Abstract
The contribution of human subtelomeric DNA and chromatin organization to telomere integrity and chromosome end protection is not yet understood in molecular detail. Here, we show by ChIP-Seq that most human subtelomeres contain a CTCF- and cohesin-binding site within ∼1–2 kb of the TTAGGG repeat tract and adjacent to a CpG-islands implicated in TERRA transcription control. ChIP-Seq also revealed that RNA polymerase II (RNAPII) was enriched at sites adjacent to the CTCF sites and extending towards the telomere repeat tracts. Mutation of CTCF-binding sites in plasmid-borne promoters reduced transcriptional activity in an orientation-dependent manner. Depletion of CTCF by shRNA led to a decrease in TERRA transcription, and a loss of cohesin and RNAPII binding to the subtelomeres. Depletion of either CTCF or cohesin subunit Rad21 caused telomere-induced DNA damage foci (TIF) formation, and destabilized TRF1 and TRF2 binding to the TTAGGG proximal subtelomere DNA. These findings indicate that CTCF and cohesin are integral components of most human subtelomeres, and important for the regulation of TERRA transcription and telomere end protection.
Keywords: histone, RNA polymerase, shelterin, subtelomere, telomere
Introduction
The ends of eukaryotic chromosomes form specialized chromatin structures that are essential for chromosome stability and genome maintenance (Zakian, 2012). The terminal TTAGGG repeats of mammalian telomeres bind to a set of proteins that are nucleated by the DNA-binding proteins TRF1, TRF2, and Pot1, and are collectively referred to as shelterin (de Lange, 2005; Palm and de Lange, 2008) or telosome (Liu et al, 2004; Ye et al, 2010). These terminal repeat binding factors regulate telomere length homoeostasis and DNA damage repair processing at the chromosome termini (Karlseder, 2003). Loss or damage of the terminal repeats can initiate a DNA damage response and trigger cellular replicative senescence (d'Adda di Fagagna et al, 2003; Deng et al, 2008). DNA damage and senescence can also be elicited by mutation or depletion of telomere repeat binding proteins (Karlseder et al, 2002). Dynamic remodelling of telomere repeat factors and telomere DNA conformation is also required for normal telomere length regulation and telomerase accessibility (Teixeira et al, 2004; Jain and Cooper, 2011; Murnane, 2011; Stewart et al, 2012).
In addition to shelterin and telomerase, telomere maintenance depends on the proper assembly and regulation of telomeric chromatin (Blasco, 2007; Schoeftner and Blasco, 2009; Ye et al, 2010). Traditionally, telomeres have been thought of as highly heterochromatic structures associated with condensed chromatin and transcriptional silencing (Loo and Rine, 1995; Perrod and Gasser, 2003; Rusche et al, 2003; Blasco, 2007). More recent studies have revealed that many eukaryotic telomeres, including human and yeast, can be transcribed, indicating that telomeric silencing is incomplete and telomere chromatin is dynamic (Azzalin et al, 2007; Azzalin and Lingner, 2008; Schoeftner and Blasco, 2008; Luke and Lingner, 2009; Arora et al, 2011). The chromatin structure of telomeres is further complicated by the variations in the subtelomeric DNA structures, suggesting that telomeric heterochromatin structure and regulation may vary among different chromosomes (Riethman et al, 2005; Riethman, 2008a, 2008b). In budding yeast, telomeric silencing is mediated by Sir proteins that interact with telomere repeat binding factor Rap1(Grunstein, 1997). In mammalian telomeres, nucleosomal arrays commonly associated with heterochromatin appear to be irregularly spaced or disrupted by telomere repeat binding factors (Wu and de Lange, 2008; Galati et al, 2012). Numerous interactions between shelterin and chromatin regulatory factors suggest that telomere repeat factors contribute to telomeric chromatin structure (Perrini et al, 2004; Garcia-Cao et al, 2004; Sugiyama et al, 2007; Deng et al, 2009; Canudas et al, 2011; Chakraborty et al, 2011). We have previously shown that TRF2 can bind directly to telomeric repeat-containing RNA (TERRA) to recruit heterochromatin proteins including ORC and HP1 and maintain histone H3K9me3 enrichment at telomeres (Deng et al, 2009). TERRA expression is itself dependent on histone H3K4 methyltransferase MLL (Caslini et al, 2009), as well as DNA methylation status and CpG-island promoter found in many subtelomeric regions (Yehezkel et al, 2008; Nergadze et al, 2009; Deng et al, 2010). In fission yeast, the expression of TERRA and other subtelomeric transcripts are subject to diverse regulation by chromatin regulatory factors (Bah et al, 2011; Greenwood and Cooper, 2011). The dynamic interplay between shelterin, telomere chromatin structure, TERRA expression, and telomere biology appears to be an essential and universal component of chromosome stability.
The chromatin organizing factor CTCF has been implicated in numerous aspects of chromosome biology, including chromatin insulation, enhancer blocking, transcriptional activation and repression, DNA methylation-sensitive parental imprinting, and DNA-loop formation between transcriptional control elements (Bushey et al, 2008; Phillips and Corces, 2009; Ohlsson et al, 2010). CTCF has been implicated in the transcriptional repression of the D4Z4 macrosatellite repeat transcript found ∼30 kb from the telomere repeats of chromosome 4q (Ottaviani et al, 2011). At D4Z4, CTCF interacts with lamin A and tethers the chromosome 4q telomere to the nuclear periphery (Ottaviani et al, 2009a, 2009b). A more general role for CTCF has been found in its ability to colocalize with cohesin subunits at many chromosomal positions (Parelho et al, 2008; Rubio et al, 2008; Stedman et al, 2008; Wendt et al, 2008). Cohesin is a multiprotein complex consisting of core subunits SMC1, SMC3, Rad21, and SCC3 (referred to as SA1 or SA2 in humans), which can form a ring-like structure capable of encircling or embracing two DNA molecules (Nasmyth and Haering, 2005; Hirano, 2006). Cohesin was originally identified as a regulator of sister-chromatid cohesion, but subsequent studies in higher eukaryotes indicate that they have functions in mediating long-distance interactions between DNA elements required for transcription regulation (Kagey et al, 2010; Dorsett, 2011). Cohesin subunit SA1 is recruited to telomere repeats by the shelterin protein Tin2, and this interaction is required for telomeric sister-chromatid cohesion and efficient telomere replication (Canudas and Smith, 2009; Remeseiro et al, 2012). Tin2 can also promote heterochromatin formation through an interaction with heterochromatin protein HP1γ, but how this relates to sister-chromatid cohesion and cohesin function is not completely clear (Canudas et al, 2011). It is also not known whether CTCF can associate with telomeres or subtelomeres in addition to binding the D4Z4 gene repeat, nor if it can interact with cohesin at these locations.
