Abstract
Chromatin organization is essential for defining transcription units and maintaining genomic integrity in eukaryotes. In this study, we found that deletion of the Schizosaccharomyces pombe Chd1 chromatin remodelers, hrp1 and hrp3, causes strong, genome-wide accumulation of antisense transcripts. Nucleosome mapping revealed a specific role for Chd1 remodelers in the positioning of nucleosomes in gene coding regions. Other mutations associated with enhanced cryptic transcription activity, such as set2Δ, alp13Δ and FACT complex subunit pob3Δ, did not, or only mildly, affect nucleosome positioning. These data indicate several mechanisms in the repression of cryptic promoter activity in eukaryotic cells.
Keywords: Chd1, chromatin remodelers, cryptic transcription, nucleosome structure, S.pombe
Introduction
The eukaryotic genome is organized into a compact and elaborate chromatin structure. The basic units are the nucleosomes, consisting of 147-base pairs (bp) DNA wrapped around an octamer of histone proteins. Nucleosomes are further organized into higher-order structures that compacts the genome and protects it against damage, but also limits the availability of the DNA to macromolecules. Chromatin-modifying activities can switch between open or closed chromatin conformations and allow regulated access to the DNA. The overall chromatin ‘landscape’ of a genomic region establishes specific functional regions, such as promoter or coding regions of transcriptional units. The definition of these units is not very strict, as demonstrated by extended transcriptional activity outside the canonical transcription units. A main source of such cryptic transcription activity is transcription initiation from cryptic promoters. As the DNA sequence requirements for RNA polymerase II (Pol II) initiation are rather loose and appear frequently in the genome, chromatin structure has a major role in distinguishing canonical promoters from cryptic promoters. Promoter regions possess a particularly open chromatin structure, including nucleosome-free regions (NFRs) [1] and the surrounding hyper-acetylated nucleosomes [2] . The chromatin in coding regions is in a more closed conformation with regularly placed nucleosomes and low levels of histone acetylation [2] . The removal of acetyl marks from histones by histone deacetylase (HDAC) complexes is crucial for the maintenance of a closed chromatin conformation within coding regions. The evolutionarily conserved Rpd3S complex in Saccharomyces cerevisiae (S. cerevisiae) is responsible for histone deacetylation within gene coding regions [3, 4] . Clr6 complex II is the S. pombe homologue of this complex [5] . Mutations in these complexes lead to increased histone acetylation levels in gene coding regions and activation of cryptic promoters [4, 6] . The Set2 histone methyltransferase is also involved in repressing cryptic promoter activity. Set2 is recruited to the elongating Pol II complex and methylates histone H3 lysine 36 (H3K36me) in gene coding regions [7] . H3K36me is thought to be the recruitment signal for the Clr6 complex II through its chromodomain-containing subunit, Alp13 (Eaf3 in S. cerevisiae) [3, 4, 6]. However, recent findings revealed that localization of the HDAC complex to coding regions is unaffected in set2Δ, suggesting that H3K36me is mainly relevant for the activity of the HDAC complex .
The elongating Pol II also recruits complexes whose main activity is the reassembly of nucleosomes following transcriptional activity on the DNA. Among these, one of the best characterized is the FACT complex. Mutations in this complex lead to decreased histone occupancy levels in transcribed genes and increased activity of cryptic promoters in these regions [9,10,11,12]. Similarly, mutations in the transcription elongation factor Spt6 [9] , or in the histone chaperone Asf1/HIR complex lead to decreased histone occupancy and increased activity of cryptic promoters [12,13,14]. Genome-wide screens also identified the chromatin remodelling enzymes Chd1 and Isw1 as factors required for repression of cryptic promoters in S. cerevisiae [12, 15] . Chd1 was also found to interact with transcription elongation complexes such as the Paf and FACT complexes [15,16,17]. A recent study from the Owen–Hughes lab reported that Isw1, Isw2 and Chd1 are responsible for the positioning of evenly spaced nucleosomes in gene coding regions in S. cerevisiae [18] . S. pombe has a slightly different subset of chromatin remodelers than S. cerevisiae. While S. pombe lacks the ISWI remodelers, Isw1 and Isw2, it contains two Chd1 remodelers, Hrp1 and Hrp3.
