Abstract
Enveloped viruses employ diverse and complex strategies for wrapping at cellular membranes, many of which are poorly understood. Here, an ultrastructural study of herpes simplex virus 1 (HSV1)-infected cells revealed envelopment in tubular membranes. These tubules were labelled by the fluid phase marker horseradish peroxidase (HRP), and were observed to wrap capsids as early as 2 min after HRP addition, indicating that the envelope had recently cycled from the cell surface. Consistent with this, capsids did not colocalise with either the trans-Golgi network marker TGN46 or late endosomal markers, but showed coincidence with the transferrin receptor. Virus glycoproteins were retrieved from the plasma membrane (PM) to label wrapping capsids, a process that was dependent on both dynamin and Rab5. Combined depletion of Rab5 and Rab11 reduced virus yield to <1%, resulting in aberrant localisation of capsids. These results suggest that endocytosis from the PM into endocytic tubules provides the main source of membrane for HSV1, and reveal a new mechanism for virus exploitation of the endocytic pathway.
Keywords: endocytosis, HSV1, Rab11, Rab5, trans-Golgi network
Introduction
Enveloped viruses exploit cellular transport pathways in their assembly and egress from the cell, and have been used to reveal fundamental aspects of cellular trafficking. Many, such as human immunodeficiency virus (HIV) and influenza virus, acquire their envelope by budding at the plasma membrane (PM) (Nayak et al, 2004; Jouvenet et al, 2006), a process that involves an interaction of virus matrix proteins with the cytoplasmic tails of envelope glycoproteins (Cosson, 1996; Barman et al, 2001; Takimoto and Portner, 2004). Others, such as hepatitis B and C viruses, are believed to acquire their envelopes by budding at internal pre-Golgi membranes where virus glycoproteins are concentrated, followed by transport through the lumen of the secretory pathway (Bruss, 2004, 2007; Roingeard et al, 2004, 2008). Larger enveloped viruses exhibit more complex processes of envelopment and egress. For example, African swine fever virus has been proposed to acquire an inner envelope from the endoplasmic reticulum (ER) and an outer envelope from the PM (Breese and DeBoer, 1966; Andres et al, 1998). The poxvirus vaccinia virus produces a triple membrane wrapped core that is exposed onto the cell surface by exocytosis and fusion of the outermost membrane with the PM (Roberts and Smith, 2008). While the source of the inner membrane is disputed, the outer two membranes have been proposed to derive from the trans-Golgi network (TGN) or endosomes (Tooze et al, 1993; Schmelz et al, 1994).
Herpes simplex virus type 1 (HSV1) is a large enveloped virus of ∼200 nm in diameter that is actively released from the cell. The HSV1 capsid is surrounded by a complex known as the tegument comprising over 20 virus-encoded proteins (Dargin, 1986; Loret et al, 2008) that is believed to link the capsid to the envelope (Mettenleiter, 2006). The complexity of the HSV1 particle and a degree of redundancy among its proteins means that the assembly pathway has been difficult to define at the molecular level, while the cell biology of envelopment has proved controversial. Unlike other large DNA viruses, genome replication and capsid assembly occur in the nucleus, and early models of morphogenesis described the inner nuclear membrane as the site of virus envelopment, with intact particles suggested to move through the secretory pathway for release at the cell surface (Nii et al, 1968; Ben-Porat and Kaplan, 1972). However, the current widely accepted morphogenesis model, termed as the envelopment-deenvelopment-reenvelopment model (Mettenleiter et al, 2006; Johnson and Baines, 2011), designates this nuclear membrane-derived envelope as a primary envelope, which is lost by fusion with the outer nuclear membrane, releasing naked capsids into the cytosol (Mettenleiter, 2006). The source of the final envelope for HSV1 has been the subject of much debate (Leuzinger et al, 2005; Mettenleiter and Minson, 2006), with a number of studies implicating multivesicular body (MVB) components in the process (Calistri et al, 2007; Crump et al, 2007; Pawliczek and Crump, 2009). However, within the relevant literature the site of HSV1 envelopment is now commonly cited to be the TGN (Johnson and Baines, 2011).
The TGN is an organelle that comprises up to three cisternae on the trans side of the Golgi apparatus, all of which are able to bud and extend tubules (De Matteis and Luini, 2008; Luini et al, 2008; Pfeffer, 2011). Proteins destined for different locations, such as late endosomes or PM, are believed to sort into different domains, and hence different tubules, of the TGN (Bard and Malhotra, 2006; Luini et al, 2008). A major marker for the TGN is the protein TGN46 and although a small proportion of TGN46 escapes to the PM followed by retrieval back through sorting endosomes, the slow rate of TGN46 exit from the TGN means that at any one time around 90% of the cellular content of this protein is located in the TGN membranes (Ponnambalam et al, 1996; Ghosh et al, 1998). The main evidence for HSV1 wrapping at this compartment comes from light microscopy showing GFP-tagged capsids close to membranes labelled positive for TGN46, with the inference that wrapping occurs at these membranes (Turcotte et al, 2005; Sugimoto et al, 2008; Remillard-Labrosse et al, 2009). The colocalisation of all glycoproteins tested with the TGN46 marker was taken as further evidence for wrapping at these membranes (Turcotte et al, 2005). In other studies investigating glycoprotein E (gE) localisation, TGN46 was seen to relocalise to the lateral membranes of HSV1-infected polarised epithelial cells together with gE, an event that happened in the absence of capsid production (McMillan and Johnson, 2001; Wisner and Johnson, 2004). Furthermore, isolated capsid-containing cellular organelles have been shown to co-stain for TGN46 by immunofluorescence (Lee et al, 2006). However, while one study has detected TGN46 in the membranes around capsids in infected human fetal axons by immunogold labelling (Miranda-Saksena et al, 2009), none of these studies have directly correlated capsid/TGN46 proximity with the presence of enveloped, infectious virus at the ultrastructural level.
Understanding virus morphogenesis and the wide range of envelopment strategies used by viruses provides important insight into fundamental aspects of cellular trafficking. In this study, we have combined detailed confocal microscopy, ultrastructural analysis, and inhibition of the endocytic pathway to show that HSV1 acquires its envelope not from the TGN or other membranes of the secretory pathway, but from endocytic tubules containing glycoproteins derived from the PM. These membranes were shown to enwrap capsids as early as 2 min after endocytosis, and inhibition of the endocytic pathway by treatment with the dynamin inhibitor dynasore or depletion of Rab5 resulted in retention of envelope glycoproteins at the cell surface and inhibition of capsid wrapping. Combined depletion of Rab11 and Rab5 further reduced virus yield. Hence, we have shown that HSV1 utilises membranes from the endocytic pathway for envelopment, revealing a new strategy for virus exploitation of cellular trafficking pathways.
Results
HSV1 capsids are wrapped in cytoplasmic tubular membranes
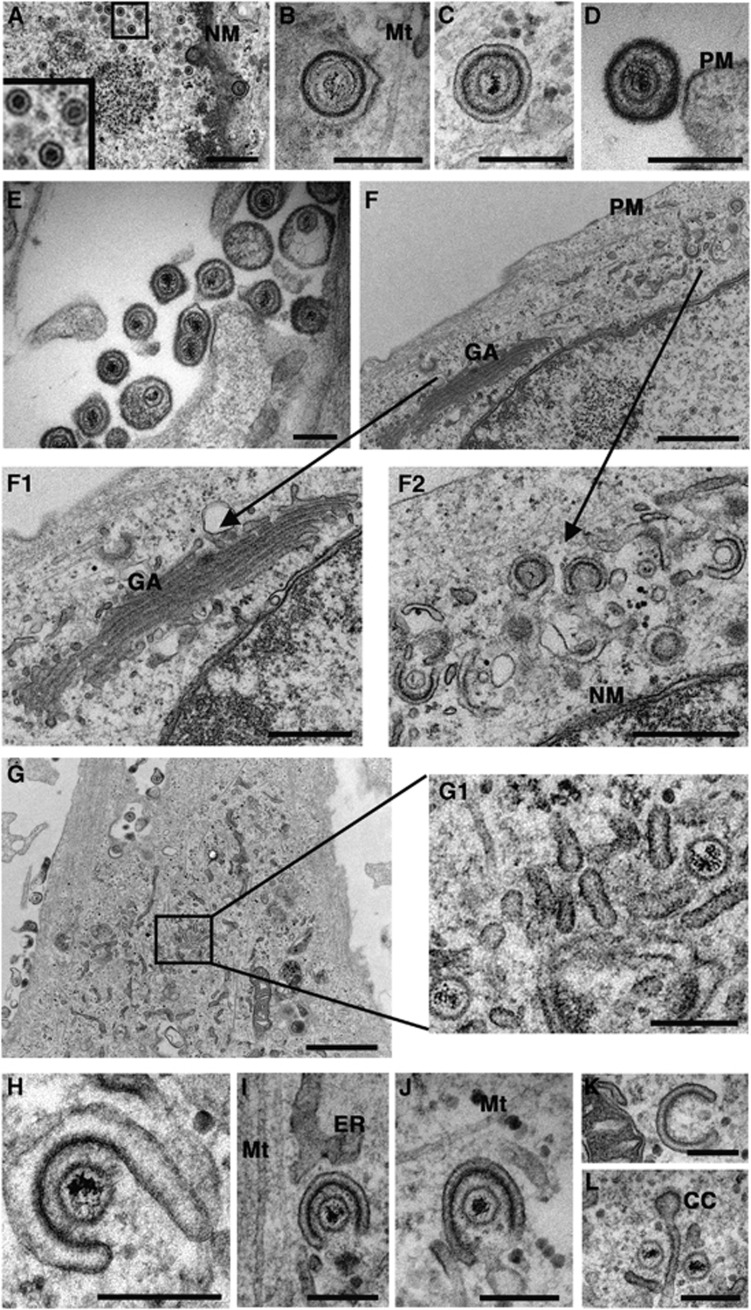
Ultrastructural studies of HSV1 were carried out in the non-transformed primary fibroblast cell line HFFF-2. HSV1 (strain 17) exhibited similar one-step replication kinetics in HFFF-2 cells compared to Vero and Hela cells—lines commonly used in HSV1 studies—with a 1000-fold increase in virus production between 6 and 12 h after infection (Supplementary Figure S1A). Furthermore, HSV1 strains s17, sc16, and HFEM all exhibited the same replication kinetics in HFFF-2 cells (Supplementary Figure S1B). Fixation of infected HFFF-2 cells for electron microscopy (EM) at 12 h revealed that all strains gave the same profiles for HSV1 morphogenesis—assembling capsids were present in the nucleus (Figure 1A); primary enveloped virions were detectable in the perinuclear space between the inner and outer nuclear membrane (Figure 1B); mature enveloped particles were observed enclosed in double membranes within the cytoplasm (Figure 1C); extracellular virions found on the cell surface were wrapped in a single membrane (Figure 1D) and some contained more than one capsid (Figure 1E). Wrapped and wrapping virions were found at a range of locations in the cytoplasm, but there were few obvious wrapping events at the trans cisternae of any Golgi complex examined (e.g., Figure 1F).