The chromosome region immediately adjacent to the terminal repeats has been referred to as the subtelomere. In humans, the distal subtelomeres consist of a variety of degenerate repeat elements with a few discrete gene transcripts interspersed at various distances from the terminal TTAGGG repeat tracts (Riethman et al, 2005; Linardopoulou et al, 2005; Ambrosini et al, 2007; Riethman, 2008a, 2008b). TERRA transcription initiates from within the subtelomeres, and a promoter containing a CpG-island and subtelomeric 29- and 61-bp repeat element has been identified in plasmid reconstitution assays (Nergadze et al, 2009). DNA methylation and DNA methyltransferases have been shown to inhibit TERRA expression since TERRA levels are highly elevated in cells where DNMTs have been genetically disrupted or depleted (Gonzalo et al, 2006; Nergadze et al, 2009), as well as in Immunodeficiency-Centromeric instability-Facial abnormalities (ICF) Syndrome cells that are genetically defective in DNA methyltransferase 3B (DNMT3B) (Ehrlich et al, 2006; Yehezkel et al, 2008; Deng et al, 2010). CTCF binding is known to be DNA methylation sensitive but it is not yet known whether CTCF associates with transcriptional regulatory elements important for TERRA regulation or telomere maintenance. Herein, we investigate the role of CTCF and cohesin at human subtelomeres and their role in regulating TERRA expression, telomere chromatin organization, and telomere DNA end protection.
Results
CTCF, cohesin, and RNAPII binding to the CpG-island promoters in human subtelomeres
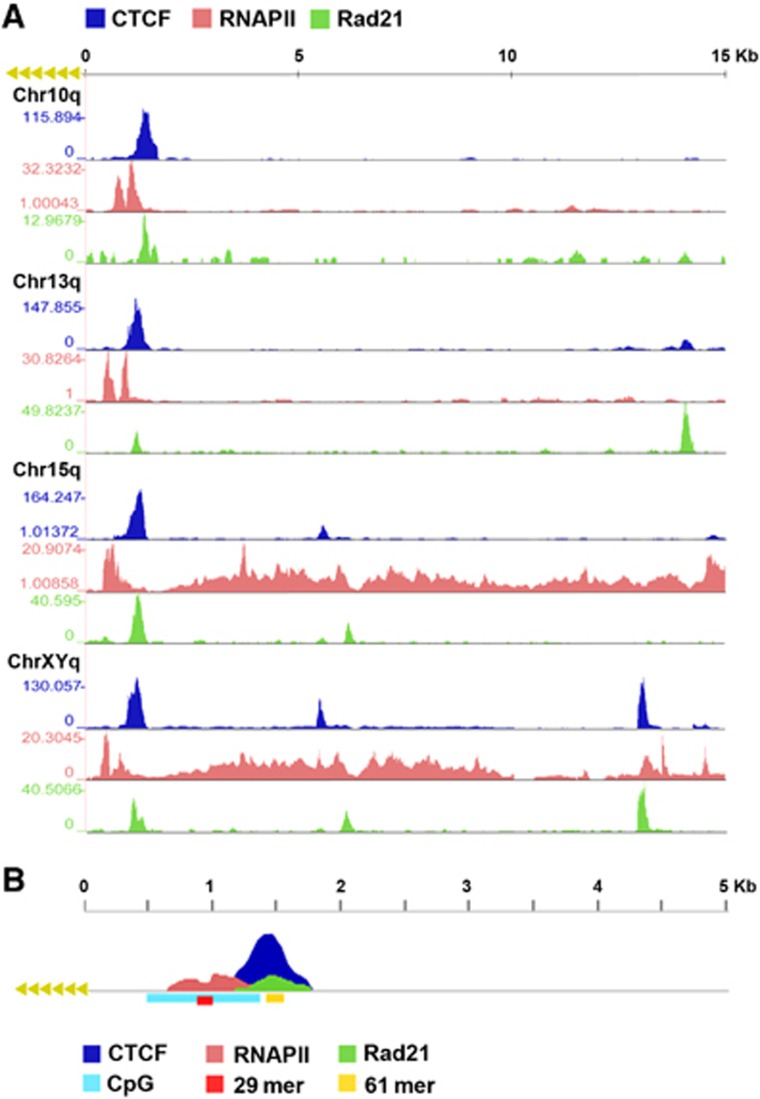
Genome-wide analyses of CTCF, cohesin, and RNA polymerase II (RNAPII) have been performed in several different cell lines from various laboratories, including those generating the human ENCODE database (Wendt et al, 2008; Cuddapah et al, 2009; Kasowski et al, 2010; Sun et al, 2011; Lee et al, 2012). In these published studies, the complete human subtelomeric DNA was not available for ChIP-Seq data mapping, with gaps immediately adjacent to the start of terminal repeat tracts for many telomeres (Riethman et al, 2004). We have generated complete assemblies of human subtelomeres for most of these chromosome ends (Stong et al, in preparation) and here we use these reference assemblies to map the read sequences from data sets, including our own, for CTCF, Rad21, SMC1, and RNAPII (Figure 1; Supplementary Figures S1 and S2; Supplementary Files S1 and S2). We found that most but not all human chromosome ends have a major CTCF-binding site within 1–2 kb from the TTAGGG repeat tracts. These CTCF sites consistently mapped to a region just upstream (centromeric) to the CpG-islands and 29 bp repeats, often overlapping 61 bp repeat element (Supplementary Figures S1 and S2). In the few exceptions to this pattern, CTCF sites were observed at positions ∼10 kb from the TTAGGG repeats (7p, XYp) or several CTCF-binding sites with relatively low peak scores (3p, 7q, 8q, and 12q) (Supplementary Figure S2). We refer to these two different subtelomeres as type I (with major CTCF peaks at ∼1–2 kb) or type II (lacking obvious CTCF peaks proximal to the telomere repeat tracts). In almost all cases, including those of type II, we observed an overlap of CTCF-binding sites with cohesin subunit Rad21 (Figure 1; Supplementary Figure S1). We confirmed that CTCF and cohesin peaks overlap in different cell lines by performing an independent CTCF and SMC1 ChIP-Seq experiment in a B-lymphoma cell line (Supplementary Figures S1 and S2). Our ChIP-Seq showed a nearly perfect overlap of CTCF and SMC1 in these cells, and a strong correlation with CTCF and Rad21 binding in multiple cell types. In contrast to CTCF and cohesin, RNAPII bound as a more diffuse peak most commonly at a position immediately telomeric to the CTCF-binding sites and more directly overlapping the CpG-islands. A schematic summary of the average binding pattern of CTCF, Rad21, and RNAPII relative to CpG-island, 29 and 61 bp repeats is shown in Figure 1B.
Figure 1.
Enrichment profiles for ChIP-Seq analysis of CTCF, cohesin, and RNAPII binding to human subtelomeres. Fragment density profiles were generated for samples and a matched IgG control as described in Materials and methods. The fold enrichment of sample over IgG is shown. (A) CTCF, RNAPII, and Rad21 binding in the first 15 kb subtelomeres of chromosome arms 10q, 13q, 15q, and XYq. The y-axis for each track is auto-scaled to highest peak in each chromosome region shown. (B) Model enrichment profile with peaks within the first 5 kb of the telomere tract. The CTCF peak is just centromeric to the CpG-island, typically centred over a 61-mer repeat. The RNA Pol II tract is centred over the 29-mer repeat. The exact position of these peaks varies with the positioning of these genomic features relative to the start of the terminal repeat tract on each chromosome arm.
