increasing availability and rapid development of high-throughput transcriptome profiling techniques, thousands of lncRNAs have been identified in mammalian genomes over the past few years [1,2,3,4,6,40]. Given the wide application of these technologies, more differentially expressed lncRNAs will probably be uncovered in several mammalian developmental and differentiation processes. Functional characterization of differentially expressed lncRNAs, however, has only been explored in a few cases. Of the identified lncRNAs, not all might be functional, and some of them might even be just transcriptional noise [19,20]. Nonetheless, mounting evidence points towards an increasing number of lncRNAs with recognized biological functions in specific physiological and pathological contexts [7,55], including the examples we described previously (Table 1). Thus, as a class of regulatory genes, lncRNAs can clearly function as versatile modulators of diverse biological processes.
Here, we discuss from our own perspectives some of the outstanding open questions in this new and rapidly expanding field (Sidebar A).
Sidebar A | In need of answers.
What fraction of mammalian transcriptomes function only as coding, non-coding or as both?
How might common sequence and structural features be used to imply functionally coherent lncRNA families?
What selective constraints shape the rapid evolution of lncRNAs?
What molecular features determine whether lncRNAs act in cis compared with in trans?
How is specific association of lncRNAs with selected protein and DNA targets achieved in vivo?
What role does local chromatin conformation have in modulating the activity of specific lncRNAs at their target sites?
How might lncRNAs be integrated into existing regulatory networks in control of cell differentiation?
How do lncRNAs contribute to mammalian organismal development in vivo?
Are the functions of lncRNAs conserved among mammals?
Technical challenges in elucidating lncRNA function
si/shRNA-mediated loss-of-function approaches are commonly used to determine the function of lncRNAs. Several caveats, however, should be taken into consideration when such techniques are used. First, every lncRNA cannot be efficiently knocked down by si/shRNA. Second, si/shRNA approaches are known to have off-target effects. Thus, additional experiments, such as phenotypic rescue by a lncRNA lacking the si/shRNA-target sequence(s), are needed to correctly interpret results from these experiments [99]. The use of a modified transcript for phenotypic rescue, however, is plagued by its own caveats; for example, it might be impractical to recapitulate the physiological expression, positional or structural genomic context of the endogenous locus, all of which seem important for lncRNA function. Third, delivery of excessive si/shRNA doses can produce spurious phenotypes by out-competing the effector machinery from its endogenous targets, as with competing endogenous RNAs [90].
For these reasons, alternative approaches must be pondered. For example, antisense oligos, which make use of endogenous RNaseH activity to cleave target transcripts, can be used as an orthogonal method for stoichiometric lncRNA knockdown; poly(A) site insertions can also provide a minimally invasive method of disrupting lncRNA transcription, and so can TALE nuclease-mediated transcriptional repression [33,100].
It should also be noted that the absence of phenotypic results from lncRNA knockdown experiments does not necessarily rule out a biological function. It remains possible that the transcription itself, but not the RNA product, exerts a regulatory function in its local context [101]. Thus, approaches such as target-specific disruption of transcription by poly(A) site insertions or by TALE-mediated repression, are needed to discriminate among these possibilities. Or, as is often the case with protein-encoding genes, several lncRNAs might have redundant functions.
Retroviral- and lentiviral-mediated ectopic expression approaches are commonly used for gain-of-function studies of lncRNAs [50,102]. One caveat of this approach is that the transcript generated from the viral LTR promoter usually contains a stretch of viral RNA. Thus, the resulting RNA is a fusion of viral RNA and lncRNA, and so any observed gain-of-function phenotypes might not necessarily be mediated by the overexpressed lncRNA. To avoid this caveat, plasmid transfection or TALE-mediated transcriptional activation of the endogenous locus can instead be used. Overexpression studies are difficult to interpret, however, as they intrinsically represent non-physiological conditions. Convincing evidence of lncRNA function will thus ultimately require dedicated structure–function analyses.
Molecular mechanisms of lncRNAs
LncRNAs can modulate gene expression through diverse mechanisms (Fig 1; [67,68,103]). Of those mechanisms implicated in mammalian cell differentiation, many seem to direct gene expression through recruitment of chromatin modifiers. This is consistent with several observations that chromatin modifiers, such as PRC2, can associate with a diversity of non-coding transcripts [11,38,98,104]. Interestingly, lncRNAs can function as scaffolds to recruit histone modification complexes [59], and lncRNAs generally have richer tissue specificity than coding genes [40]. It thus seems tempting to speculate that one main, although not exclusive, function of lncRNA during mammalian cell differentiation is to promote, in a cell-type-specific manner, the assembly of select combinations of ubiquitously expressed chromatin modifiers, which in turn govern the chromatin state of specific target genomic regions [11]. However, case-by-case analysis is required to dissect in detail how specific association with chromatin-modifying partners is achieved in vivo, what sequence properties enable lncRNAs to target these partners to specific areas in the genome and what role local chromatin conformation might have in modulating these interactions.