Figure 1.
HSV1 capsids are wrapped in tubular membranes in HFFF-2 cells. HFFF-2 cells were infected at a multiplicity of 2 with HSV1, fixed and processed 12 h after infection, and imaged by EM. Panels F1/F2, and G2 are expanded regions of panels F and G respectively. NM, nuclear membrane; GA, Golgi apparatus; PM, plasma membrane; Mt, microtubule; Ca, capsid; ER, endoplasmic reticulum; CC, clathrin coat. In (A, F1, F2) scale bar=500 nm. In (B–E, G1, H–L), scale bar=200 nm. In (F, G), scale bar=1 μm.
Large numbers of free capsids were found clustered together with tubular membranes at the periphery of cells (Figure 1G). Numerous examples of capsids in the process of envelopment were found enwrapped by curved tubules with an electron dense concave surface that we believe to be tegument (Figure 1H–J). Frequently, these wrapping profiles were close to microtubules (Figure 1I and J). Furthermore, similar curved tubular profiles lacking capsids were also found suggesting that the formation of these curved membranes may not be dependent on the presence of a capsid (Figure 1K). Strikingly, in some fields there were capsids clustered with tubules that carried a curved clathrin-like coat at one end (Figure 1L).
Wrapping of HSV1 capsids in PM-derived tubules
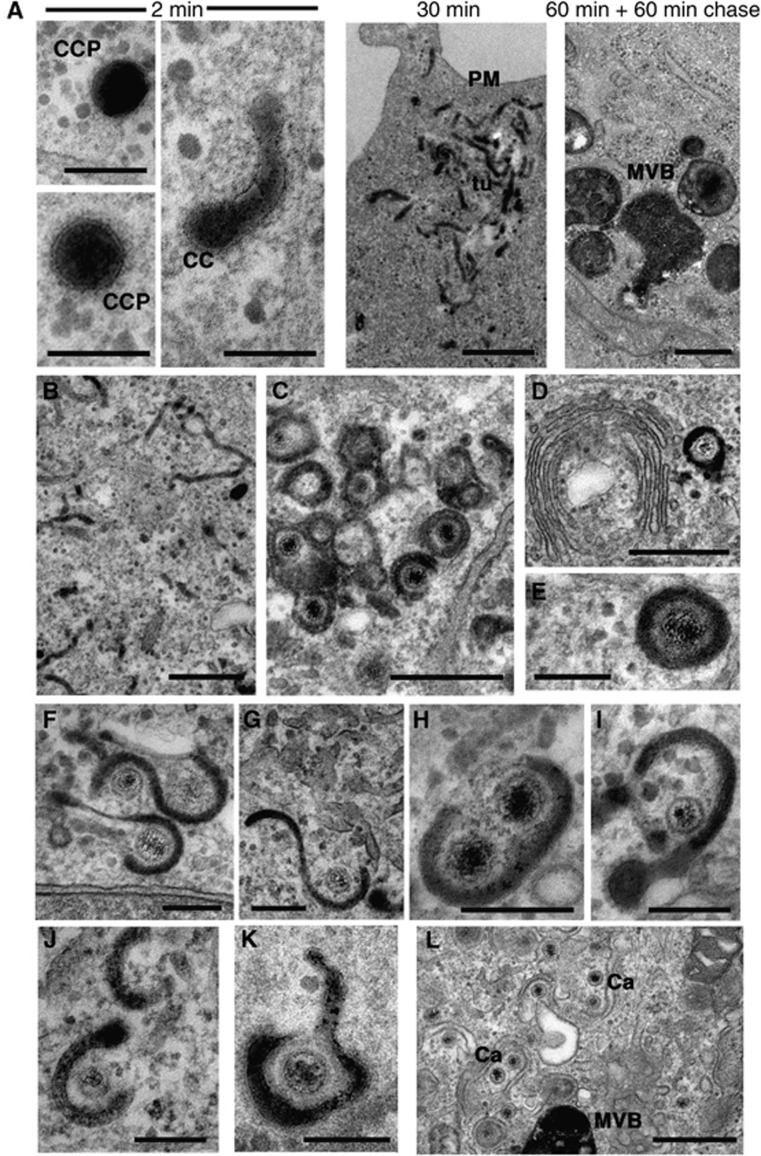
Curved clathrin coats as seen on tubules by EM are commonly found on vesicles budding from the PM, which are destined for the early endosome, or tubules budding off the TGN, which are destined for the late endosome (Bard and Malhotra, 2006; Doherty and McMahon, 2009). Additionally, flat clathrin coats have been described on tubules budding from the sorting endosome complex in a retromer-dependent fashion for transport back to the TGN (Bonifacino and Hurley, 2008). A less well-reported location of a curved clathrin coat is on tubules leaving the recycling endosome destined for the cell surface (van Dam and Stoorvogel, 2002; van Dam et al, 2002). To distinguish endocytic tubules from those that may originate from the TGN, cells were incubated with horseradish peroxidase (HRP) to label fluid phase endocytic events. Following endocytosis, HRP is sorted into MVBs at the early endosome, with some recycling back to the cell surface via recycling endosomes (Gonatas et al, 1980; Griffiths et al, 1989; Gu et al, 1997; D’Souza-Schorey et al, 1998). With the exception of specialised secretory cells (Orci et al, 1986), retrograde trafficking of HRP to the TGN is generally undetectable and therefore any HRP-positive clathrin coat containing tubules would be unlikely to have originated from the TGN. To first test the ability of HFFF-2 cells to take up HRP, uninfected cells were incubated with 10 mg/ml HRP in the cell medium for various times ranging from 2 to 60 min, or 60 min followed by a 60-min chase with HRP-free medium. Samples were stained with DAB after fixation to identify HRP-positive lumina within the cell. After 2 min, HRP was shown to label clathrin-coated pits at the cell surface (Figures 2A, 2 min, CCP) and some tubules carrying a clathrin coat (Figure 2A, 2 min, CC). As incubation time progressed, clusters of tubules were evident at the cell periphery (Figure 2A, 30 min, tu) and in the centre of the cell (data not shown). After a 60-min chase HRP was chased into characteristic MVBs (Figure 2A, MVB) as shown by others (Gonatas et al, 1980). To next determine if any of these HRP-containing structures provided the wrapping membranes of HSV1, infected cells were initially incubated with HRP continuously for 60 min. The same tubular structures detected in uninfected cells were also present in infected cells (Figure 2B). Moreover, the tubules wrapping around capsids in these infected cells were positive for HRP (Figure 2C). Analysis of higher magnification images revealed a range of HRP-positive wrapping profiles (Figure 2D–I), including examples of two capsids wrapping in a single membrane (Figure 2H). Wrapping profiles where a clathrin coat was present on the wrapping tubule were found with ease (Figure 2I). In agreement with other studies (Gonatas et al, 1980; Griffiths et al, 1989) none of the trans lumina of the Golgi cisternae or emanating tubules on the trans side of any Golgi complex were labelled with HRP, even when capsids wrapped in highly HRP-positive membrane were observed in close proximity to the Golgi (Figure 2D). Furthermore, HRP labelling experiments in Vero and HeLa cells (Supplementary Figure S2) showed that these cells also contained HRP-positive tubules that were seen to enwrap cytoplasmic capsids. Hence, the wrapping of HSV1 in endocytic membranes demonstrated in HFFF-2 cells appears to be a general mechanism for HSV1 maturation and is not cell-type specific.
Figure 2.
HSV1 capsids are wrapped by tubules derived from the plasma membrane. (A) Uninfected HFFF-2 cells were labelled for 2 or 30 min with 10 mg/ml HRP, or 60 min followed by 60 min chase. After fixation, cells were stained with DAB and processed for imaging by EM. (B–J) HFFF-2 cells were infected at a multiplicity of 2 with HSV1 and labelled 11 h after infection with 10 mg/ml HRP for 60 min (B–I), 10 min (J), 2 min (K) or 60 min followed by 60 min chase (L). PM, plasma membrane; ER, endoplasmic reticulum; NM, nuclear membrane; GA, Golgi apparatus; MVB, multivesicular body; Mt, microtubule; CC, clathrin coat; CCP, clathrin-coated pit; Tu, HRP-positive tubule. In (A) (2 min) and (E–K), scale bar=200 nm. In (A) (30 and 60 min+60 min chase) and (B–D, L), scale bar=500 nm.
HRP is known to accumulate in early structures for 5–10 min after addition and in pulse-chase experiments to progress to MVBs within 2 h (Oliver et al, 1989; Gu et al, 1997). To correlate virus wrapping in HRP-positive tubules with these kinetics, infected HFFF-2 cells were labelled with HRP for 2, 10, 30, or 60 min before fixation and processing. In each case, 100 capsids were scored for the presence of an HRP-positive lumen (Table I). After as little as 2 min labelling, 1 in 50 capsids was already wrapped in HRP-positive membranes (Figure 2K) with over 25% scoring positive by 10 min (Figure 2J), and by 30 min numbers had reached nearly 90%. Considering these HRP-positive wrapping membranes had to not only undergo endocytosis but also encounter a capsid to enwrap in the timescale of these incubations, these results suggest that the membranes wrapping HSV1 capsids are recently derived from the PM.
Table 1. Incorporation of HRP into HSV1 wrapping membranes.
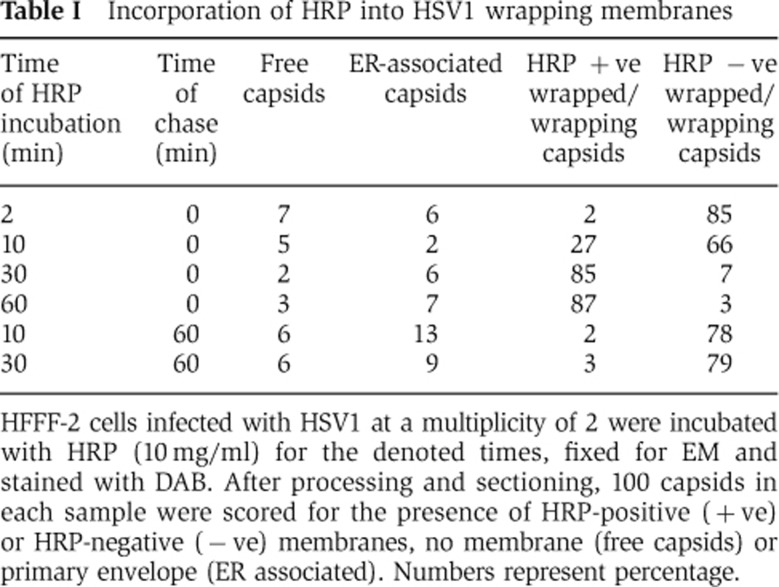
HFFF-2 cells infected with HSV1 at a multiplicity of 2 were incubated with HRP (10 mg/ml) for the denoted times, fixed for EM and stained with DAB. After processing and sectioning, 100 capsids in each sample were scored for the presence of HRP-positive (+ve) or HRP-negative (−ve) membranes, no membrane (free capsids) or primary envelope (ER associated). Numbers represent percentage.