Due to the complex duplications in subtelomeric sequence, we permitted multimapping signals weighted according to the number of perfect subtelomeric mapping sites to contribute, along with uniquely mapping reads, to subtelomeric ChIP-seq signals. We found that the remaining unique signals recapitulated the ChIP-Seq peak positions in most cases when multiple mappings were eliminated (Supplementary Figure S1E), suggesting that most of the binding sites can be uniquely assigned to specific subtelomeres. Some unique signals are lost, as expected for perfect duplications. This was sometimes the case with the 29-mer repeats over which RNAPII signal is centred and which a portion of the CTCF and cohesin read peaks was formed at many subtelomeres. Supplementary Figure S2D illustrates this effect for the example subtelomeres shown in the Supplementary Figure S1E. At the same time, Supplementary Figure S2D also shows the clear enrichment of RNAPII ChIP-seq reads mapping to the 29-mer variable number tandem repeat (VNTR) over the IgG controls, a true binding peak that would have been missed if multimapping signal contributions were disallowed.
CTCF binds directly upstream of the CpG-island and 29 repeat element found in subtelomeres
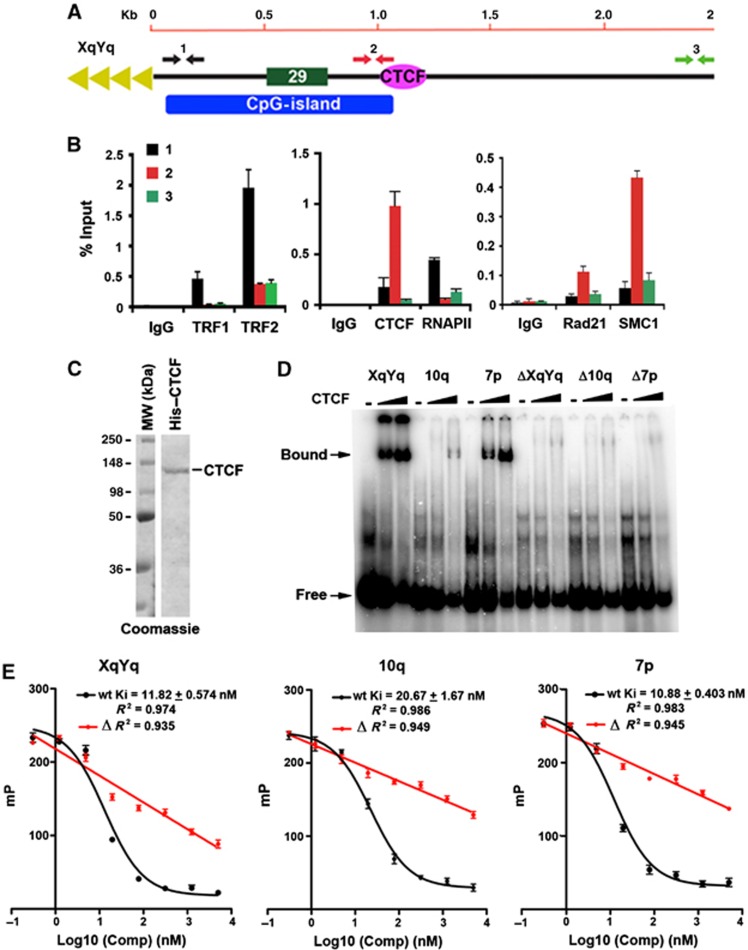
To verify the ChIP-Seq data for CTCF, cohesin, and RNAPII, we performed conventional ChIP-qPCR with primers spanning the first 3 kb of the XYq (Figure 2A and B) and 10q (Supplementary Figure S3) subtelomeres in a B-cell lymphoma-derived cell line used for ChIP-seq. As a control, we assayed TRF1 and TRF2 ChIP. As expected, TRF1 and TRF2 were enriched at positions closest to the TTAGGG repeats (primer set 1). ChIP assays with CTCF and cohesin subunits Rad21 and SMC1 revealed strong enrichment at the CpG-islands (primer set 2), while RNAPII was enriched at regions closest to the TTAGGG repeats (primer set 1), consistent with ChIP-Seq data indicating that RNAPII bound to broad peaks between CTCF–cohesin and the TTAGGG repeats. To determine if CTCF bound directly to subtelomere DNA, we assayed the ability of purified recombinant CTCF protein to bind candidate recognition sites in vitro by electrophoretic mobility shift assay (EMSA) (Figure 2C and D). Human CTCF protein was expressed and purified from baculovirus (Figure 2C). Candidate CTCF-binding sites from the ChIP-seq peaks in subtelomere XYq, 10q, and 7p, as well as control oligonucleotides containing substitution mutations in the putative CTCF consensus sites, ΔXYq, Δ10q, and Δ7p, were synthesized as 46 mers for EMSA probes (Supplementary Table S3). Purified CTCF protein bound efficiently to the XqYq and 7p probes, less efficiently to 10q probe, but not to the mutated ΔXYq, Δ10q, and Δ7p probes (Figure 2D), indicating that these subtelomere ChIP-Seq peaks contain bonafide CTCF recognition sites. The relative binding affinities of these subtelomeric CTCF-binding sites was further quantified by a fluorescence polarization based competitor assay (Figure 2E). The wild-type CTCF-binding sites from XYq, 10q, and 7p showed robust competition against a FAM6-labelled probe containing a CTCF-binding site with high similarity to the consensus motif as defined previously (Kim et al, 2007). Inhibitory constants (Ki) for each binding sites were equal to 11.82, 20.67, and 10.88 nM, respectively. On the other hand, the mutant ΔXYq, Δ10q, and Δ7p probes show linear relationship to increasing competitor with no plateau, suggesting a nonspecific inhibition of CTCF binding (Figure 2E). These findings indicate that the subtelomeric CTCF-binding sites have relatively high affinities for CTCF in vitro.
Figure 2.
Identification of CTCF-binding site elements in the 61-bp element of human subtelomeres. (A) Schematic of the type I subtelomere showing the relative positions of the 29- and 61-bp repeat element, CpG-island, and TTAGGG terminal repeats. (B) ChIP-qPCR for TRF1, TRF2, CTCF, RNAPII, Rad21, and SMC1 relative to IgG controls using primers for the XYq subtelomere at positions close (∼150 bp) to TTAGGG repeat (black), at CpG-island (red), or ∼3 kb from terminal repeats (green). Bar graph represents the average value of percentage of input for each ChIP from three independent PCR reactions (mean±s.d.). (C) Purified recombinant CTCF protein analysed by Coomassie staining of SDS–PAGE gel. (D) EMSA with CTCF protein binding to DNA oligonucleotide probes containing putative binding sites from subtelomere XYq, 10q, or 7p, as well as with oligonucleotides containing point mutations in CTCF recognition sites designated ΔXYq, Δ10q, and Δ7p. Free probe and bound probe were indicated with arrow. (E) Inhibitory constants (Ki) were calculated by titrating the same DNA probes used in EMSA against a FAM6-labelled probe with a known dissociation constant and measuring changes in CTCF binding via fluorescence polarization. Mutant (Δ) sites show a linear binding isotherm over the same concentration range of competitor, suggesting only nonspecific competition.