In this study, we attempted to identify factors that have a role in repressing cryptic transcription activity in S. pombe. We screened a deletion library for chromatin-related factors and tested their effect on cryptic transcription. We found that deletion of the S. pombe Chd1 chromatin remodelers, hrp1 and hrp3, results in a marked increase in cryptic transcription. To determine the underlying molecular mechanism, we mapped genome-wide nucleosome position and histone acetylation patterns in the Chd1-deficient strain. These experiments uncovered a specific role for Chd1 remodelers in maintaining the highly ordered nucleosome structure within transcription units. We extended our analysis to other mutations also known to enhance cryptic transcription activity. Although these mutants accumulate cryptic transcripts very similarly to the Chd1-deficient strain, our data showed that the underlying mechanisms are remarkably different.
Results and discussion
Chd1-deficient cells accumulate cryptic RNA transcripts
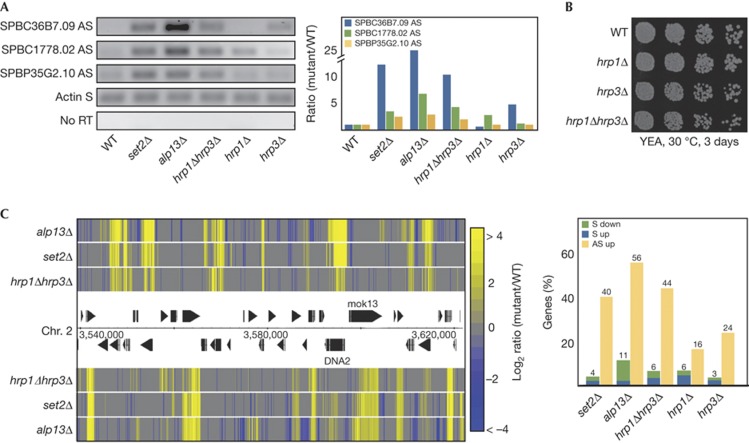
To further understand the role of chromatin in defining transcription units, we sought to identify mutations that activate cryptic transcription. To monitor cryptic transcription, we quantified the levels of antisense (AS) transcripts for selected genes in the S. pombe genome. AS transcript levels are very low in wild-type (WT) strain but increase substantially in mutants causing cryptic transcription; therefore, they serve as a sensitive indicator of cryptic transcription. We isolated total RNA from deletion mutants of chromatin-related factors (S. pombe deletion library—Bioneer Corporation, Daejeon, South Korea) and performed strand-specific reverse transcription PCRs. As positive controls, we used strains lacking Set2 and Clr6 complex II subunit Alp13. Both mutants are known to accumulate cryptic transcripts [6] . Among the mutant strains that were tested, we observed that deletion of hrp3 or hrp1 also leads to increased levels of AS transcripts (Fig 1A). Hrp1 and Hrp3 are two members of the Chd1 subfamily of ATP-dependent chromatin remodelling factors in S. pombe. We also tested AS transcript levels in a hrp1Δhrp3Δ double-deletion strain (from herein also referred to as Chd1-deficient strain). The combination of the two deletions demonstrated a strong increase in the levels of the monitored AS transcripts (Fig 1A). Despite the strong AS transcript accumulation, the hrp1Δhrp3Δ strain did not demonstrate an obvious growth defect (Fig 1B). We also analysed genome-wide expression profiles in the above-mentioned mutant strains using high-resolution tiling microarrays (Fig 1C). The Chd1-deficient strain revealed strong, genome-wide AS transcript accumulation with 44% of the analysed genes showing significantly increased AS levels. Interestingly, changes observed in the sense transcripts were rather minor, with only 6% of the gene transcripts significantly up- or downregulated. Single deletions of hrp3 or hrp1 also showed moderate AS transcript accumulation (24% and 16%, respectively, of genes analysed). We also confirmed that deletion of set2 or alp13 leads to genome-wide AS transcript accumulation (40% and 56% of the genes, respectively) [6] . Comparison of these expression analyses revealed a remarkable similarity between the AS accumulation profile of the Chd1-, Alp13- and Set2-deficient strains (Fig 1C). This result suggests that Chd1-type chromatin remodelers, Hrp1 and Hrp3, are responsible for the repression of cryptic promoter activity in coding regions, similar to the previously characterized Set2–Clr6 complex II pathway.