As previous studies have implicated MVB membranes in HSV1 envelopment (Calistri et al, 2007), we used HRP uptake pulse-chase experiments (Figure 2A) to examine HSV wrapping under conditions where only MVB membranes were positive for HRP. Infected HFFF-2 cells were labelled with HRP for 10 or 30 min followed by a 60-min incubation in media lacking HRP, and capsids scored for the presence of HRP-positive membranes. The 60-min chase resulted in a reduction of capsids associated with HRP-positive membranes from 27 and 85% in the 10 and 30 min incubations, respectively, to around 2% after the chase step (Table I; Figure 2L). These results indicate that chasing HRP out of early endocytic membranes into late MVB structures results in the vast majority of wrapping membranes being HRP negative, suggesting that MVB membranes are unlikely to be the source of HSV1 envelope in HFFF-2 cells. Furthermore, immunogold labelling of the late endosomal marker LAMP2 demonstrated that although these structures were readily detectable by EM and were heavily labelled with gold particles, capsids were never seen in proximity to these structures, and membranes wrapping capsids were not LAMP2 positive (Supplementary Figure S3).
HSV1 capsids do not colocalise with TGN46-positive membranes in any of three cell lines
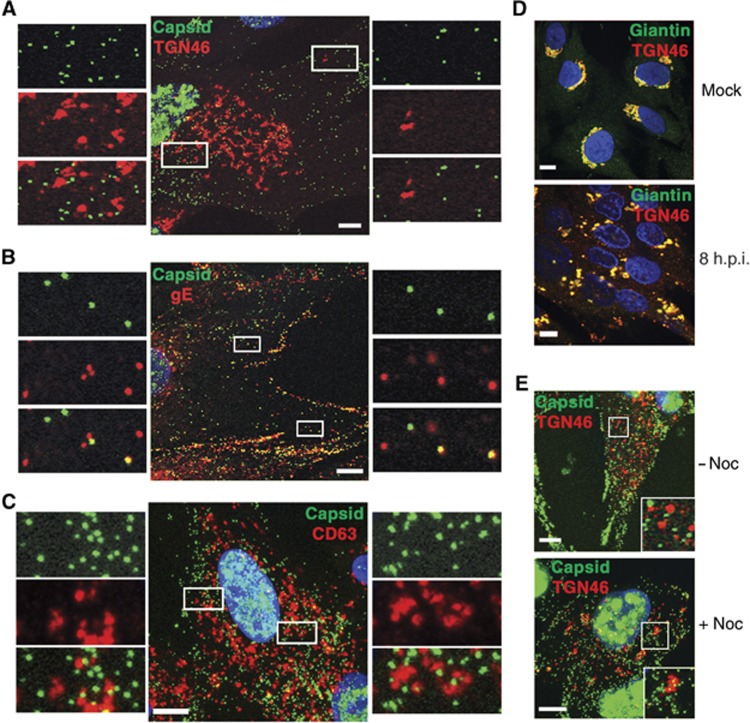
These results were contrary to the accepted model of HSV1 wrapping at the TGN (Johnson and Baines, 2011). In the previous publications that reported the TGN as the site of HSV1 capsid envelopment, an antibody to the cellular antigen TGN46 was used to define the membranes of the TGN (Turcotte et al, 2005; Sugimoto et al, 2008; Remillard-Labrosse et al, 2009). In light of our results above, HFFF-2 cells were infected with HSV1 expressing the capsid protein VP26 as a GFP-fusion protein (GFP-VP26 virus) and examined 8 h after infection by co-staining for either TGN46 or the envelope glycoprotein gE. Co-staining of the MVB marker CD63 was used to compare the relative localisation of capsids to late endosomal membranes. Confocal microscopy revealed that there was little colocalisation between capsids and TGN46 either in the area of the Golgi or at the cell periphery (Figure 3A). In addition, immunogold labelling of TGN46 indicated that although the antibody detected trans cisternae of the Golgi stacks in uninfected cells, this positivity was reduced in late infected cells, and TGN46 was not detected in the PM or membranes wrapping capsids (Supplementary Figure S4). By contrast, a large number of individual capsids colocalised with gE throughout the cytoplasm confirming at least a proportion were enveloped (Figure 3B). These light microscopy images correlated with immunogold EM localisation of virus glycoprotein (in this case gD) where gD was detected in the Golgi apparatus, at the PM, on membranes wrapping capsids in the cytoplasm, and on extracellular virions (Supplementary Figure S5). As anticipated from our findings above, there was no obvious colocalisation between capsids and CD63 by immunofluorescence (Figure 3C). Co-staining of infected HFFF-2 cells for both TGN46 and the Golgi integral membrane protein giantin indicated that the TGN46 signal remained with the Golgi in these cells at this time, despite fragmentation of the Golgi stacks (Figure 3D), confirming that the antigen remained an appropriate marker for the TGN at least until 8 h after infection. Furthermore, because the Golgi/TGN is held close to the nucleus by its relationship with microtubules, treatment of infected cells with the microtubule depolymerising drug nocodazole resulted in increased fragmentation of the TGN46 signal. Significantly, there was no obvious clustering of capsids with TGN46 in these conditions, suggesting that capsid localisation at a perinuclear location may be more dependent on the presence of intact microtubules than the presence of the Golgi/TGN membranes (Figure 3E).
Figure 3.
HSV1 capsids colocalise with glycoprotein E but not TGN46 or CD63 in HFFF-2 cells. Cells grown on coverslips were infected with the GFP-VP26 virus (green) (A–C, E) or s17 (D) at a multiplicity of 2, and fixed 8 h after infection. Cells were stained with antibody against (A, E) TGN46 (red), (B) gE (red), (C) CD63 (red) or (D) TGN46 (red) and giantin (green), and nuclei stained with DAPI (blue) before imaging with a Zeiss LSM510 Meta confocal microscope. In (E), nocodazole was added at a concentration of 2 μg/ml for 30 min before fixing to depolymerise the microtubule network (+noc). Scale bar=10 μm.
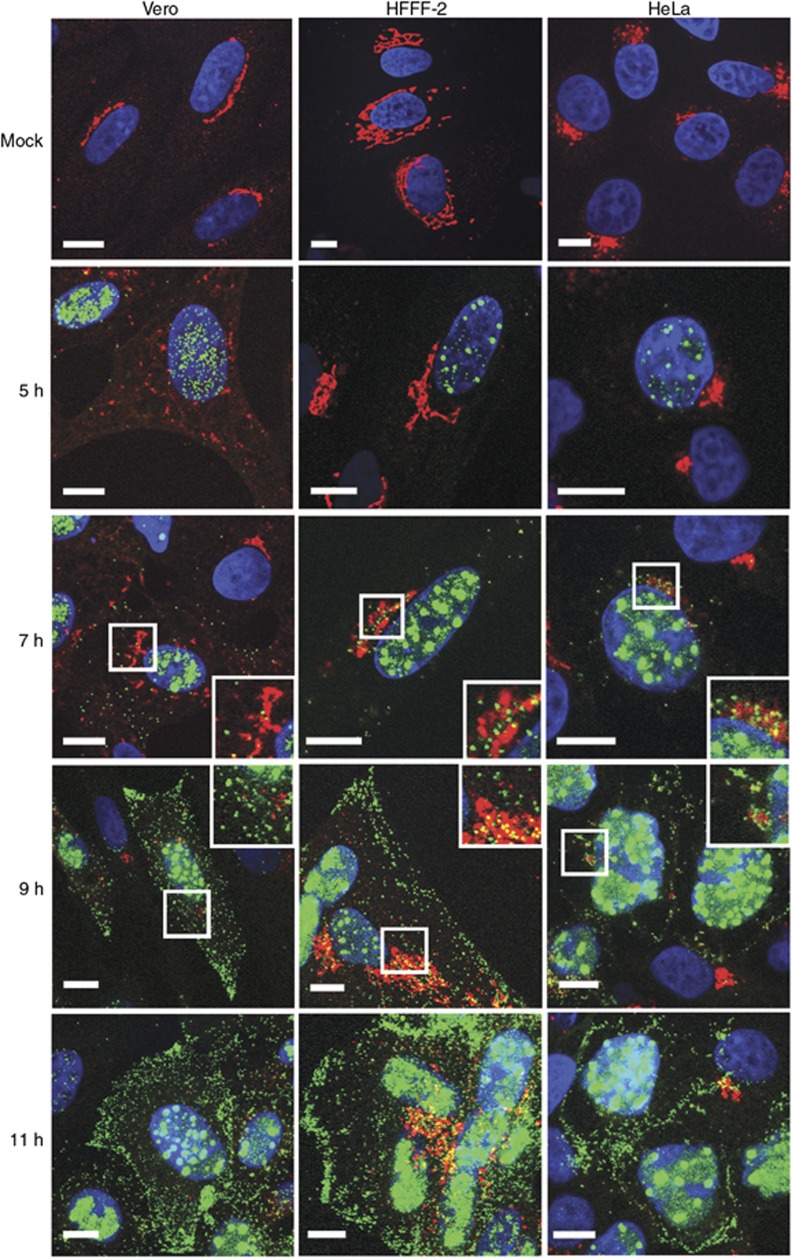
HSV1 capsid localisation in relation to the TGN46 antigen was compared at various times after infection between HFFF-2, and Vero and HeLa cells. As for HFFF-2 cells, TGN46-positive signal remained relatively intact in HeLa cells during the early part of infection (Figure 4, compare mock and 5 h). However as early as 5 h after infection, when GFP capsids were just detectable in the nucleus of all cell lines, TGN46 was dispersed through the cytoplasm of Vero cells (Figure 4, compare mock and 5 h). This dispersal was clearly not due to virion egress from the TGN as capsids were not yet in the cytoplasm at that time. Furthermore, for the rest of the time course when capsids appeared in the cytoplasm, there was no correlation of capsids and TGN46 in Vero cells. When capsids were initially detected in the cytoplasm of HFFF-2 and HeLa cells a few capsids clustered close to TGN46 labelling (Figure 4, 7 h). As cytoplasmic capsid numbers increased, there was a similar dispersal of the TGN46 signal in HeLa cells without any colocalisation (Figure 4, 9 h). Although some GFP capsids were clustered around the partially maintained TGN46 signal in HFFF-2 cells, the entire cytoplasm was full of capsids at this time making it difficult to ascertain if the proximity to TGN46 was in any way specific. At the latest time point, capsids decorated the PM in all cell lines (Figure 4, 11 h).