An expanding tool-box of molecular approaches is rapidly becoming available to address these and other questions regarding mechanisms of lncRNA function. Investigating these typically begins by first establishing their subcellular localization. Cellular fractionation followed by RNA detection can be a cost-effective method to broadly distinguish between nuclear-acting and cytoplasmic lncRNAs. In addition, direct visualization of lncRNA by RNA FISH can provide a high-resolution picture of localization to even smaller subcellular compartments, such as paraspeckles or the nucleolus [31,33,34,35], or even to specific chromosomal regions [32,105], and can also be used to examine multimerization potential and co-localization with RNA or protein partners [38,106]. Importantly, de novo protein partners can be identified through RNA-mediated pull-downs [56,102]. Several powerful assays have also been developed to determine the genomic binding sites of nuclear-acting lncRNAs [107,108].
These and other assays will greatly facilitate the exploration of lncRNA function in mammalian cell differentiation at the molecular mechanistic level. Judging by the constant development and broad application of these assays, we predict that such exploration will greatly advance in the coming years.
Integrating lncRNAs to known regulatory networks
The differentiation of mammalian cells is exquisitely controlled at every stage by complex networks that respond to developmental and environmental cues. The aforementioned examples argue that lncRNAs are probably integrated as key components of these regulatory networks, together with transcription factors and microRNAs. Precisely how they should be integrated can be answered by first exploring their regulatory relationship with other components (Fig 3). Expression of many mammalian lncRNA-modulating cell differentiation programmes is indeed controlled by key transcription factors in charge of those programmes. We have noted the case of lncRNAs involved in ES cell pluripotency maintenance, which are driven by known pluripotency-associated factors [10,11,98]. Interestingly, some lncRNAs seem also to bind physically to transcription factors [98], suggesting the possibility that lncRNA transcription regulation networks might use some of the same motifs as those of mRNA networks, such as autoregulation and feedback loops. Further progress in identifying the global binding sites of key transcription factors during cell differentiation, as well as the protein interactome of lncRNAs, will help in reconstructing transcription regulation networks involving lncRNAs. Simply intersecting such data sets with transcriptome profiling will be of great use in identifying lncRNAs as candidates for functional studies.
Figure 3.
Integrating lncRNAs to known regulatory networks of mammalian cell differentiation. A first step towards integrating lncRNA functions with those of microRNAs, transcription factors and chromatin modifiers during differentiation of mammalian cells is to explore their mutual regulatory relationships. Some examples of these relationships are depicted. lncRNAs (red RNAs and red arrows) can regulate microRNAs as target site decoys, directly bind to transcription factors as target mimics or as allosteric regulators, and participate in assembly of chromatin-modifying complexes as structural components and recruiters to genomic targets. microRNAs (green RNAs and arrows) post-transcriptionally regulate RNAs from transcription factor, chromatin modifier or lncRNA loci by directly base-pairing to short stretches of RNA. Transcription factors (blue proteins and arrows) can regulate transcription of all the other regulators by directly binding to their promoters. Similarly, chromatin modifiers (orange proteins and arrows) also regulate transcription of the other network components through chromatin modification. Regulatory relationships between microRNAs and chromatin modifier are not depicted. lncRNA, long non-coding RNA.
Several studies have also proposed that certain lncRNAs and microRNAs can regulate each other at the post-transcriptional level [63,64,65]. However, global identification of lncRNA targets of microRNAs still remains largely unexplored. Thus, identifying microRNA and lncRNA with complementary expression patterns during cell differentiation might generate candidate lncRNA–microRNA regulatory pairs to be tested for integration into regulatory networks. Such studies might not only serve to define such new networks, but also to expand our understanding of how they contribute to mammalian cell differentiation.
In comparing the role of lncRNAs with those of other factors involved in mammalian cell differentiation, it is important to note that, similar to many microRNAs, the biological effects of many lncRNAs are relatively mild—about a 1.5–2-fold change in the expression of target loci upon lncRNA perturbation. This might be partly due to limitations in achieving efficient knockdown of lncRNAs by si/shRNA approaches. Alternatively, it might indicate that lncRNAs mainly act to fine-tune target gene expression, as with microRNAs. In vivo knockout models of lncRNAs might thus be required to discriminate between these two possibilities, as we discuss below.
Compared to known transcription factors and microRNA regulators of cell differentiation, lncRNAs can use a diversity of mechanisms to modulate gene expression at several levels (Fig 1). Thus, during cell differentiation lncRNAs might co-operate with both transcription factors and microRNAs to ensure precise gene expression at both the transcriptional and post-transcriptional levels.
In vivo functions of lncRNAs
Although perturbation of many lncRNAs can result in phenotypic changes during differentiation of in vitro-cultured cells, our knowledge of the in vivo functions of lncRNAs is still limited in mammals (Sidebar A). Several lncRNA-altered mice have been generated to address this question. Although some of these mice seem to have alterations in certain cellular and physiological processes [95], whether lncRNAs, particularly those that can affect cellular differentiation in vitro, contribute to organismal development in vivo remains largely unanswered. Pioneering studies in non-mammalian vertebrate models are only beginning to establish that this might indeed be the case [58,116]. Surprisingly, however, knocking out MALAT1—a highly expressed lncRNA conserved from zebrafish to humans—in mice results in no detectable developmental defects [111,117,118]. Similarly, a mouse deleted for the entire HoxC cluster, which contains HOTAIR, showed no obvious developmental defects [112]; whereas in cultured human somatic cells, as noted, HOTAIR has been shown to be involved in repressing genes in the HoxD cluster [56]. Although differences between in vitro and in vivo studies might be attributable to potential redundant pathways that compensate for lncRNA alterations in vivo, they also highlight the need for further investigation of additional lncRNAs under informative physiological conditions.