Figure 1.
hrp1Δhrp3Δ cells accumulate AS transcripts. (A) Gel electrophoresis (left panel) of strand-specific reverse transcription and PCR amplification of the indicated S and AS transcripts in WT and mutant strains. Total RNA from the indicated strains was reverse transcribed using strand-specific primers, and the resulting complementary DNA was amplified by PCR. Controls lacking reverse transcriptase were used to exclude DNA contamination (no RT). The chart (right panel) indicates quantification of the PCR transcripts normalized to the amount of actin S transcripts (WT set to 1). (B) Growth of indicated strains in a series of fivefold dilutions on YEA plates. The cells were incubated at 30 °C for 3 days. (C) Heat map (left panel) showing a portion of the S. pombe genome demonstrating relative levels of transcripts from the forward (upper panel) and reverse (lower panel) directions in the indicated strains compared with WT. Colours represent up- or downregulation of gene expression on a log2 scale. The chart (right panel) shows genome-wide statistics of changes in S and AS transcript levels for the indicated strains compared with WT. AS, antisense; Chr., chromosome; RT, reverse transcriptase; S, sense; WT, wild-type.
Nucleosomes are not depleted in Chd1-deficient cells
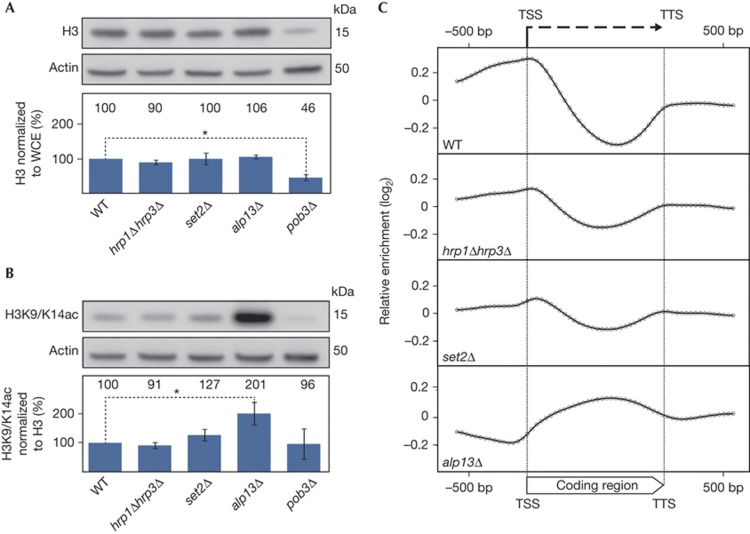
Previous studies showed that genome-wide nucleosome depletion can activate cryptic transcription in yeast [12, 14] . This prompted us to check nucleosome occupancy in the Chd1-deficient strain. Accurate quantification of nucleosome occupancy is very challenging because genome-wide techniques only provide relative enrichment values, and cannot detect changes affecting the whole genome. Instead, we determined total histone H3 levels in WT and mutant strains. As nearly all histones are incorporated into the chromatin, alteration of the total H3 content of a cell is a good indicator of changes in genome-wide nucleosome occupancy [19, 20]. Western blot signals for bulk H3 levels from total cell extracts were quantified and normalized to the amount of total protein in the extract (Fig 2A; see signals normalized to actin in supplementary Fig S1A online). Quantification of western blot signals are limited in their precision and are dependent on the method of normalization. Therefore, our data are not a precise measurement but rather an indication of the changes in H3 content in the mutant strains compared with WT. Deletion of the FACT complex subunit pob3 resulted in a significant decrease in bulk H3 levels, confirming previous findings that nucleosome occupancy is substantially decreased in FACT complex mutants [11, 21] . In contrast, alp13Δ, set2Δ and hrp1Δhrp3Δ strains showed no significant change in their bulk H3 content. Although we cannot exclude a slight decrease in nucleosome occupancy in the hrp1Δhrp3Δ strain, a substantial nucleosome depletion such as the one observed in the pob3Δ strain was not detected in this mutant.
Figure 2.