Figure 4.
Relative localisation of virus capsids and TGN46 in HSV1-infected Vero, HeLa, and HFFF-2 cells. Cells grown on coverslips were infected with the GFP-VP26 virus (green) at a multiplicity of 2, and fixed at 5, 7, 9, or 11 h after infection. Cells were stained for TGN46 (red) and nuclei stained with DAPI (blue) before imaging with a Zeiss LSM510 Meta confocal microscope. Scale bar=10 μm.
Glycoprotein D undergoes endocytosis to HSV1 wrapping membranes
If, as suggested by the HRP uptake studies, capsids are wrapped by tubules derived from the PM, then glycoproteins would need to be targeted to the PM prior to capsid envelopment. Studies carried out here show that in all cell lines examined, gE was obvious in cytoplasmic membranes, including a juxtanuclear location in HFFF-2 and HeLa cells, prior to capsids leaving the nucleus (Supplementary Figure S6, 5 h). By the time a small number of capsids were in the cytoplasm, gE was already located at the cell periphery of all three cell lines (Supplementary Figure S6, 7 h), suggesting that the envelope protein was transported through the secretory pathway to the PM prior to the process of capsid envelopment. As the numbers of cytoplasmic capsids increased they were directed towards the cell periphery where, in contrast to the result with TGN46, many colocalised with gE (Supplementary Figure S6, 9 and 11 h).
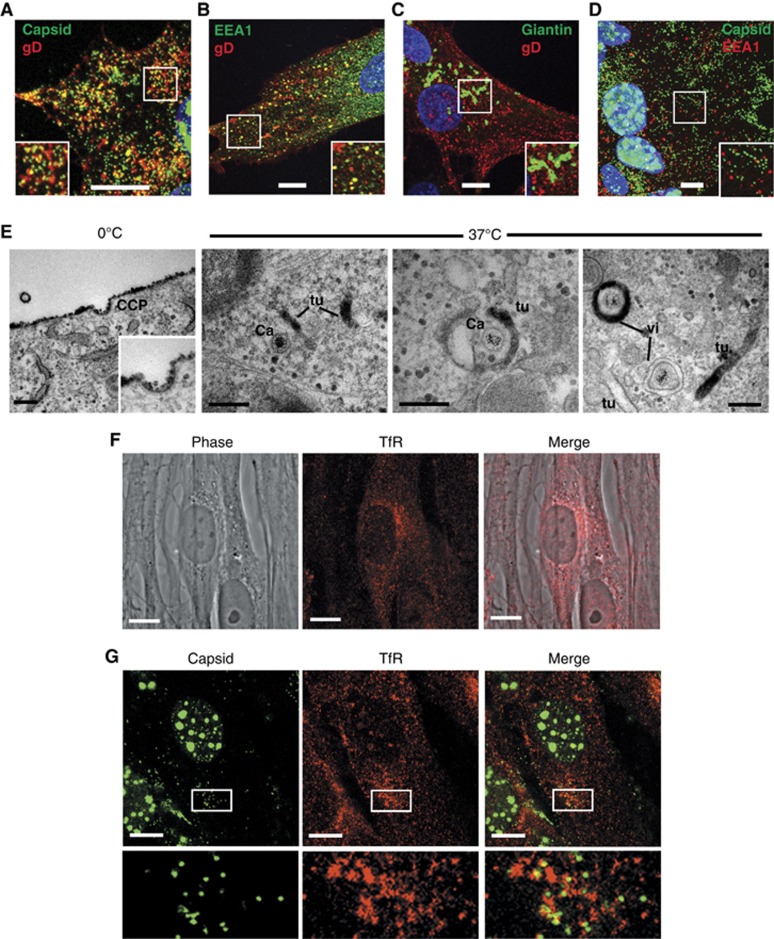
For the virus envelope to be derived by endocytosis from the PM, it would be anticipated that all glycoproteins must undergo endocytosis. While both gE and gB have been shown to contain signals for and undergo endocytosis (Alconada et al, 1999; Beitia Ortiz de Zarate et al, 2004), and gM may affect the endocytosis of other glycoproteins lacking retrieval signals by an unknown mechanism (Crump et al, 2004), we reasoned that if our model was correct then it would be possible to detect the retrieval of any glycoprotein from the PM. Hence, to examine glycoprotein endocytosis in our system, we used a monoclonal antibody available to us that recognises the extracellular domain of gD. Cells infected with the GFP-VP26 virus were incubated at 4°C with an anti-gD monoclonal antibody, and following extensive washing to remove unbound antibody, the cells were incubated for 30 min at 37°C to allow entry of the antibody into the cell via concomitant endocytosis of gD. Secondary antibody was added to permeabilised cells, and immunofluorescence revealed that while the gD antibody was located only at the cell surface in cells incubated at 4°C (data not shown), in cells incubated at 37°C, the gD antibody had been endocytosed, and localised in multiple membrane structures throughout the cytoplasm (Figure 5A). Moreover, a large number of these gD-positive membranes correlated with cytoplasmic capsids. The same experiment carried out with Wt HSV1-infected cells and stained for the early endosome marker EEA1 showed that a proportion of retrieved gD was in the early endosome while a substantial amount was elsewhere in the cytoplasm (Figure 5B). By contrast, staining for giantin to label the Golgi/TGN revealed no detectable levels of endocytosed gD population trafficking back to these membranes (Figure 5D). Interestingly, in spite of some gD localising to the early endosome (Figure 5B) capsids did not obviously colocalise with EEA1 suggesting that although the retrieved gD may traffic through the early endosome, the individual wrapping membranes were likely to be derived from structures distinct from EEA1-positive structures (Figure 5C). Furthermore, analysis of ultrathin sections of infected HFFF-2 cells by both immunofluorescence and immunogold revealed that EEA1-positive structures were very infrequent in the thickness of these sections, and thus by definition difficult to detect (Supplementary Figure S7). As wrapping capsids are highly abundant in the cytoplasm of infected cells, and do not label for EEA1, we conclude that the wrapping membranes are not continuous with EEA1-positive early endosomes.
Figure 5.
Glycoprotein D is endocytosed from the plasma membrane to HSV1 wrapping membranes. HFFF-2 cells grown on coverslips were infected with (A) the GFP-VP26 virus or (B, C) Wt HSV1 at a multiplicity of 2. Ten hours after infection, gD antibody uptake experiments were carried out by incubating cells for 30 min on ice with anti-gD monoclonal antibody followed by 30 min at 37°C to allow uptake. Cells were fixed and permeabilised then labelled with (B) anti-EEA1 (green), or (C) anti-TGN46 (green), and counterstained with Alexa 568 anti-mouse secondary antibody to detect anti-gD antibody (red) and DAPI to detect nuclei (blue). (D) HFFF-2 cells infected with the GFP-VP26 virus were fixed at 10 h and labelled with anti-EEA1 (red) and DAPI to detect nuclei (blue). (E) HFFF-2 cells were infected with HSV1 and at 10 h gD antibody uptake assay carried out as described above with the inclusion of a 30-min incubation on ice with an HRP-tagged F(ab′)2 fragment of anti-mouse IgG prior to incubation at 37°C. Cells were fixed and stained for HRP prior to processing for EM. (F) Uninfected HFFF-2 cells were labelled with anti-transferrin receptor (red). (G) HFFF-2 cells infected with GFP-VP26 virus (green) were fixed 10 h after infection and labelled with anti-transferrin receptor antibody (red). All immunofluorescence images were acquired with a Zeiss LSM510 Meta confocal microscope. In (A–D, F–G), scale bar=10 μm. In (E), scale bar=200 nm.
Glycoprotein D antibody uptake was also assessed at the ultrastructural level by including an HRP-tagged F(ab′)2 fragment of anti-mouse IgG in the uptake assay, followed by processing for EM and HRP staining as described above. After incubation at 4°C alone, the gD antibody remained bound at the cell surface and was detected in clathrin-coated pits, but following incubation at 37°C it was retrieved into endocytic tubules and membranes wrapping HSV1 capsids, confirming that gD from the PM ended up in the virus envelope (Figure 5D).
To examine the localisation of capsids in relation to membranes of the endocytic recycling pathway, we carried out immunofluorescence of HFFF-2 cells with an antibody to the transferrin receptor (TfR), a well-characterised marker for this pathway. Labelling of uninfected cells indicated that the level of TfR present in this cell type was relatively low, but that it was detectable particularly in accumulations beside the nucleus which are likely to be close to the microtubule organising centre (MTOC) (Figure 5F). In cells infected with HSV1 expressing GFP-VP26, capsids clustered in close proximity to these accumulations of TfR-labelled membranes (Figure 5G). Hence, of the four compartments that we have examined for proximity to trafficking capsids—TGN, early endosome, late endosome, and TfR-positive recycling membranes—the TfR correlated most convincingly with the cytoplasmic capsids.