Acknowledgments
W.H. is a Merck Fellow of the Life Sciences Research Foundation. H.F.L. was supported by National Institutes of Health grants 2R01 DK0467618, 2 P01 HL032262 and R01 DK068348.
Footnotes
The authors declare that they have no conflict of interest.
References
- Guttman M et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P et al. (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Kapranov P et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Carninci P et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Carninci P et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD et al. (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J et al. (2010) Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M et al. (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T et al. (2010) Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356 [DOI] [PubMed] [Google Scholar]
- Pachnis V, Brannan CI, Tilghman SM (1988) The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 7: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI et al. (1990) The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT et al. (2005) A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Mercer TR et al. (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME et al. (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M et al. (2008) Ripples from neighbouring transcription. Nat Cell Biol 10: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Struhl K (2007) Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- De Santa F et al. (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MI et al. (2007) Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR (2007) Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Berretta J, Morillon A (2009) Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 10: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A (2009) The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS (2007) Raising the estimate of functional human sequences. Genome Res 17: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G (2007) Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S et al. (2004) Genomic analysis of mouse retinal development. PLoS Biol 2: E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ et al. (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Nagano T et al. (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Redrup L et al. (2009) The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT et al. (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H et al. (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM et al. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T et al. (2006) Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J et al. (2009) Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5: e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M et al. (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN et al. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ (2011) Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 12: 459–463 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Brockdorff N et al. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Duret L et al. (2006) The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Lin MF et al. (2008) Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput Bio 4: e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M (2011) PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Knopf PM, Rich A (1963) A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci USA 49: 122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT et al. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH, Rout M (2011) Nuclear export dynamics of RNA–protein complexes. Nature 475: 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W et al. (2011) Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25: 2573–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2011) Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12: 815–826 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2004) RNA regulation: a new genetics? Nat Rev Genet 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS (2008) Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS (2007) The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29: 288–299 [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361 [DOI] [PubMed] [Google Scholar]
- Rinn JL et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S et al. (2011) The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell 43: 1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang KC et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC et al. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V et al. (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH et al. (2012) LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA et al. (2011) In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L et al. (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M et al. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C (2012) Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453 [DOI] [PubMed] [Google Scholar]
- Young RA (2011) Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J et al. (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 12: 542–553 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K (2010) Induced pluripotency: history, mechanisms, and applications. Genes Dev 24: 2239–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R (2010) Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143: 508–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S et al. (2010) Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M et al. (2012) Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26: 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger MR et al. (2006) A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci USA 103: 5781–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Kloter U, Suga H (2009) Evolution of the Hox gene complex from an evolutionary ground state. Curr Top Dev Biol 88: 35–61 [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904 [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP (2008) MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 9: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liu S, Zhu H (2011) The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol 11: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S (2012) The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol 9: 703–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R et al. (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71: 6320–6326 [DOI] [PubMed] [Google Scholar]
- Kondo M et al. (2003) Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol 21: 759–806 [DOI] [PubMed] [Google Scholar]
- Wagner LA et al. (2007) EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 109: 5191–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X et al. (2009) A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113: 2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K et al. (2010) A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 115: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Gautel M (2011) Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12: 349–361 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I et al. (2011) ceRNAs: miRNA target mimic mimics. Cell 147: 1431–1432 [DOI] [PubMed] [Google Scholar]
- Gong C et al. (2009) SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR et al. (2010) Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y (2008) Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun 372: 691–696 [DOI] [PubMed] [Google Scholar]
- Feng J et al. (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM et al. (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S (2010) The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N et al. (2012) The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA 18: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW (2012) Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 31: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr (2012) Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science 337: 421–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L et al. (2012) Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 3: 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Huarte M et al. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM (2012) Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40: 6391–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J et al. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B et al. (2010) Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D et al. (2012) Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat Methods 9: 360–362 [DOI] [PubMed] [Google Scholar]
- Chu C et al. (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD et al. (2011) The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA 108: 20497–20502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon FE et al. (2010) Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology 151: 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S et al. (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S et al. (2012) Malat1 is not an essential component of nuclear speckles in mice. RNA 18: 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D (2011) Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC et al. (2011) Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 7: e1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F et al. (2010) Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499 [DOI] [PubMed] [Google Scholar]
- Ripoche MA et al. (1997) Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ulitsky I et al. (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B et al. (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M et al. (2012) Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. Biol RNA 9: 1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]



 [Left to right] Wenqian Hu, Juan R Alvarez-Dominguez and Harvey F Lodish
[Left to right] Wenqian Hu, Juan R Alvarez-Dominguez and Harvey F Lodish