Coding regions are not hyper-acetylated in hrp1Δhrp3Δ. (A) WCE were subjected to SDS–polyacrylamide gel electrophoresis and H3 levels were monitored by western blot. The relative H3 levels were quantified and normalized to the total protein content of the WCE (chart, lower panel) and WT was set to 100%. (B) Western blot analysis of the same WCEs using a polyclonal antibody recognizing both acetylated H3K9 and H3K14 (H3K9/K14ac). The relative H3K9/K14ac levels were quantified and normalized to levels of H3, with WT set to 100% (chart, lower panel). For (A) and (B), data represent the means of three independent experiments and error bars represent s.d. *Indicates statistical significance using Student’s t-test (P<0.05). (C) Composite plot of ChIP-on-chip analyses using the anti-H3K9/K14ac antibody in WT and indicated mutant strains. H3K9/K14ac relative enrichment values were determined in a genome-wide ChIP-on-chip experiment with 2,320 genes represented on the array. Each gene was divided into 30 parts (bins) and the average relative enrichment value was determined for each bin. In addition, 600 bp from both the flanking 5′ and 3′ regions were included in the analysis. The average of the log2 values of each of these bins (geometric mean) was calculated and plotted on a log2 scale, where each dot represents a bin. bp, base pairs; ChIP, chromatin immunoprecipitation; TSS, transcriptional start site; TTS, transcriptional termination site; WCE, whole-cell extracts; WT, wild-type.
ORFs are not hyper-acetylated in the hrp1Δhrp3Δ strain
As the pattern of AS transcript accumulation in hrp1Δhrp3Δ is very similar to those of alp13Δ and set2Δ, we wondered whether this similar phenotype might be the result of convergent molecular mechanisms. Current models suggest that both set2 deletion and mutations in the Clr6 complex II lead to increased histone acetylation in gene coding regions, resulting in enhanced cryptic promoter activity [4] . To test whether the increased cryptic transcription activity in the Chd1-deficient strain is also coupled to enhanced histone acetylation, we analysed bulk H3 acetylation levels in WT and mutant strains. We used a polyclonal antibody against acetylated H3 lysine 9 (H3K9ac) and H3 lysine 14 (H3K14ac), two known targets of the HDAC Clr6 complex II. The signals were normalized to the H3 content of the strains (Fig 2B; normalization to total protein extract or actin in supplementary Fig S1B online). Similar to published results, bulk H3K9/K14ac levels were remarkably increased in the HDAC subunit deletion alp13Δ compared with the WT strain [5] . In contrast, we did not observe significant changes in hrp1Δhrp3Δ, indicating an HDAC-independent mechanism in the activation of cryptic transcription in this mutant. Remarkably, in the set2Δ strain we could detect only a moderate increase in bulk H3 acetylation.
To understand the acetylation pattern of the mutants in more detail, we performed chromatin immunoprecipitation (ChIP) experiments to map the genome-wide distribution of H3K9/K14ac in WT and mutant strains. Composite plot of H3K9/K14ac signals for 2,320 genes shows the characteristic acetylation pattern in WT strain, with highly acetylated promoter regions and hypo-acetylated coding regions (Fig 2C). Deletion of alp13 results in the redistribution of acetylated histones with dramatically increased levels in coding regions (Fig 2C). In contrast, the genome-wide distribution of acetylated histones in hrp1Δhrp3Δ and set2Δ is similar to WT, with hyper-acetylated promoter regions and hypo-acetylated coding regions (Fig 2C). The reduced ratio of acetylation levels in the promoter region to levels in the coding region might indicate a modest increase of acetylation in the coding regions, but can also be the consequence of generally decreased acetylation levels. This result further supports an HDAC-independent mechanism for activating spurious transcription in the Chd1-deficient strain. The unexpected finding that deletion of set2, the sole H3K36 methyltransferase in S. pombe, had a significantly different effect on H3 acetylation levels than deletion of alp13 indicates that H3K36 methylation is not, or is only partly, responsible for the recruitment and activity of the HDAC Clr6 complex II in S. pombe.