Depletion of early and recycling endosome Rab GTPases inhibits virus production and wrapping
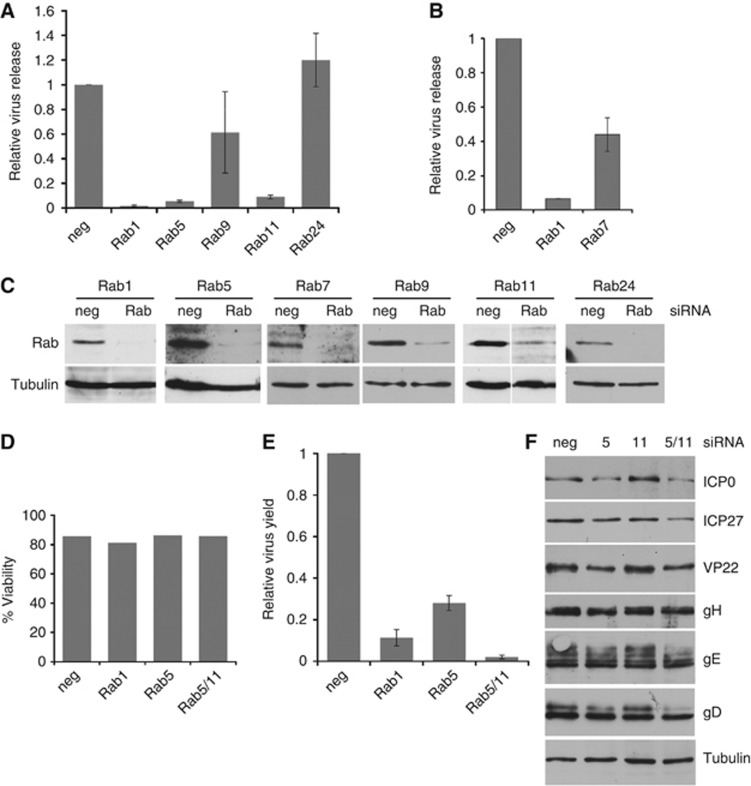
Cargo movement through the endocytic pathway is regulated at different stages by a number of Rab GTPases including Rabs 5, 7, 9, and 11 (Stenmark, 2009). To determine the role of these in HSV1 envelopment, siRNA depletion experiments were carried out in HeLa cells. Depletion of Rab1, required for ER-to-Golgi transport was included as a positive control, as it has recently been shown to be important for HSV1 glycoprotein movement through the secretory pathway (Zenner et al, 2011). Depletion of Rab24, an ER localised Rab involved in autophagosome formation (Chua et al, 2011), was also included because it was not anticipated to be involved in envelopment. In each case, siRNAs to all isoforms of individual Rabs were co-transfected to ensure all redundant forms were depleted. HeLa cells transfected with siRNAs were infected 2 days later and released virus harvested from the cell medium 16 h after infection. As shown by others, knockdown of the ER-to-Golgi regulator Rab1 reduced virus yield to <2% of the negative siRNA control (Figure 6A). By contrast, depletion of Rab24 had no effect on virus yield. Among the endosomal Rabs that were tested, depletion of Rab5 or Rab11 significantly reduced virus yield while depletion of Rab9 (Figure 6A) or Rab7 (Figure 6B) both reduced virus yield by two-fold or less. Efficient knockdown of all these proteins was demonstrated by western blotting (Figure 6C), and the viability of the siRNA-treated cells was demonstrated to be unaffected in any of the knockdowns that reduced virus yield (Figure 6D). Interestingly, Rabs 5 and 11, identified as being required for efficient virus production, are involved in trafficking from the PM to the EE and through recycling endosomes back to the PM, respectively (Bucci et al, 1992; Ullrich et al, 1996). By contrast Rabs 7 and 9, which showed limited involvement in virus production, are involved in maturation of early endosomes to MVBs and trafficking from MVBs to the TGN, respectively (Lombardi et al, 1993; Rink et al, 2005). These results provide further evidence that the late endocytic pathway is not involved in virus production.
Figure 6.
Rab GTPases 5 and 11 are required for efficient HSV1 production. (A, B) HeLa cells transfected with siRNAs as denoted were infected 48 h later with HSV1 at a multiplicity of 5, extracellular virus harvested 16 h later and titre measured. Graphs show titres in relation to those obtained for the negative siRNA (taken as 1), and values are shown as mean±standard error of the mean from three independent experiments. (C) Total cell extracts of siRNA-transfected HeLa cells were analysed by western blotting with antibodies as labelled. (D) HeLa cells transfected with siRNAs for Rab1, Rab5, or Rab11 were assessed for viability. Graph denotes viability in relation to non-transfected cells (taken as 100%). (E) HeLa cells transfected with siRNAs as denoted were infected 48 h later with HSV1 at a multiplicity of 5, and the titre of cell-associated virus was measured 16 h later. Graph shows titres in relation to those obtained for the negative siRNA (taken as 1) and values are shown as mean±standard error of the mean from three independent experiments. (F) Total cell extracts of siRNA transfected, infected HeLa cells, harvested at 16 h, were analysed by western blotting with antibodies as labelled.
Immunofluorescence of Rab-depleted cells for the early endosome marker EEA1, a binding partner of Rab5, indicated that in Rab5-depleted cells EEA1 was no longer localised in early endosome structures providing further confirmation that Rab5 had been successfully depleted (Supplementary Figure S8, EEA1). By contrast, Rab7 or Rab11 depletion had no obvious effect on EEA1 localisation (Supplementary Figure S8, EEA1). In addition, immunofluorescence of Rab5-depleted cells for the late endosomal MVB protein CD63 indicated that MVB localisation was subtly altered in the absence of Rab5 from being distributed throughout the cytoplasm to clustering close to the nucleus (Supplementary Figure S8, CD63). While Rab11 depletion had no effect on CD63 localisation, Rab7 depletion caused an apparent increase in the number and size of MVBs present throughout the cytoplasm (Supplementary Figure S8, CD63), in agreement with studies from others (Vanlandingham and Ceresa, 2009). Hence, although Rab7 depletion had affected the characteristics of the late endocytic pathway, its limited effect on virus yield confirmed that this part of the endocytic pathway had little involvement in HSV1 envelopment.
Given the significant defect in virus production in either the Rab5 or Rab11-depleted cells we focused on the activity of these two Rabs in HSV1-infected cells. Early endosome localised Rab5 is involved in initial entry steps into the cell as well as onward sorting and transport out of the early endosome (Jovic et al, 2010). In contrast, Rab11 is localised in recycling endosomes located around the MTOC and is required for movement from early to recycling endosomes, and transport from recycling endosomes to cell surface (Ullrich et al, 1996; Trischler et al, 1999). To determine if the effect of their knockdown was at the level of virus release or virus envelopment the knockdown experiments were repeated and the level of cell-associated virus was measured. In both cases, there was a significant reduction in cell-associated virus indicating that the block in virus replication was truly at the level of envelopment rather than release from the cell (Figure 6E). In addition, we carried out a double knockdown of both Rab5 and Rab11 and showed that this combined depletion reduced virus yield to only 1% of the negative control—making it even more effective in reducing virus production than Rab1 depletion (Figure 6E). The relative virus gene expression, as measured by western blotting, was also similar under these knockdown conditions (Figure 6F) confirming that virus entry and gene expression were unaffected by the depletion of these Rabs. Taken together, these results suggest that Rabs 5 and 11 located on early and recycling endosomes respectively play a critical role in the morphogenesis of HSV1.
Localisation of virus envelope proteins in Rab5 and Rab11-depleted cells
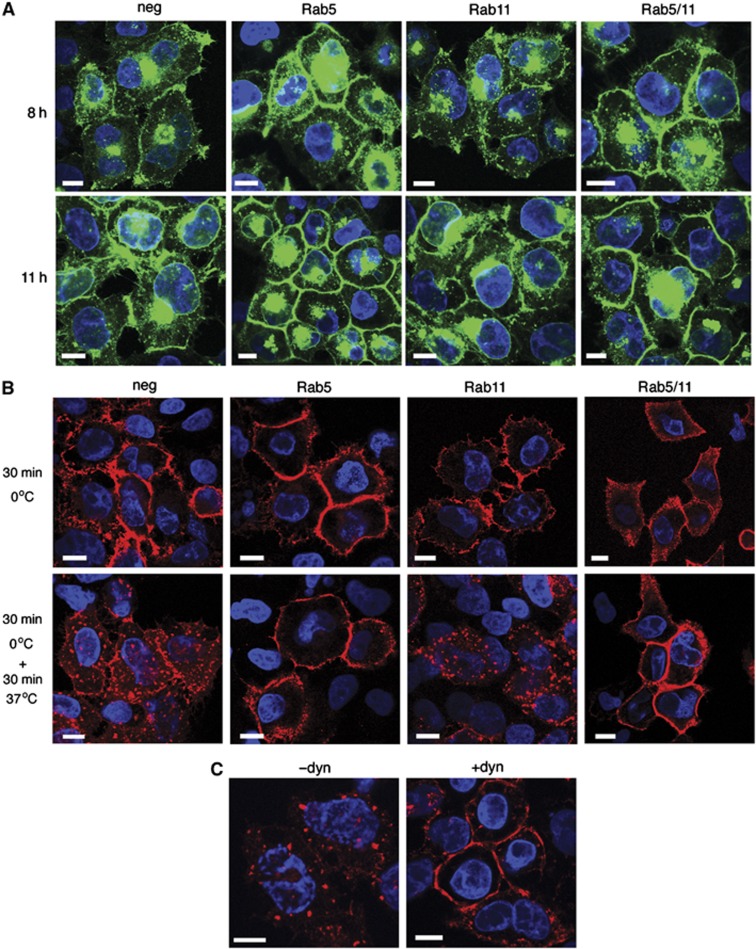
To determine the effect of Rab5 and/or Rab11 depletion on virus glycoprotein trafficking, siRNA-transfected HeLa cells were infected with HSV1 expressing the glycoprotein gD as a GFP-fusion protein, and examined 8 or 11 h later. In cells transfected with the negative siRNA, gD-GFP was localised at a juxtanuclear position together with specific sites on the cell surface, progressing to more intense cell surface fluorescence at 11 h (Figure 7A, neg). However, in Rab5-depleted cells, there was a significant enrichment of gD-GFP at the PM as early as 8 h that was more obvious by 11 h (Figure 7A, Rab5). gD-GFP localisation in Rab11-depleted cells was indistinguishable from negative siRNA-transfected cells (Figure 7A, Rab11) while the appearance in the double depleted cells was similar to depletion of Rab5 alone (Figure 7A, Rab5/11).
Figure 7.
Rab5 and dynamin are required for retrieval of glycoprotein D from the plasma membrane. (A) HeLa cells on coverslips transfected with siRNAs as denoted were infected 48 h later with HSV1 expressing gD-GFP (green) at a multiplicity of 2, fixed 8 or 11 h later and stained with DAPI to detect nuclei (blue). (B) HeLa cells on coverslips transfected with siRNAs as denoted were infected 48 h later with HSV1, and gD antibody uptake experiments were carried out 8 h later. After fixation, cells were stained with anti-mouse secondary antibody to detect anti-gD antibody (red) and DAPI to detect nuclei (blue). (C) HeLa cells infected with HSV1 were incubated for 30 min with dynasore 8 h later (+dyn), or left untreated (−dyn). gD antibody uptake was carried out and cells processed as for (B). Scale bar=10 μm.
Antibody uptake experiments were next carried out to establish if the increased concentration of gD-GFP at the PM in Rab5-depleted cells was due to inhibition of glycoprotein retrieval by endocytosis. In cells transfected with the negative siRNA, the gD antibody was recruited as before into cytoplasmic membranes (Figure 7B, neg). However, in cells in which Rab5 had been depleted either alone or in combination with Rab11, gD antibody uptake was inhibited and remained at the cell surface (Figure 7B, Rab5 and Rab5/11). By contrast in Rab11-depleted cells, gD endocytosis appeared similar to that in the negative control. Hence, the increased level of gD-GFP at the cell surface of cells depleted of Rab5 can be attributed to a greatly reduced rate of glycoprotein endocytosis from the PM.