Chromatin organization of human subtelomeres
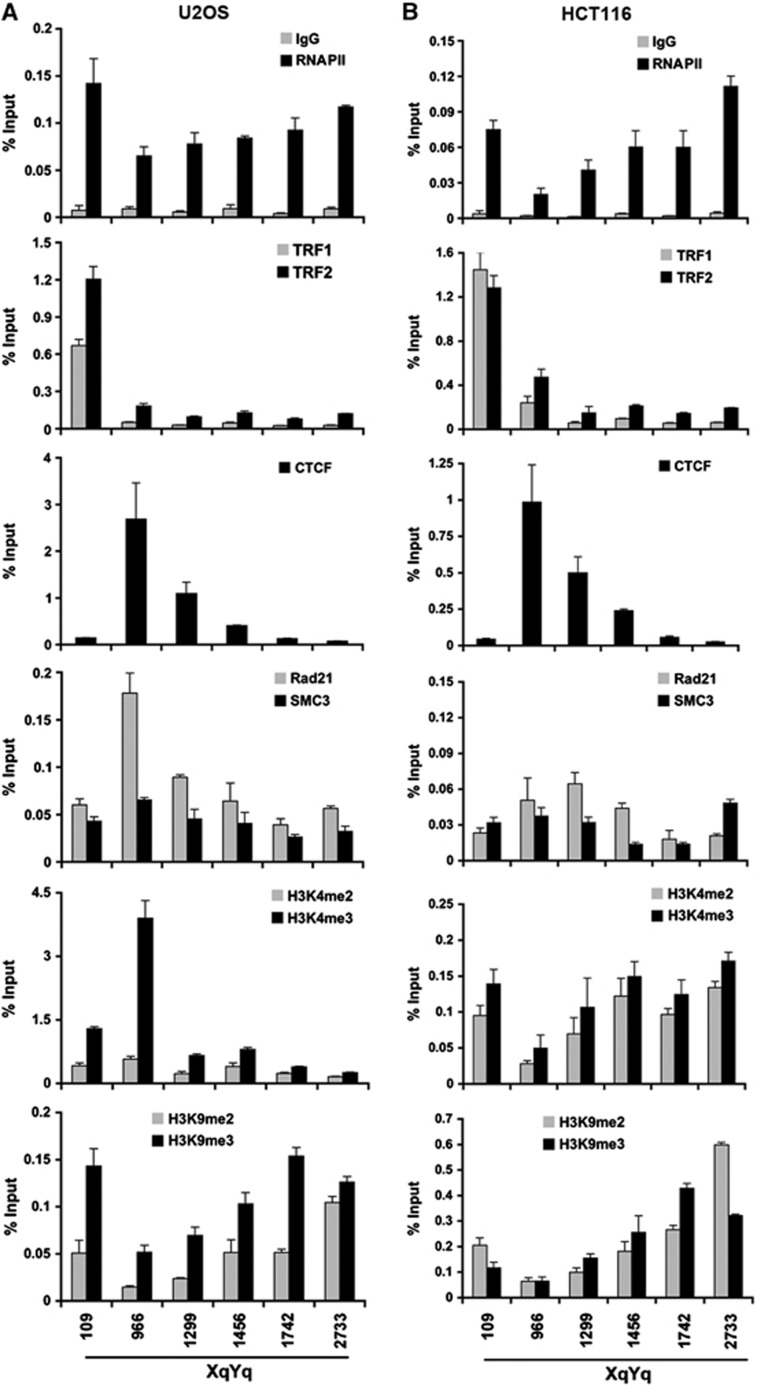
To investigate the potential role of CTCF in subtelomere chromatin organization, we assayed chromatin factor binding and histone modification patterns at several positions across the subtelomeres for XYq (Figure 3) and 10q (Supplementary Figure S4). We assayed ChIP binding patterns for TRF1, TRF2, CTCF, Rad21, SMC3, H3K4me2, H3K4me3, H3K9me2, H3K9me3, RNAPII, and IgG. We compared these binding patterns in two commonly used cell lines that utilize different telomere lengthening mechanisms and TERRA expression levels, namely ALT positive U2OS, which have high TERRA expression, and telomerase positive HCT116 cells, which have relatively low TERRA expression. ChIP-qPCR analysis revealed that TRF1 and TRF2 interact primarily with the region closest to the TTAGGG repeats in both cell types. CTCF was enriched at position centred at 966 bp from the TTAGGG in both cell types. Rad21 colocalized with CTCF in U2OS, but was more diffusely localized in HCT116, while SMC3 appeared more diffusely distributed in both cell types. Interestingly, histone H3K4me3 was highly enriched at CTCF site in U2OS, but somewhat depleted in HCT116. H3K9me3 was enriched at the TTAGGG repeats in U2OS, but also detected at more telomere-distant regions in both U2OS and HCT116 cells. RNAPII was elevated at the regions close to the TTAGGG sites in both cell types, but also distributed at more telomere-distant subtelomere regions (similar patterns were observed at other subtelomeres, including 13q and 15q) (Supplementary Figure S5). Notably, the highest subtelomeric RNAPII signals centromeric to the CpG-island corresponded to the positions of gene bodies for the WASH subtelomeric gene family (Linardopoulou et al, 2007) and the RPL23a pseudogene family (Fan et al, 2002), both of which are transcribed towards the telomere with their 3′ ends within 2–6 kb of the start of the (TTAGGG)n tract (Riethman, 2008a). Taken together, these findings suggest that subtelomeric histone modification patterns vary between cell types, but that CTCF binding remains invariant.
Figure 3.
Chromatin organization of human subtelomeres. (A, B) Conventional ChIP-qPCR was used to assay CTCF, Rad21, SMC3, TRF1, TRF2, histone H3K4me2 and me3, H3K9me2 and me3, and RNAPII binding at various nucleotide positions relative to the TTAGGG repeat tract (position 0) in the XYq subtelomere for either U2OS (A) or HCT116 (B) cell lines. Bar graph represents the average value of percentage of input for each ChIP from three independent ChIP experiments (mean±s.d.).