Hrp1 and Hrp3 organize nucleosomes in ORFs
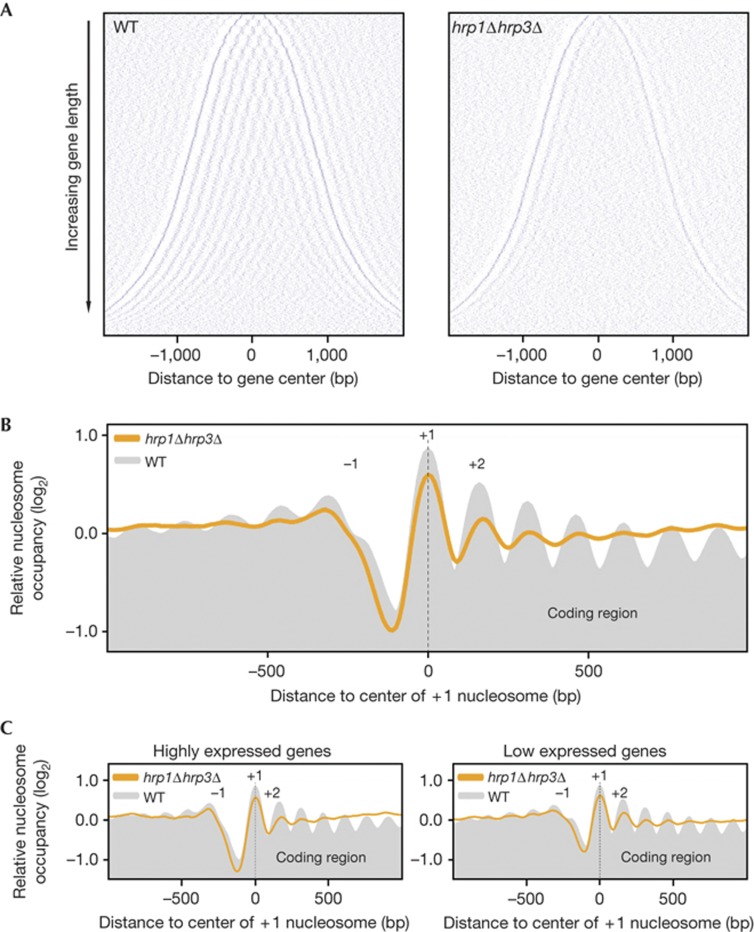
As ATP-dependent chromatin remodelling enzymes are known to have a major role in the correct positioning of the nucleosomes, we decided to map nucleosome positions in WT and Chd1-deficient strains. After a mild crosslinking and MNase treatment, we isolated and labelled the DNA from the mono-nucleosome fraction, and hybridized it to DNA microarrays covering 75% of the S. pombe genome at 10-bp resolution. Normalized hybridization signals were then used to determine the relative nucleosome occupancy levels. The lack of the linker histone H1 is compensated by very short linker DNA segments in S. pombe, which explains the median nucleosome distance of 150 bp in the WT dataset, and it is in agreement with previously published results [22] . WT cells show the classical nucleosome organization pattern seen in S. cerevisiae and in higher eukaryotes: a pronounced NFR upstream of the transcriptional start site, flanked by the highly positioned +1 and −1 nucleosomes (Fig 3B; supplementary Fig S2A online), and an organized nucleosome array in the coding regions. This structure is particularly evident on the two-dimensional plot (Fig 3A), which reveals a remarkably regular nucleosome organization pattern in the entire coding region of the genes. In contrast, the Chd1-deficient strain showed disorganized nucleosome arrays in coding regions (Fig 3A,B). This effect is not the result of a significantly decreased number of nucleosomes, as the quantity of nucleosomes detected is only mildly reduced (−8%) compared with WT. The composite plot reveals that the organization of the NFR and the surrounding +1 and −1 nucleosomes are similar to WT, but the subsequent peaks in the coding regions quickly disappear from the plot (Fig 3B). The resulting plateau in the composite plot is due to irregularly placed nucleosomes, which do not adhere to a position trend in the averaged dataset. We also detected a 10-bp increase in the median distance between nucleosomes. When nucleosome spacing is irregular, nucleosomes generally occupy a slightly longer DNA segment. A 10-bp increase in the average nucleosome distance would lead to the loss of roughly one nucleosome per gene for genes of average length. Extrapolating these numbers to the S. pombe genome would yield a ∼6–8% loss in nucleosomes, which is compatible with our bulk H3 quantification results in this mutant. Our expression profiling experiments in the Chd1-deficient strain showed that the lack of an organized nucleosome structure in coding regions has only a minor effect on gene expression but results in significantly elevated cryptic transcription activity from coding regions. We also carried out nucleosome mapping in the hrp1Δ and hrp3Δ single-deletion strains (supplementary Fig S2B,C online). We could not detect a remarkable change in nucleosome organization in the hrp1Δ strain compared to the WT. In contrast, hrp3Δ showed significantly disturbed nucleosome organization in coding regions, although this was less severe than in the hrp1Δhrp3Δ double-deletion strain. This suggests that the two Chd1-type chromatin remodeler enzymes, Hrp1 and Hrp3, have redundant functions in nucleosome organization in S. pombe.