This inhibition of gD retrieval in Rab5-depleted cells was similar to cells treated with the inhibitor of dynamin-dependent endocytosis, dynasore (Figure 7C, +dyn). It is noteworthy that long-term exposure to dynasore greatly reduced the numbers of capsids transported from the nucleus into the cytoplasm, indicating that dynasore may inhibit some stage of primary envelopment/deenvelopment through the nuclear membrane. Because of this feature, we were unable to determine the effect of dynasore on virus yield. However, EM of HRP uptake in HeLa cells pretreated for only 30 min with dynasore prior to incubation with HRP reduced the number of HRP-positive wrapped capsids by 70% (Table II). Unlike gD, HRP was still taken into cells in which dynamin-dependent endocytosis had been inhibited (data not shown) a result that was not unexpected as HRP is taken up by all mechanisms of fluid phase endocytosis including dynamin-independent pathways (Doherty and McMahon, 2009).
Table 2. Inhibition of dynamin-specific endocytosis reduces the number of HRP-positive wrapping membranes.
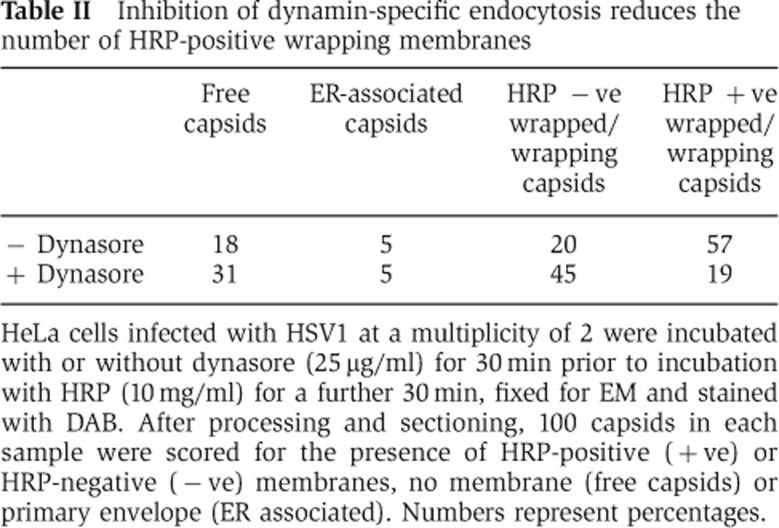
HeLa cells infected with HSV1 at a multiplicity of 2 were incubated with or without dynasore (25 μg/ml) for 30 min prior to incubation with HRP (10 mg/ml) for a further 30 min, fixed for EM and stained with DAB. After processing and sectioning, 100 capsids in each sample were scored for the presence of HRP-positive (+ve) or HRP-negative (−ve) membranes, no membrane (free capsids) or primary envelope (ER associated). Numbers represent percentages.
Virus capsids are mislocalised in Rab5-depleted cells
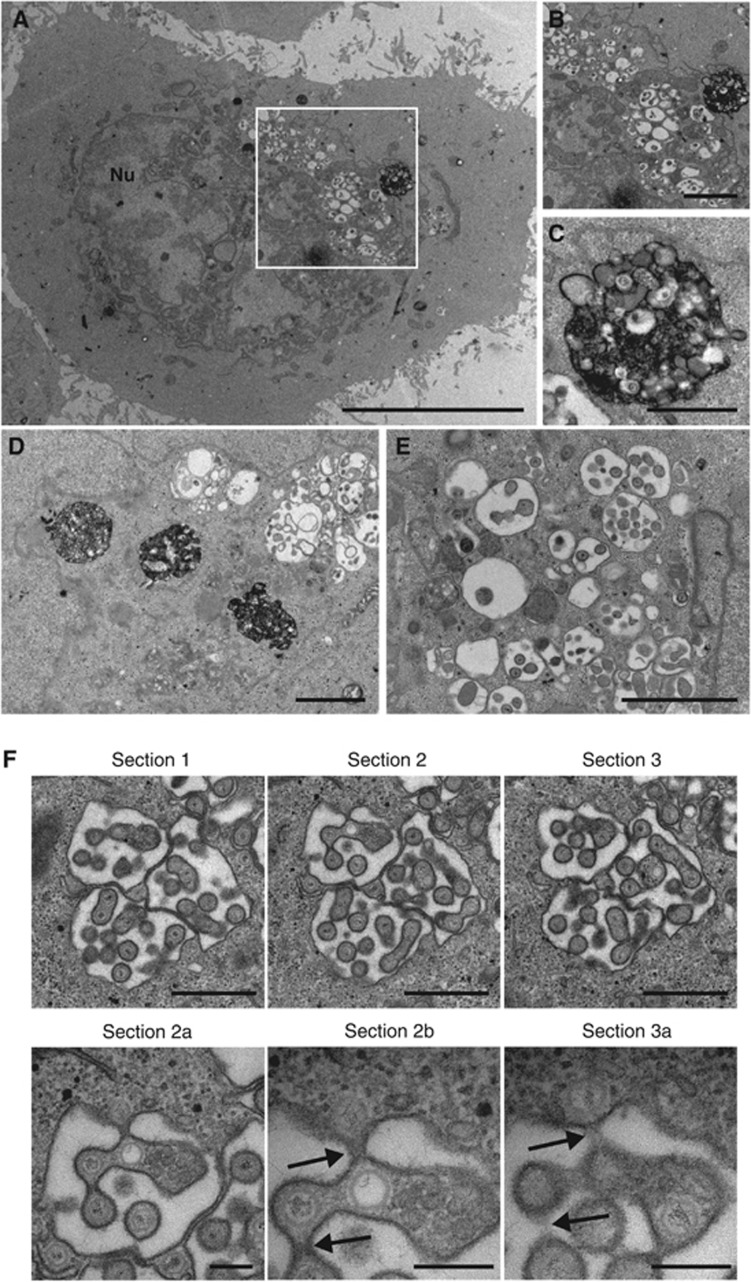
To determine the effect of Rab5 depletion on capsid wrapping, HRP uptake experiments were carried out, resulting in cell profiles exemplified in Figure 8A. Similar to dynasore-treated cells, HRP uptake was not blocked in Rab5-depleted cells. However, rather than numerous HRP-positive tubules in the cytoplasm, large, aberrant HRP-containing structures (around 2 μm in diameter) were present that were similar in appearance to the much smaller MVBs observed following a 60-min chase of HRP in non-depleted cells (Figure 8A–D—compare to MVB in Figure 2A). The size and localisation of these MVBs is consistent with the altered CD63 localisation we observed by immunofluorescence (Supplementary Figure S8, CD63).
Figure 8.
Rab5 is required for HSV1 envelopment. HeLa cells transfected with siRNAs for Rab5 were infected 48 h later with HSV1 at a multiplicity of 2, and labelled for 30 min with 10 mg/ml HRP 11 h later. Cells were fixed, stained with DAB and processed for EM. (A) An overview of a representative infected, Rab5-depleted cell. (B) Magnified image of juxtanuclear membranes. (C) Magnified image of HRP containing membrane-bound structure. (D) A second example of large HRP-positive structures next to capsids clustering at membranes. (E) Distended membranes with associated capsids. (F) Three serial sections of a cell exhibiting capsids associated with dilated membranes. Sections 2a and 2b are magnified images of section 2, showing the continuum between the individual particles and the cytoplasm (arrowed) that is not obvious in section 3 or the magnified section 3a. In (A), scale bar=10 μm, (B, D), scale bar=2 μm. In (C, E, F) (sections 1–3) scale bar=1 μm. In (F) (sections 2a, 2b & 3a), scale bar=200 nm.
In the Rab5-depleted infected cells, numerous capsids were observed both in and out of the nucleus, but it was rare to find capsids wrapping in HRP-positive membranes, or released from the cell surface. Many cytoplasmic capsids were associated with the lumina of dilated membranes, which we believe to be the cisternae of the Golgi stacks (Figure 8A, B). Closer examination of these profiles using serial sections revealed that although these particles looked to be within these lumina, they were in fact present in long finger-like projections into the Golgi that maintained a continuum with the cytoplasm and hence these capsids were not enveloped (Figure 8F).
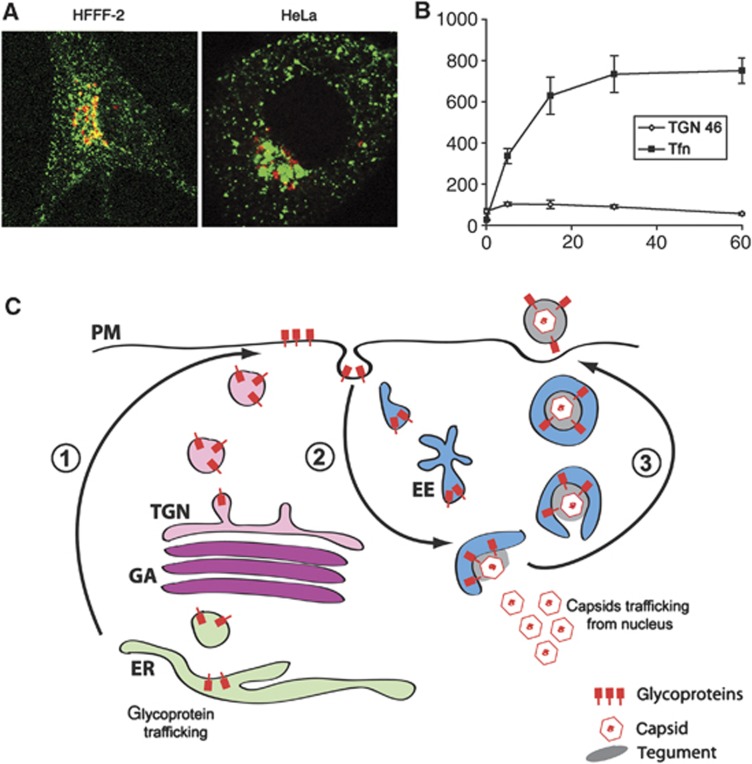
Taken together, our data suggest that HSV1 uses the retrieval of PM as a source of envelope. The abundance of membrane cycling from and to the PM in comparison to TGN46-positive membranes is readily demonstrated by the uptake of fluorescent transferrin, a well-characterised marker of clathrin-mediated endocytosis, into early and recycling endosomes (Figure 9A). Quantification of transferrin-containing membranes showed that they reached over 10 times the abundance of TGN46-containing membranes (Figure 9B). Moreover, the clustering of the transferrin-containing recycling endosomes in proximity to TGN46 was reminiscent of capsids clustering around the TGN. In light of these results, we propose a new model for virus envelopment involving retrieval of virus membrane proteins from the cell surface into relatively abundant cytoplasmic tubular membranes in the endocytic network (Figure 9C).
Figure 9.