CTCF binding stimulates TERRA promoter driven transcription in an orientation-specific manner
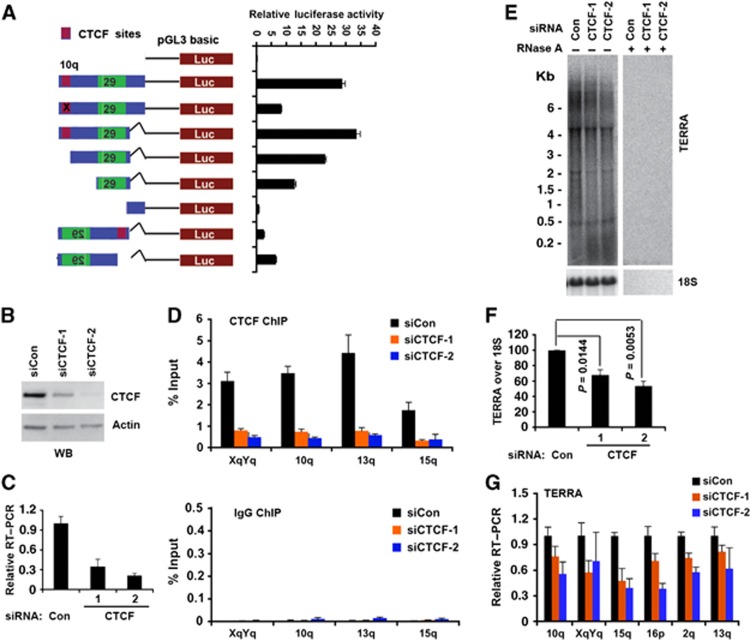
The subtelomere CpG-islands and 29 bp repeat have been implicated in the control of TERRA transcription initiation (Nergadze et al, 2009). To test if the CTCF-binding sites in the CpG-island contributes to transcription control, we used a luciferase reporter plasmid to assay promoter activity of the 10q CpG-island and 29 bp repeat elements (Figure 4A). When the entire CpG-island and 29 bp repeat were tested in the TERRA orientation, we found relatively robust luciferase activity. Point mutations that disrupt CTCF binding reduced luciferase activity ∼4-fold, while a truncation of the CTCF site reduced luciferase activity only ∼1.5-fold. Deletion of the 29-bp repeats completely eliminated luciferase activity, indicating that these elements constitute the major promoter activity. When the promoter region was tested in the reverse orientation, low levels of luciferase activity was detected, and deletion of CTCF resulted in an ∼2.5-fold increase in promoter activity. These findings suggest that CTCF supports promoter activity of the 29-bp repeat element for RNAPII oriented towards the TTAGGG repeats and TERRA transcription.
Figure 4.
CTCF function in TERRA transcription. (A) Luciferase reporter constructs containing 10q subtelomere sequence with point mutations in CTCF site (red) or deletion mutations or orientation changes. CTCF-binding site was shown in red and 29 bp element was shown in green. Bar graph represents the average value of relative luciferase activity to Renilla control from three independent transfections (mean±s.d.). (B) Western blot of U2OS cells transfected with siRNA control, siCTCF-1, or siCTCF-2 and assayed with anti-CTCF or anti-actin at 4 days post-transfection. (C) qRT–PCR of U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 relative to actin mRNA. Relative RT–PCR represents the value calculated by ΔΔCT methods relative to siControl and Gapdh. Bar graph represents the average value from three independent CTCF depletion experiments (mean±s.d.). (D) ChIP-qPCR of CTCF (top panel) or control IgG (lower panel) in U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 at the CTCF-binding sites in chromosome XYq, 10q, 13q, and 15q presented as percentage of input DNA. (E) Northern blot analysis of TERRA in U2OS cells transfected with siControl, siCTCF-1, or siCTCF-2 with control 18S (lower panel) or RNase A treatment (right panel). Numbers on the left show the position of RNA markers in Kb. (F) Quantification of at least three independent Northern blot assays, a representative is shown in (E). Bar graph represents TERRA signal intensity relative to 18S signal, and relative intensity for siRNA control was set at 100. P-value was calculated by paired two-tailed Student’s t-test (n=3). (G) qRT–PCR of TERRA from individual telomeres at 10q, XYq, 15q, 16q, 2q, and 13q treated with siControl, siCTCF-1, and siCTCF-2 in U2OS cells. Relative RT–PCR represents the value calculated by ΔΔCT methods relative to siControl and Gapdh. Bar graph represents the average value from three independent CTCF depletion experiments (mean±s.d.).
To determine if CTCF contributes to endogenous TERRA transcription, we first used siRNA targeting CTCF. We identified two different siRNAs that could effectively deplete CTCF during a transient transfection of siRNA in U2OS cells (Figure 4B and C) and HCT116 (Supplementary Figure S6). ChIP assays indicated that CTCF protein was depleted from subtelomeres (Figure 4D). Depletion of CTCF led to an ∼2-fold decrease in steady-state levels of TERRA as measured by northern blot assay (Figure 4E and F; Supplementary Figures S6 and S7). CTCF depletion led to a loss of TERRA expression at most subtelomeres examined by chromosome-specific RT–qPCR (Figure 4G; Supplementary Figures S6 and S7), although TERRA derived from type II subtelomeres (e.g., 7p, XYp) that lack proximal CTCF-binding sites were less responsive to CTCF depletion (Supplementary Figure S7C). These findings are consistent with the transient transfection assay, and suggest that CTCF is required for the positive regulation of TERRA transcription.
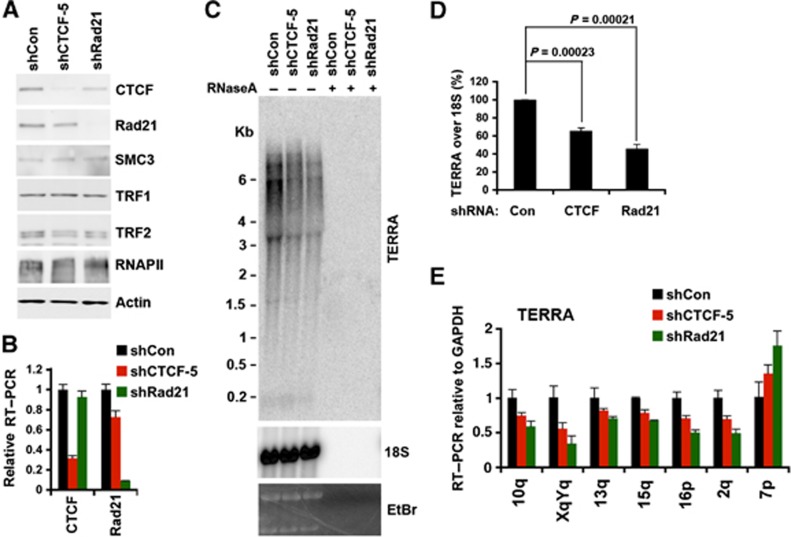
To further investigate the role of CTCF and cohesin at telomeres, we generated selectable shRNA lentivirus expression vectors targeting CTCF and Rad21 (Figure 5; Supplementary Figures S8 and S9). U2OS (Figure 5; Supplementary Figure S8) or HCT116 (Supplementary Figure S9) cells were transduced and selected for puromycin resistance, and then assayed 6 days post-infection. CTCF was partially depleted (∼70–80%), while Rad21 was mostly depleted (>90%), as measured by western blot (Figure 5A; Supplementary Figures S8A and S9A) and by RT–PCR (Figure 5B). Consistent with siRNA depletion, shRNA targeting CTCF caused a reduction in TERRA as measured by northern blot (Figure 5D; Supplementary Figure S8), as well as by chromosome-specific qRT–PCR (Figure 5E; Supplementary Figure S9B). While most telomeres showed a reduction in TERRA, a few chromosomes, including type II subtelomeres, showed either no effect or a slight increase in TERRA expression after CTCF or Rad21 depletion (Figure 5E; Supplementary Figure S9B). In general, Rad21 depletion produced an even greater reduction in TERRA levels (Figure 5C–E; Supplementary Figure S9B), suggesting that CTCF and cohesin cooperate to promote TERRA transcription at type I telomeres containing promoter proximal CTCF-binding sites.