Figure 3.
The regularly organized nucleosome arrays in gene coding regions are disrupted in the hrp1Δhrp3Δ strain. (A) Two-dimensional plots of nucleosomes along 3,778 genes in WT and hrp1Δhrp3Δ strains. Positions of the first and last nucleosomes were determined for each gene in an independent experiment with the WT strain. These annotations were used for the analysis of all data sets. Each row represents a gene; genes were sorted vertically (shortest at the top and longest genes at the bottom) according to the distance between the first and last nucleosome of the gene and were aligned at the mid point. Blue dots correspond to the centre of identified nucleosomes. (B) Composite plots of relative nucleosome occupancy for the WT (grey shading) and the hrp1Δhrp3Δ mutant (orange line). Using the same annotation from the independent WT data set, 3,778 genes were aligned at their first annotated nucleosome and the average of their log2 nucleosome occupancy data (geometric mean) was plotted. (C) The genes represented in (B) were sorted according to gene expression levels, and composite plots were generated for the highest expressed 25% of genes (left) and the lowest expressed 25% of genes (right). bp, base pairs; WT, wild-type.
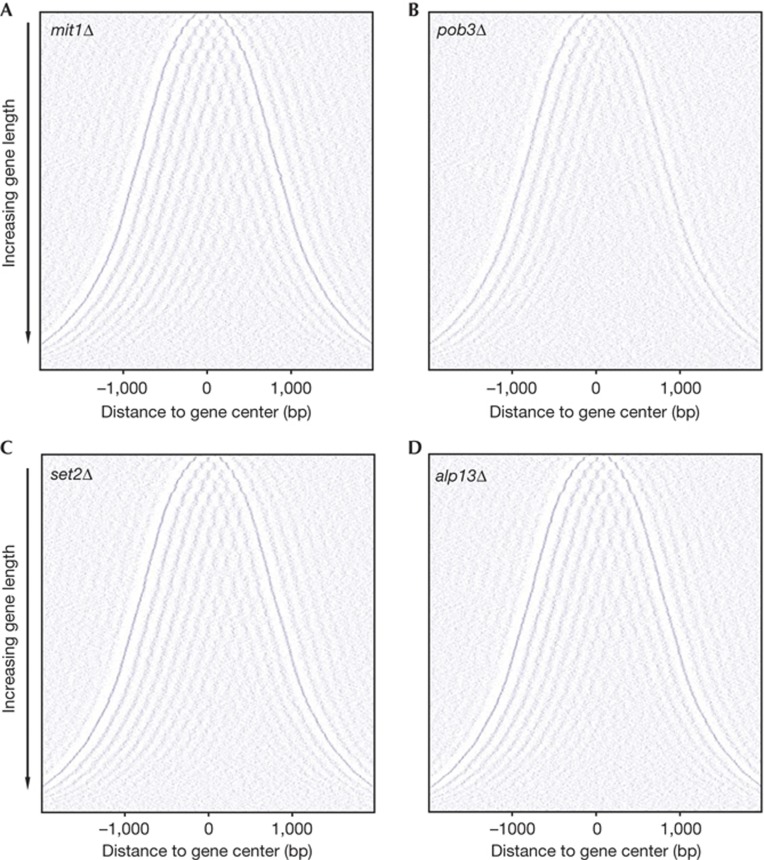
Next, we analysed the effects of transcription levels on nucleosome organization in WT and Chd1-deficient strains. We sorted genes according to their expression levels, and plotted composite plots for the highest- or lowest-25% expressed genes (highly or lowly expressed genes) (Fig 3C). Both groups of genes were equally affected in the hrp1Δhrp3Δ strain, resulting in irregularly organized nucleosomes in the coding regions. We concluded that the activity of Hrp1 and Hrp3 is essential in the establishment of regularly spaced nucleosome arrays in coding regions independently of the transcriptional status of the gene. A recent study in S. cerevisiae found a similar role for Chd1 together with the ISWI chromatin remodelers Isw1 and Isw2. S. pombe does not posses ISWI chromatin remodelers, but two Chd1 remodelers. Considering that S. pombe and S. cerevisae are evolutionarily very distant species, these results indicate an evolutionarily conserved role for Chd1 chromatin remodelers in the establishment of regularly spaced nucleosome structure in gene coding regions. Mit1, a member of the Chd3–Chd4 subfamily of chromatin remodelers, was also reported to function in the nucleosome organization of euchromatic gene coding regions [22] . Although we cannot exclude a minor effect, we did not observe significant changes in the chromatin structure of gene coding regions in mit1Δ (Fig 4A; supplementary Fig S3A online). This underlines the specific role of the Chd1chromatin remodelers in nucleosome organization in gene coding regions.