Endocytic membranes as a source of HSV1 envelope. (A) HFFF-2 and HeLa cells were incubated with 200 ng/ml Cy3-transferrin (green) for 30 min prior to fixing and staining for TGN46 (red). (B) HeLa cells were treated as described in (A) for times ranging from 0 to 60 min. Using LSM510 software, TGN46 and transferrin-positive areas were measured in projected Z stacks relative to area of the cell. The results are the average of five cells for each time point. (C) Model for virus envelopment via endocytosis. (1) Virus glycoproteins are processed in the Golgi/TGN and exported to the cell surface. (2) The glycoprotein-containing plasma membrane is endocytosed and transported through the early endosome to produce wrapping tubules in a pathway dependent on dynamin, and Rabs 5 and 11. (3) Glycoprotein-containing tubules wrap cytoplasmic capsids forming virions with a double membrane. Fusion of the outer membrane at the cell surface results in release of a single-membrane virion outside the cell. EE, EEA1-positive early endosome; ER, endoplasmic reticulum; GA, Golgi apparatus; PM, plasma membrane; TGN, trans-Golgi network.
Discussion
Enveloped viruses have been shown to use various membranes of the secretory pathway or the PM as sites of virus wrapping. However, many of these processes are poorly defined. One major issue when investigating the source of virus envelope is the fact that many trafficking compartments, including the ERGIC, the Golgi, the TGN, late and recycling endosomes are organised by microtubule-based minus-end directed motor proteins such as dynein, and tend to cluster in close proximity to the MTOC adjacent to the nucleus. Light microscopy often cannot distinguish between these compartments and does not offer high enough resolution to determine the relationship between specific membranes and individual enveloping particles. By contrast, EM studies can provide high-resolution images of capsids enveloping at internal membranes, but are limited in their ability to identify them.
In the case of HSV1, the mechanism of envelopment has been contested for many years. Various models have been proposed, ranging from envelopment at the inner nuclear membrane (Nii et al, 1968; Ben-Porat and Kaplan, 1972) to envelopment at all possible internal membranes (Leuzinger et al, 2005), but the TGN is now favoured as the site of envelope acquisition (Granzow et al, 2001; Turcotte et al, 2005; Sugimoto et al, 2008; Johnson and Baines, 2011). However, based on the results presented here we propose a new model for HSV1 envelopment in which a population of Rab5 and 11 regulated endocytic tubules derived from the PM is the source of the final envelope (Figure 9C). We propose that virus envelope proteins are first processed through the Golgi/TGN, exported to the PM, and retrieved by endocytosis into the endocytic system to provide wrapping membranes for capsids. Egress would then result from natural recycling of vesicles, such as those used by the transferrin receptor, to the cell surface. In support of our endocytic model, many viral glycoproteins are exported to and accumulate at the cell surface in advance of capsid production (Hutchinson et al, 1992; Whiteley et al, 1999; Wisner and Johnson, 2004), while several such as gB and gE have endocytic motifs in their cytoplasmic tails that result in their retrieval (Alconada et al, 1999; Beitia Ortiz de Zarate et al, 2004). The role of glycoprotein endocytosis in virus replication has not been previously ascertained, and although studies on pseudorabies virus gE have concluded that endocytosis is not required for gE assembly into the virus (Tirabassi and Enquist, 1998, 1999), other work on human cytomegalovirus (HCMV) and varicella zoster virus suggest that endocytosis may be prerequisite for glycoprotein incorporation into the envelope (Radsak et al, 1996; Maresova et al, 2005), a conclusion that would support the envelopment model presented here for HSV1. Furthermore, others have demonstrated wrapping of HCMV capsids in endocytic membranes (Tooze et al, 1993; Fraile-Ramos et al, 2002). Our gD antibody uptake experiments show that PM gD is retrieved into the cytoplasm and is incorporated into wrapped virions, in spite of not returning to the Golgi/TGN. When considering the normal trafficking routes taken by cellular proteins, this lack of retrieval to the TGN is not surprising given that the majority of cell surface glycoproteins are recycled back to the PM following endocytosis, with only very few making the specialised journey back to the TGN (Pfeffer, 2009; Huotari and Helenius, 2011). Furthermore, the absence of LAMP2 or EEA1 markers from wrapping virions, and the tubular appearance of wrapping membranes, with no evidence for continuity of these membranes with vesicular domains, supports our conclusion that capsids do not acquire their envelopes from early or late endosomes, but rather are enwrapped in endocytic tubules.
The dependency of virus production on Rabs 5 and 11, but not Rabs 7 or 9, is compelling evidence for envelopment in the endocytic network, as these Rabs function in a sequential fashion in the recycling of membrane proteins to and from the cell surface. These studies also suggest that the endocytosis of virus glycoproteins is Rab5 and dynamin dependent. Although there are many entry pathways into the cell (Mayor and Pagano, 2007; Doherty and McMahon, 2009), all of which would be labelled by HRP uptake in our studies here, Rab5 and dynamin are involved in only a few of them including clathrin-mediated endocytosis and caveolae. Hence, inhibition of Rab5 or dynamin would not significantly inhibit HRP uptake from the cell surface, but would specifically block glycoprotein uptake and subsequent virus wrapping. EM studies showed that such backing up of the glycoproteins at the PM resulted in aberrant association of capsids with Golgi membranes, presumably due to a build-up of glycoproteins within the secretory pathway, but importantly our infectivity studies showed that these did not represent infectious virus (Figure 6D). Hence even though envelope proteins were making it all the way through to the PM, virion production was greatly inhibited.
While the presence of curved clathrin coats on some HSV1 wrapping membranes does not necessarily reflect that all envelopes started life with a clathrin coat, it is an indicator that at least some of them originate from sites of clathrin coat production. The major sites for such coats within the endocytic network are clathrin-coated pits at the cell surface and budding tubules on recycling endosomes (van Dam and Stoorvogel, 2002; van Dam et al, 2002; Doherty and McMahon, 2009), but as yet we cannot differentiate between these sites as a source of HSV1 wrapping membranes. Nonetheless, the fact that it took up to 30 min incubation in HRP to saturate the HSV1 wrapping membranes with HRP (Table I) may suggest that the virus uses the system of recycling endocytic membranes as a source for its envelope, allowing wrapping to occur anywhere in the cell where a capsid encounters a glycoprotein-containing endocytic membrane travelling on a microtubule. Indeed, it has been shown that extracellular virions contain TfR, a major marker for this compartment (Loret et al, 2008). Interestingly, Rab5 depletion resulted in the appearance of large HRP-positive MVB-like structures that may reflect a block in cargo sorting out of the early endosome and a subsequent block in onward trafficking through the recycling network. This is an attractive model for virus wrapping, as the membranes of this compartment are much more abundant and readily available throughout the cell than the smaller TGN compartment (Figure 9A and B). Hence, the profound effect of double depletion of Rabs 5 and 11 on virus production likely reflects the involvement of these two Rabs in the sequential trafficking of virus glycoproteins through this network and suggests that the virus exploits the normal trafficking of PM in its envelopment strategy.
The ability to wrap capsids at different sites within the cell—in the cell interior or the cell periphery, depending on where the capsid meets the envelope—may provide an explanation for contradictory studies on HSV1 envelopment in infected neurons, where it is still unclear if progeny capsids are wrapped before moving in an anterograde direction along the axon, or if they are wrapped after being transported to the distal region of the axon (Antinone and Smith, 2006; Snyder et al, 2006). If the model presented here were to translate to infected neurons then the site of wrapping may simply be dictated by the relative rate of capsid movement in an anterograde direction and tubule movement in a retrograde direction along the axon, properties that might differ depending on the experimental conditions. It is therefore noteworthy that recent studies have suggested that capsids moving in the anterograde direction along neuronal axons are both wrapped and unwrapped (Negatsch et al, 2010; Wisner et al, 2011).
Our results provide evidence for a new virus envelopment strategy that may be applicable to other enveloped viruses. They also add HSV1 to a growing number of viruses, including HIV, influenza A and hepatitis C that are now known to have a dependency on endocytic Rabs for their morphogenesis (Bruce et al, 2010; Manna et al, 2010; Caillet et al, 2011). Indeed, the unusual process of HSV1 envelopment may be akin to the process used by viruses that bud from the PM (Nayak et al, 2004; Jouvenet et al, 2006), as HSV1 could be considered to bud into membrane derived from the cell surface. With this new understanding of HSV1 envelopment it will now be possible to use this virus as a novel model for investigating the interplay between the cellular factors involved in various aspects of endocytosis.
Materials and methods
General reagents such as antibodies, cells and viruses, and standard methods are described in Supplementary data.
siRNAs and transfection
Silencer Select siRNA duplexes (see Supplementary data) from Ambion were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions for reverse transfection. The final concentration of siRNA (2 duplexes per isoform) was 5 nM per isoform. The Silencer Select negative siRNA number 1 was used as a negative control.
Viability assays
Cell viability was assessed by flow cytometry using LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen). Flow cytometry was carried out using a BD CyAn machine and analysed using Summit 4.3 software.
Antibody uptake assay
Culture supernatant of anti-gD monoclonal antibody LP14 was diluted 1 in 50 in DMEM without serum at 4°C, and added to cells on coverslips. The cells were left on ice for 30 min, washed twice with fresh media, then incubated at 37°C for 30 min before fixation for IF. For EM studies of the same process, cells were additionally incubated with anti-mouse F(ab′)2 fragment HRP conjugate for 30 min prior to raising temperature to 37°C for 30 min.
Transmission electron microscopy
Cells were fixed in 0.5% glutaraldehyde in 200 mM sodium cacodylate buffer for 30 min, washed in buffer and secondarily fixed in reduced 1% osmium tetroxide, 1.5% potassium ferricyanide for 60 min. The samples were washed in distilled water and stained overnight at 4°C in 0.5% magnesium uranyl acetate, washed in distilled water and dehydrated in graded ethanol. The samples were then embedded flat in the dish in Epon resin. Resin filled stubs were placed on embedded cell monolayers and polymerised. Ultrathin sections (typically 50–70 nm) were cut parallel to the dish and examined in a FEI Tecnai electron microscope with CCD camera image acquisition. Endocytic events were labelled by incubating cells with media containing HRP (Serva) at a concentration of 10 mg/ml for times ranging from 2 to 60 min. After fixation all samples except 2 min labelled samples were washed in buffer and stained with metal enhanced DAB substrate kit (Thermo Scientific) for 30 min, then washed in buffer and processed.
Immunoelectron microscopy
Cells were fixed in 250 mM HEPES buffer pH 7.4 containing 4% paraformaldehyde (w/v) for 10 min on ice and 20 min at 20°C in 8% paraformaldehyde, then scraped, pelleted and fixed for a further 60 min at 20°C. Cell pellets were washed in PBS, frozen in 2.1 M sucrose/PBS and stored under liquid nitrogen. Cryosections were prepared by the Tokuyasu method (Tokuyasu, 1980), and immunolabelled with relevant antibodies. Mouse monoclonal antibodies were detected with rabbit anti-mouse (Cappel/ICN) and either 6 or 9 nm protein A-gold particles (Slot and Geuze, 1985). Sheep antibody was detected with a directly conjugated donkey anti-sheep 6 nm gold (Abcam).