Figure 5.
shRNA depletion of Rad21 and CTCF decreases TERRA expression. (A) Western blot of U2OS cells selected for lentivirus transduction with shControl, shCTCF, or shRad21, and assayed for expression levels of CTCF, Rad21, SMC3, TRF1, TRF2, RNAPII, and actin at 6 days post lentiviral infection. (B) qRT–PCR of U2OS cells for CTCF or Rad21 mRNA expression after transduction with shControl, shCTCF, or shRad21 lentivirus. Relative RT–PCR represents the value calculated by ΔΔCT methods relative to shControl and Gapdh. Bar graph represents the average value from three independent lentiviral infection experiments (mean±s.d.). (C) Northern blot analysis of TERRA in U2OS cells transduced with shControl, shCTCF, or shRad21 relative to control 18S or eithidium stain (lower panels), and control RNase A treatment (+). Equal amount of total RNA (∼7.5 mg) isolated from transfected cells at 6 days post lentiviral infection was used for each sample. (D) Quantification of at least three northern blots as represented in (C). Bar graph represents TERRA signal intensity relative to 18S signal, and relative intensity for shRNA control was set at 100. P-value was calculated by paired two-tailed Student’s t-test (n=4). (E) qRT–PCR for TERRA RNA with chromosome-specific primers at 10q, XYq, 13q, 15q, 16p, 2q, and 7p relative to Gapdh mRNA in U2OS cells transduced with shControl (black), shCTCF (red), or shRad21 (green). Relative RT–PCR represents the value calculated by ΔΔCT methods relative to shControl and Gapdh. Bar graph represents the average value from three independent lentiviral infection experiments (mean±s.d.).
CTCF is required for cohesin, TRF, and RNAPII recruitment to telomeres and subtelomeres
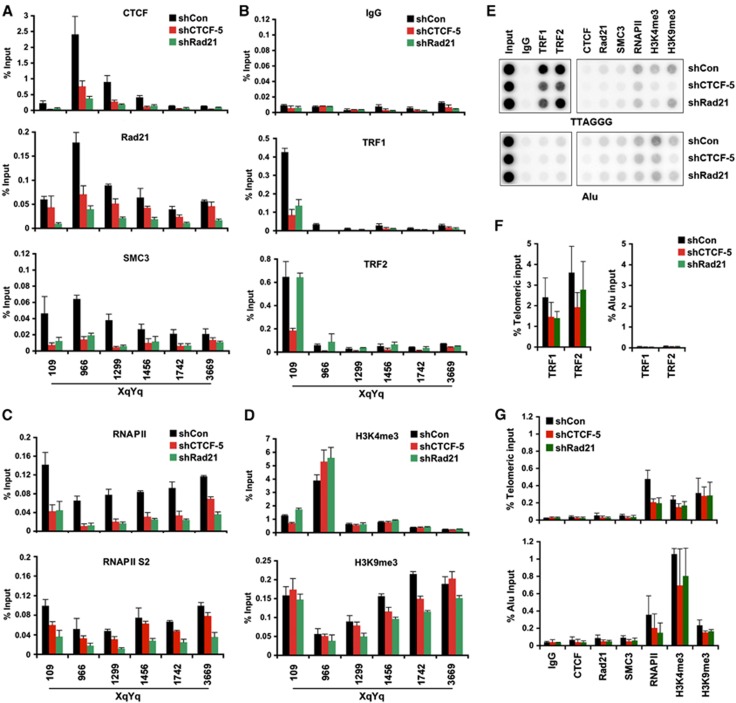
To begin to address the function of CTCF and cohesin at subtelomeres and in TERRA transcription regulation, we assayed the effect of CTCF and Rad21 shRNA depletion on chromatin factor binding and histone modification patterns at subtelomeres (Figure 6; Supplementary Figure S10). CTCF and Rad21 shRNA were shown to cause a loss of CTCF and Rad21 binding to subtelomeres, as expected (Figure 6A; Supplementary Figure S10A). We also found that CTCF and Rad21 depletion caused a loss of SMC3 binding, indicating that CTCF is required for cohesin loading at subtelomeres, and that Rad21 is an essential component of the cohesin complex required for chromatin assembly. Somewhat surprisingly, we found that Rad21 depletion led to a loss of CTCF binding, suggesting that cohesin is important for stabilizing CTCF binding to subtelomeric DNA. Interestingly, we also observed a consistent loss of TRF1 and TRF2 binding in CTCF-depleted cells, and a partial loss of TRF1 binding in Rad21-depleted cells (Figure 6B; Supplementary Figure S10B). The loss of SMC3, TRF1, and TRF2 binding in CTCF-depleted cells correlates to the efficiency of CTCF depletion by two shCTCF, indicating that the decreased binding of these factors at subtelomeres is not due to an off-target effect of shRNA (Supplementary Figure S11). Nor did CTCF depletion cause a global loss of TRF1 or TRF2 protein levels, indicating that the effects on telomere binding are not due to changes in TRF1 or TRF2 abundance (Figure 5A).
Figure 6.
CTCF and Rad21 depletion leads to a loss of RNAPII and TRF binding at telomere and subtelomere. (A) ChIP-qPCR for CTCF, Rad21, or SMC3 were shown at various positions of the XYq subtelomere relative to the TTAGGG repeat tracts (position 0). U2OS cells were transduced with shControl (black), shCTCF (red), or shRad21 (green) and assayed by ChIP at 6 days post-infection. Bar graph represents the average value of input (%) for each ChIPs from three independent experiments (Mean±s.d.). (B–D) Same as in (A), except that TRF1, TRF2, or control rabbit IgG (B), RNAPII or RNAPII serine 2 (S2) phosphorylation (C), or histone H3 K4 and K9 tri-methylation (D) were assayed by ChIP-qPCR in infected U2OS cells. (E) U2OS cells were infected with lentivirus expressing shControl, shCTCF, or shRad21 and assayed by ChIP for TRF1, TRF2, CTCF, Rad21, SMC3, RNAPII, H3K4me3, and H3K9me3 at 6 days post-infection. ChIP DNA were dot-blotted, and assayed by hybridization with either 32P-labelled (TAACCC)4 or 32P-labelled Alu probe. (F, G) Quantification of dot-blots for shControl (black), shCTCF (red), and shRad21 (green) relative to either telomeric input (top panel) or Alu input (bottom panel). Bar graph represents average values of percent input for each ChIP (mean±s.d.) from three independent ChIP experiments, a representative shown in (E).
To investigate the potential effects of CTCF or Rad21 depletion on transcription, we assayed RNAPII binding in the subtelomere. We found that depletion of CTCF and Rad21 led to a substantial (∼80%) loss of RNAPII binding as well as RNAPII serine 2 (S2) phorporylation at all positions across the subtelomere (Figure 6C; Supplementary Figure S10C). This suggests that CTCF and Rad21 enhance TERRA transcription through facilitating RNAPII binding and transcriptional elongation.