Figure 4.
Two-dimensional plots of nucleosomes along 3,778 genes in the indicated mutant strains. Nucleosome positioning is unaffected in mit1Δ (A), set2Δ (C) and alp13Δ (D) strains, and is moderately affected in the pob3Δ strain (B). See Figure legend 3A for details. bp, base pairs.
Nucleosome arrays are mostly unaffected in pob3Δ
Mutations in the FACT complex can lead to significantly decreased histone levels and a concomitant reduction of genome-wide nucleosome occupancy [14, 21] . Our western blot analysis confirmed substantial nucleosome depletion in the FACT complex subunit deletion strain, pob3Δ (Fig 2A). Nucleosome mapping experiments in pob3Δ showed that substantially decreased nucleosome occupancy had only a modest effect on nucleosome positions in gene coding regions (Fig 4B; supplementary Fig S3B online). We detected a mild, 10-bp increase in median nucleosome distance in gene coding regions, but the regularly organized nucleosome pattern remained mostly intact in the pob3Δ strain. Similar conclusions were presented by Celona et al [21] in a study investigating the deletion of the FACT complex subunit Nhp6 in S. cerevisiae and its mammalian homologue, Hmgb1.
This result also suggests that the marked loss of regularly positioned nucleosomes in hrp1Δhrp3Δ is not a result of a reduction in the detection sensitivity of nucleosome positions due to decreased nucleosome occupancy levels. Nucleosome occupancy is not, or only mildly, reduced in the hrp1Δhrp3Δ strain, but the normal nucleosome positioning is diminished. In contrast, the pob3Δ strain exhibits strong nucleosome depletion, but only mildly irregular positioning of nucleosomes. Overall, these data indicate that the main role of the Chd1-type chromatin remodelers is to rearrange randomly positioned nucleosomes to equally spaced nucleosome arrays in gene coding regions.
Nucleosome arrays in alp13Δ and set2Δ are unaffected
The AS transcript accumulation profile in the hrp1Δhrp3Δ strain is very similar to those of the set2Δ or alp13Δ strains. We ruled out the possibility that Hrp1 and Hrp3 function upstream of the Clr6 HDAC complex, as hrp1Δhrp3Δ did not show increased histone acetylation. An alternative possibility is that Hrp1 and Hrp3 function downstream of Set2 and the Clr6 complex II. In this hypothesis, set2Δ and/or alp13Δ strains would also show irregular nucleosome spacing in gene coding regions, similar to the Chd1-deficient strain. We could not detect significant changes in the nucleosome organization of these mutant strains compared to the WT. Two-dimensional plots (Fig 4C,D) and composite plots (supplementary Fig S3C,D online) of set2Δ and alp13Δ showed regularly positioned nucleosomes in coding regions. Although cryptic promoters are derepressed in these mutants, the nucleosome structure and occupancy are not affected. These results also indicate that Chd1 remodelers repress cryptic transcription independently of Set2 and the Clr6 complex II.