Supplementary Material
Acknowledgments
We thank Helena Browne for the HSV1 gD and gH antibodies and the gD-GFP plasmid used to construct the gD-GFP recombinant HSV1. We also thank David Johnson and Steve Rice for the HSV1 gE and ICP27 antibodies, respectively, and Prashant Desai for the GFP-VP26 plasmid. GE, HLJ, CGL, and MH were funded by the Medical Research Council, CLS is a Wellcome Trust Research Student and GLS is a Wellcome Trust Principal Research Fellow.
Author contributions: GE, MH, and GLS designed the experiments. MH, GE, HLJ, CLS, and CGL performed the experiments. GE wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alconada A, Bauer U, Sodeik B, Hoflack B (1999) Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol 73: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres G, Garcia-Escudero R, Simon-Mateo C, Vinuela E (1998) African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol 72: 8988–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinone SE, Smith GA (2006) Two modes of herpesvirus trafficking in neurons: membrane acquisition directs motion. J Virol 80: 11235–11240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Malhotra V (2006) The formation of TGN-to-plasma-membrane transport carriers. Annu Rev Cell Dev Biol 22: 439–455 [DOI] [PubMed] [Google Scholar]
- Barman S, Ali A, Hui EK, Adhikary L, Nayak DP (2001) Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res 77: 61–69 [DOI] [PubMed] [Google Scholar]
- Beitia Ortiz de Zarate I, Kaelin K, Rozenberg F (2004) Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J Virol 78: 1540–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T, Kaplan AS (1972) Studies on the biogenesis of herpesvirus envelope. Nature 235: 165–166 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hurley JH (2008) Retromer. Curr Opin Cell Biol 20: 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese SS Jr, DeBoer CJ (1966) Electron microscope observations of African swine fever virus in tissue culture cells. Virology 28: 420–428 [DOI] [PubMed] [Google Scholar]
- Bruce EA, Digard P, Stuart AD (2010) The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol 84: 5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss V (2004) Envelopment of the hepatitis B virus nucleocapsid. Virus Res 106: 199–209 [DOI] [PubMed] [Google Scholar]
- Bruss V (2007) Hepatitis B virus morphogenesis. World J Gastroenterol 13: 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70: 715–728 [DOI] [PubMed] [Google Scholar]
- Caillet M, Janvier K, Pelchen-Matthews A, Delcroix-Genete D, Camus G, Marsh M, Berlioz-Torrent C (2011) Rab7A is required for efficient production of infectious HIV-1. PLoS Pathog 7: e1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri A, Sette P, Salata C, Cancellotti E, Forghieri C, Comin A, Gottlinger H, Campadelli-Fiume G, Palu G, Parolin C (2007) Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J Virol 81: 11468–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CE, Gan BQ, Tang BL (2011) Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 68: 3349–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P (1996) Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J 15: 5783–5788 [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM (2004) Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J Gen Virol 85: 3517–3527 [DOI] [PubMed] [Google Scholar]
- Crump CM, Yates C, Minson T (2007) Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol 81: 7380–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ (1998) ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol 140: 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargin D (1986) The structure and assembly of herpes viruses. InElectronmicroscopy of Proteins, Virus Structure Harris JR, Horne RW (eds)Vol. 5: pp359–437London: Academic Press [Google Scholar]
- De Matteis MA, Luini A (2008) Exiting the Golgi complex. Nat Rev Mol Cell Biol 9: 273–284 [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT (2009) Mechanisms of endocytosis. Annu Rev Biochem 78: 857–902 [DOI] [PubMed] [Google Scholar]
- Fraile-Ramos A, Pelchen-Matthews A, Kledal TN, Browne H, Schwartz TW, Marsh M (2002) Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3: 218–232 [DOI] [PubMed] [Google Scholar]
- Ghosh RN, Mallet WG, Soe TT, McGraw TE, Maxfield FR (1998) An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J Cell Biol 142: 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatas J, Stieber A, Olsnes S, Gonatas NK (1980) Pathways involved in fluid phase and adsorptive endocytosis in neuroblastoma. J Cell Biol 87: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC (2001) Egress of alphaherpesviruses: comparative ultrastructural study. J Virol 75: 3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Back R, Marsh M (1989) A quantitative analysis of the endocytic pathway in baby hamster kidney cells. J Cell Biol 109: 2703–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Aniento F, Parton RG, Gruenberg J (1997) Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes. J Cell Biol 139: 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30: 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC (1992) A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol 66: 2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Baines JD (2011) Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9: 382–394 [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD (2006) Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 4: e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M, Sharma M, Rahajeng J, Caplan S (2010) The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol 25: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GE, Murray JW, Wolkoff AW, Wilson DW (2006) Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J Virol 80: 4264–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzinger H, Ziegler U, Schraner EM, Fraefel C, Glauser DL, Heid I, Ackermann M, Mueller M, Wild P (2005) Herpes simplex virus 1 envelopment follows two diverse pathways. J Virol 79: 13047–13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR (1993) Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J 12: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret S, Guay G, Lippe R (2008) Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol 82: 8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luini A, Mironov AA, Polishchuk EV, Polishchuk RS (2008) Morphogenesis of post-Golgi transport carriers. Histochem Cell Biol 129: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Aligo J, Xu C, Park WS, Koc H, Heo WD, Konan KV (2010) Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresova L, Pasieka TJ, Homan E, Gerday E, Grose C (2005) Incorporation of three endocytosed varicella-zoster virus glycoproteins, gE, gH, and gB, into the virion envelope. J Virol 79: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Pagano RE (2007) Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol 8: 603–612 [DOI] [PubMed] [Google Scholar]
- McMillan TN, Johnson DC (2001) Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol 75: 1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC (2006) Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet Microbiol 113: 163–169 [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H (2006) Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol 9: 423–429 [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC, Minson T (2006) Egress of alphaherpesviruses. J Virol 80: 1610–1611author reply 1611–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saksena M, Boadle RA, Aggarwal A, Tijono B, Rixon FJ, Diefenbach R, Cunningham AL (2009) Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J Virol 83: 3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DP, Hui EK, Barman S (2004) Assembly and budding of influenza virus. Virus Res 106: 147–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negatsch A, Granzow H, Maresch C, Klupp BG, Fuchs W, Teifke JP, Mettenleiter TC (2010) Ultrastructural analysis of virion formation and intraaxonal transport of herpes simplex virus type 1 in primary rat neurons. J Virol 84: 13031–13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii S, Morgan C, Rose HM (1968) Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol 2: 517–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C, Tolbert CL, Waters JF (1989) Internalization of horseradish peroxidase isozymes by pancreatic acinar cells in vitro. J Histochem Cytochem 37: 49–56 [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Brown D, Perrelet A (1986) Transport of horseradish peroxidase from the cell surface to the Golgi in insulin-secreting cells: preferential labelling of cisternae located in an intermediate position in the stack. EMBO J 5: 2097–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawliczek T, Crump CM (2009) Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol 83: 11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR (2009) Multiple routes of protein transport from endosomes to the trans Golgi network. FEBS Lett 583: 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR (2011) Entry at the trans-face of the Golgi. Cold Spring Harbor Perspect Biol 3: pii: a005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S, Girotti M, Yaspo ML, Owen CE, Perry AC, Suganuma T, Nilsson T, Fried M, Banting G, Warren G (1996) Primate homologues of rat TGN38: primary structure, expression and functional implications. J Cell Sci 109: Part 3675–685 [DOI] [PubMed] [Google Scholar]
- Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hubinger A, Reschke M (1996) Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch Virol 141: 557–572 [DOI] [PubMed] [Google Scholar]
- Remillard-Labrosse G, Mihai C, Duron J, Guay G, Lippe R (2009) Protein kinase D-dependent trafficking of the large Herpes simplex virus type 1 capsids from the TGN to plasma membrane. Traffic 10: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122: 735–749 [DOI] [PubMed] [Google Scholar]
- Roberts KL, Smith GL (2008) Vaccinia virus morphogenesis and dissemination. Trends Microbiol 16: 472–479 [DOI] [PubMed] [Google Scholar]
- Roingeard P, Hourioux C, Blanchard E, Brand D, Ait-Goughoulte M (2004) Hepatitis C virus ultrastructure and morphogenesis. Biol Cell 96: 103–108 [DOI] [PubMed] [Google Scholar]
- Roingeard P, Hourioux C, Blanchard E, Prensier G (2008) Hepatitis C virus budding at lipid droplet-associated ER membrane visualized by 3D electron microscopy. Histochem Cell Biol 130: 561–566 [DOI] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G (1994) Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol 68: 130–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ (1985) A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol 38: 87–93 [PubMed] [Google Scholar]
- Snyder A, Wisner TW, Johnson DC (2006) Herpes simplex virus capsids are transported in neuronal axons without an envelope containing the viral glycoproteins. J Virol 80: 11165–11177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y (2008) Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J Virol 82: 5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Portner A (2004) Molecular mechanism of paramyxovirus budding. Virus Res 106: 133–145 [DOI] [PubMed] [Google Scholar]
- Tirabassi RS, Enquist LW (1998) Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol 72: 4571–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirabassi RS, Enquist LW (1999) Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol 73: 2717–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT (1980) Immunochemistry on ultrathin frozen sections. Histochem J 12: 381–403 [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Reis B, Radsak K, Kern H (1993) Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol 60: 163–178 [PubMed] [Google Scholar]
- Trischler M, Stoorvogel W, Ullrich O (1999) Biochemical analysis of distinct Rab5- and Rab11-positive endosomes along the transferrin pathway. J Cell Sci 112: Part 244773–4783 [DOI] [PubMed] [Google Scholar]
- Turcotte S, Letellier J, Lippe R (2005) Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79: 8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG (1996) Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol 135: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam EM, Stoorvogel W (2002) Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol Biol Cell 13: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam EM, Ten Broeke T, Jansen K, Spijkers P, Stoorvogel W (2002) Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J Biol Chem 277: 48876–48883 [DOI] [PubMed] [Google Scholar]
- Vanlandingham PA, Ceresa BP (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 284: 12110–12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley A, Bruun B, Minson T, Browne H (1999) Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol 73: 9515–9520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisner TW, Johnson DC (2004) Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids. J Virol 78: 11519–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisner TW, Sugimoto K, Howard PW, Kawaguchi Y, Johnson DC (2011) Anterograde transport of herpes simplex virus capsids in neurons by both separate and married mechanisms. J Virol 85: 5919–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenner HL, Yoshimura S, Barr FA, Crump CM (2011) Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J Virol 85: 8012–8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.