In addition to these factors, we assessed the effects of CTCF and Rad21 depletion on histone modification patterns in the subtelomere DNA (Figure 6D; Supplementary Figure S10D). We observed only a small increase in histone H3K4me3 at the CTCF-binding site upon CTCF and Rad21 depletion, but no significant change in histone H3K9me3 occupancy. We also assayed the effect of CTCF and Rad21 depletion on interactions with the telomere repeat DNA using dot-blot analysis of ChIP DNA (Figure 6E–G). We found that CTCF and Rad21 depletion produced a small, but significant decrease of TRF1 and TRF2 (for CTCF only) at telomere repeat DNA and a similar loss of RNAPII, but no significant change in histone H3K4me3 or H3K9me3 (Figure 6E–G). As expected, we did not observe significant binding of CTCF, Rad21, and SMC3 binding at telomeric repeats (Figure 6E and G). In addition, we did not observe any obvious changes in the global MNase I digestion pattern at telomere repeats or the XYq subtelomere (Supplementary Figure S12), suggesting that CTCF and cohesin depletion did not alter gross nucleosome assembly at telomeres or subtelomeres. Taken together, these observation suggest that CTCF and cohesin stabilize, either directly or indirectly, both shelterin and RNAPII interactions with subtelomeric DNA.
Depletion of CTCF or Rad21 leads to telomere DNA damage foci formation
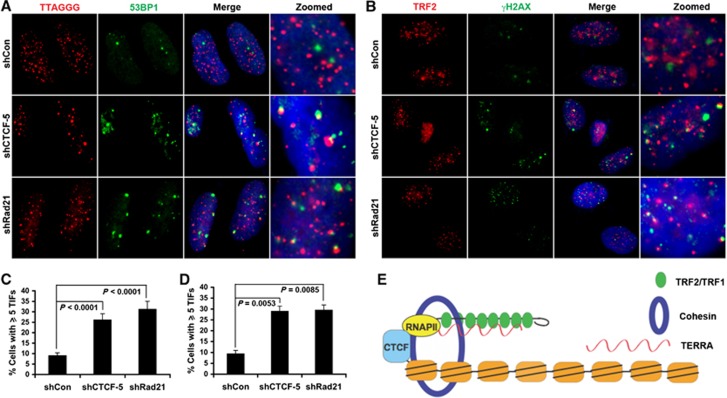
The loss of TRF1 or TRF2 from subtelomeric and telomere repeat DNA may be predicted to elicit a DNA damage response due to telomere uncapping. We therefore assayed the effect of CTCF or Rad21 depletion on telomere dysfunction-induced foci (TIFs) (Figure 7; Supplementary Figure S13). We found that shRNA-mediated depletion of CTCF and Rad21 caused ∼3-fold increase in γH2AX and 53BP1-associated TIFs. Similar results were observed with siRNA depletion of CTCF (Supplementary Figure S13). CTCF and Rad21 depletion did not have any apparent effect on telomere repeat length, as measured by Southern blot of restriction enzyme fragments (Supplementary Figure S14). These findings indicate that CTCF and Rad21 depletion induces a DNA damage response at telomeres that is not due to the loss of telomere repeat DNA length, and supports a model that CTCF and cohesin protect telomere ends by regulating RNAPII recruitment, TERRA expression, and shelterin interactions with the subtelomere (Figure 7E).
Figure 7.
CTCF or Rad21 depletion leads to an increase in telomere-associated DNA damage foci (TIFs). (A) U2OS cells were transduced with shControl, shCTCF, or shRad21 and assayed by immuno-FISH with TTAGGG PNA probe (red) and antibody to 53BP1 (green) at 6 days post-infection. Dapi (blue), merge, and zoom images are shown to the right. (B) Immunofluorescence with anti-TRF2 (red) or γH2AX (green) in U2OS cells transduced with shControl, shCTCF, or shRad21. Merged and zoomed images are shown to the right. (C) Quantification of telomere-associated DNA damage foci as represented in (A). The bar graph is the mean and s.d. derived from quantification of >300 nuclei from multiple independent TIF assays (n=5). P-value was calculated by two-tailed Student’s t-test. (D) Quantification of telomere-associated DNA damage foci as represented in (B). The bar graph is the mean and s.d. derived from quantification of >100 nuclei from three independent TIF assays. P-value was calculated by two-tailed Student’s t-test. (E) Model depicting CTCF and cohesin as integral components of type I human subtelomeres. CTCF–cohesin function in recruitment of RNAPII to the CpG-island promoter, TERRA transcription regulation, and stabilization of telomere end capping.
Discussion
A foundation for a chromatin atlas of the human subtelomeres
Genome-wide studies on chromatin structure and histone modification patterns have been incomplete near human telomeres, due both to remaining gaps in the reference sequence adjacent to the start of the (TTAGGG)n tracts and to subtelomeric segmental duplication families near many telomeres. In this work, we provide a foundation for a more complete analysis of the human genome by examining regions of the human subtelomeres that had previously not been included in human genome-wide studies. Using new sequence data to complete most of the gaps adjacent to (TTAGGG)n tracts and stringent read mapping criteria (both described in detail in Stong et al, in preparation), we have established a human subtelomere map and genome browser for next-generation DNA sequence analyses, including ChIP-Seq and RNA-Seq. Here, we mapped several ChIP-Seq data sets to the most distal parts of human subtelomeres (Figure 1; Supplementary Figures S1 and S2; Supplementary Files S1 and S2). We focused on CTCF and cohesin subunits because of their general importance in chromosome organization throughout vertebrate evolution. We found that CTCF and cohesin colocalized at a position immediately adjacent to the CpG-islands implicated in TERRA promoter regulation (Nergadze et al, 2009) (Figures 1 and 2). We confirmed this binding by generating a new experimental data set for CTCF and SMC1 ChIP-Seq in a B-lymphoma cell lines. In addition, we mapped RNAPII binding and found that it localized more broadly across the subtelomeres, but had an average enrichment at the telomeric side of the CpG-island promoter for TERRA expression. CTCF and cohesin bound just centromeric to the CpG-island, and were further investigated for their role in TERRA expression and telomere end protection. The genome browser and methods established for mapping next-generation sequence data to the subtelomere provides a foundation for building a more complete atlas of epigenetic marks and chromatin organization at human subtelomeres.
CTCF recruits RNAPII to subtelomeres
Our data implicate CTCF and cohesin as positive regulators of TERRA transcription and RNAPII recruitment to the TERRA promoter region. CTCF has been shown to physically and functionally interact with largest subunit of RNAPII (Chernukhin et al, 2007). More recent studies have shown that CTCF can modulate RNAPII activity through regulation of RNAPII large subunit CTD phosphorylation (Kang and Lieberman, 2009; Fay et al, 2011; Kang and Lieberman, 2011). The phosphorylation status of RNAPII has been shown to correlate with its activity in promoter assembly (S5 phosphorylation) and transcriptional elongation (S2 phosphorylation) (Selth et al, 2010; Nechaev and Adelman, 2011). CTCF has been shown both to modulate RNAPII CTD phosphorylation status, as well as to colocalize with RNAPII pausing sites within the bodies of gene transcripts (Wada et al, 2009). We found that depletion of CTCF or cohesin subunit Rad21 results in a loss of RNAPII and RNAPII S2 binding at the TERRA promoter region, as well as at positions throughout the entire subtelomere (Figure 6; Supplementary Figure S10). ChIP experiments revealed that RNAPII is enriched at the CTCF–cohesin binding site in the CpG-island, but is also broadly distributed in some subtelomeres (Figures 1 and 2). The localization of RNAPII at the CpG correlates with the transcriptional promoter activity observed in vivo and in genetic mutations on transient reporter plasmids (Figure 4). Consistent with the findings of Azzalin and colleagues, we found that the 29-bp repeat was the dominant promoter element for TERRA in transient assays (Nergadze et al, 2009). We confirmed that CTCF bound to a specific site in the 61-bp repeats located centromeric to the CpG-island and 29 bp repeat element. Site directed mutation of the CTCF site revealed that CTCF contributes as a positive acting factor in TERRA transcription. However, when the TERRA promoter was positioned in the reverse orientation, CTCF deletion resulted in an increase in transcription activity, suggesting that CTCF functions to restrict RNAPII transcribing towards the centromere. Since recent studies suggest that RNAPII can be oriented in both directions at many promoters (Rhee and Pugh, 2012), we suggest that CTCF plays a role in both recruiting RNAPII to TERRA promoters, as well as orientating RNAPII towards telomeric transcription.