Our study shows that there are several parallel mechanisms responsible for the maintenance of a repressive chromatin environment in gene coding regions. Disturbing any of these mechanisms results in activation of cryptic promoter sequences and accumulation of cryptic, non-coding RNAs. We showed that in the Chd1-deficient strain, nucleosomes are irregularly positioned in gene coding regions. We propose that the lack of organized nucleosome arrays can result in areas of nucleosome-free DNA, which can expose cryptic promoter sequences and lead to cryptic transcription. Other mutations, such as the deletion of the FACT complex subunit pob3, are associated with decreased nucleosome occupancy. Nucleosome positions are mostly unaffected in these mutants, but the time that these positions remain unoccupied is significantly longer. This temporarily allows transcription initiation events from exposed cryptic promoters. Interestingly, our study shows that deletion of the HDAC subunit alp13 does not affect nucleosome positioning or occupancy, although cryptic transcription is derepressed. We speculate that in alp13Δ, the increased histone acetylation in coding regions recruits bromodomain-containing chromatin remodelling factors, such as the RSC complex, which might temporarily establish NFRs, thereby exposing cryptic promoter sequences. Nucleosome occupancy and positions were also unaffected in the set2Δ strain; however, levels of acetylation were significantly lower than in the alp13Δ strain. This suggests that there might be more, unknown mechanisms that are important in the repression of cryptic promoters in gene coding regions, and in the maintenance of genomic integrity.
Methods
See supplementary Table S1 online for a list of strains used in this study. Microarrays used for gene expression profiling contain alternating probes for both the forward and reverse DNA strand. This allows us to distinguish between sense and AS transcripts.
Western blot and ChIP experiments were performed using antibodies against histone H3 (ab1791, Abcam), H3K9/K14ac (pAB-005-044, Diagenode) and actin (MAB1501R, Millipore). All microarray experiments were repeated at least two times, with the exception of nucleosome mapping experiments with no detectable difference compared to the WT (alp13Δ, set2Δ and mit1Δ) and the single deletions hrp1Δ and hrp3Δ. Full methods and any associated references are available in the supplementary methods online.
Accession codes . The microarray data from this publication have been submitted to the Gene Expression Omnibus (GEO) database and assigned the following accession number: GSE40872.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We would like to thank Jutta Worsch for her excellent technical assistance and Emmalene Bartlett for proof reading the manuscript. This work was supported by a grant from the Ministry of Science, Research and the Arts of Baden-Wuerttemberg. Y.Z. was supported by China Scholarship Council (CSC).
Author contributions: B.P.H. and T.F. designed the experiments, analysed the data and wrote the manuscript. B.P.H., K.B. and Y.Z. performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Jiang C, Pugh BF (2009) Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 3: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 5: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC et al. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive rpd3 complex. Cell 4: 593–605 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ et al. (2005) Histone H3 methylation by set2 directs deacetylation of coding regions by rpd3s to suppress spurious intragenic transcription. Cell 4: 581–592 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Xiao G, Noma K, Malikzay A, Bjerling P, Ekwall K, Kobayashi R, Grewal SI (2003) Alp13, an MRG family protein, is a component of fission yeast clr6 histone deacetylase required for genomic integrity. EMBO J 11: 2776–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI (2007) Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol 5: 372–380 [DOI] [PubMed] [Google Scholar]
- Buratowski S, Kim T (2010) The role of cotranscriptional histone methylations. Cold Spring Harb Symp Quant Biol 75: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG (2010) Phosphorylated Pol II CTD recruits multiple hdacs, including rpd3c(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 2: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 5636: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2003) The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 22: 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M (2009) Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol Cell 3: 377–383 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F (2008) Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 11: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI (2011) Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell 1: 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M-C (2012) The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem 3: 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA (2010) Histone H3K4 and K36 methylation, chd1 and rpd3s oppose the functions of Saccharomyces cerevisiae spt4-spt5 in transcription. Genetics 2: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DE, Stokes DG, Perry RP (1999) CHD1 interacts with SSRP1 and depends on both its chromodomain and its atpase/helicase-like domain for proper association with chromatin. Chromosoma 1: 10–25 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 20: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T (2011) A role for snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 6050: 1758–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Paik J, Verreault A (2006) The emergence of regulated histone proteolysis. Curr Opin Genet Dev 2: 112–118 [DOI] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK (2010) Elevated histone expression promotes life span extension. Mol Cell 5: 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celona B et al. (2011) Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol 6: e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Strålfors A, Yuan GC, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 2: 251–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.