CTCF and cohesin stabilize TRF binding to subtelomeres
We found that TRF1 and TRF2 binding to the subtelomeric DNA and telomere repeat DNA was reduced in cells where CTCF and cohesin were depleted (Figure 6; Supplementary Figure S10). This result was somewhat surprising since neither CTCF nor cohesin are known to physically interact with TRFs. Furthermore, it seems unlikely that factors binding in the subtelomere would affect telomere repeat factor binding at the terminal repeats. CTCF and cohesin depletion had no effect on the total abundance of TRF1 or TRF2, as determined by western blot (Figure 5A; Supplementary Figure S8), but it is possible that additional indirect effects of depletion may contribute to the loss of TRF1 and TRF2 binding at telomeres. A more plausible explanation is that CTCF and cohesin influence RNAPII binding and local histone modifications, which are required for proper maintenance of subtelomeric chromatin and its association with shelterin. In support of this, Canudas and Smith have shown that a cohesin subunit, SA1, can interact with a shelterin component, Tin2, to promote telomere sister-chromatid cohesion and telomeric heterochromatin (Canudas and Smith, 2009). TRF1 and TRF2 have been shown to bind efficiently to TERRA RNA and these interactions may be important for forming and stabilizing higher-ordered chromatin structure necessary for telomere–subtelomere communication. Interactions between the telomere repeat ends and the subtelomere may be required for T-loop formation, and it is possible that CTCF and cohesin may stabilize this type of higher-ordered chromatin structure (de Lange, 2004). We also suspect that CTCF and cohesin binding influences histone modifications and DNA methylation patterns that may regulate TRF1 and TRF2 interactions with subtelomeric DNA or chromatin. In this respect, CTCF and cohesin may provide a chromatin barrier function that keeps the subtelomeric nucleosomes from obstructing shelterin assembly at the telomere repeat tracts.
CTCF and cohesin prevent telomere DNA damage signalling
CTCF and Rad21 depletion resulted in an increase in the colocalization of γH2AX and 53BP1 with telomere repeat DNA foci (Figure 7; Supplementary Figure S13). Previous studies have shown that loss of TRF2 or TRF1 binding induces DNA damage foci (TIFs) at telomeres (Takai et al, 2003). We have previously shown that depletion of TERRA levels can also result in telomere DNA damage response foci formation (Deng et al, 2009). Furthermore, telomere sister-chromatin cohesion has been shown to be mediated by Tin2 interaction with the SA1 subunit of cohesin (Canudas and Smith, 2009), while CTCF has been shown to bind SA2 (Xiao et al, 2011). SA1 and SA2 are thought to form mutually exclusive and functionally distinct cohesin complexes, with SA1 functioning within the telomere repeats and SA2 functioning at more centromeric regions of the chromosome (Canudas and Smith, 2009). These observations lead us to propose that the fundamental role of CTCF–cohesin binding at subtelomeres is to recruit RNAPII to maintain TERRA promoter activity and subtelomere chromatin architecture, which are both necessary for telomere end protection. The molecular basis for RNAPII recruitment can be attributed to the physical interaction between CTCF and RNAPII (Chernukhin et al, 2007). CTCF and cohesin, which have both been implicated in DNA-loop formation, may also help to establish or stabilize the telomere T-loop structure (Figure 7E). While the primary biochemical function of CTCF and cohesin at the TERRA promoter can be explored in future studies, our findings show that cohesin work coordinately with CTCF to recruit RNAPII dependent TERRA expression and protect telomere ends.
Materials and methods
Cells
U2OS cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. HCT116 cells was grown in McCoy’ 5A medium supplemented with 10% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C. Lymphoblastoid cells (LCLs) and B lymphoma-derived cells (BCBL1) were cultured in RPMI 1640 medium supplemented with 15% fetal bovine serum and antibiotics in a 5% CO2 incubator at 37°C.
ChIP-Seq data
ChIP-Seq was performed using 1 × 107 BCBL1 cells per assay with either rabbit anti-cellular SMC1 (Bethyl A300-055A-3) or CTCF antibody (Millipore 07–729), or control rabbit IgG (Santa Cruz Biotechnology), using Illumina-based sequencing as described (Lu et al, 2012).
The public CTCF data are from ENCODE data series GSE19622 (Lee et al, 2012). The RNAPII data are from data series GSE19484 (Kasowski et al, 2010). The Rad21 data are from ENCODE/HAIB data series GSE32465. All three data used were from LCL lines.
Mapping ChIP-Seq to human subtelomeres
The human subtelomere reference assemblies used for the mapping studies represent the most distal 15 kb of DNA sequence adjacent to the (TTAGGG)n terminal repeat tract for the indicated telomeres. Each assembly is oriented with the telomere end on the left with nucleotide position 1 corresponding to the first (CCCTAA) of the tract, which continues to the left of this position but was truncated for mapping consistency purposes. Some of these sequences were available in HG19 (Riethman et al, 2004) whereas others were assembled by merging new fosmid sequence data with HG19 to bridge remaining gaps. In several instances, structural variants corresponding to alternative subtelomere alleles were also included in the set of subtelomere assemblies used here because they differed substantially from the original reference telomere. The full set of subtelomere assemblies is described in detail in Stong et al (in preparation). All of the sequences in the described orientation are available in FASTA format in Supplementary File S1.
Reads were mapped to the subtelomere reference using bowtie (Langmead et al, 2009). Many subtelomeres are duplicon rich with duplicon-specific nucleotide sequence similarities ranging from 90 to 99% between individual members of duplicon families that occur on separate subtelomeres (Linardopoulou et al, 2005; Riethman et al, 2005; Ambrosini et al, 2007; Descipio et al, 2008). To deal with this issue, we required a perfect match to retain a read, and all perfect matches of a given read to positions within the reference assemblies were recorded. Multiply mapping reads were dealt with as described previously (Faulkner et al, 2008), by assigning weights to reads such that multiple mapping positions sum to one read. Mapping likelihood was added to the reads as the inverse of the number of mapping positions. Picard (picard.sourcefourge.net) was used to mark and remove pcr duplicates. Coverage maps were then constructed using the mapping likelihood as a weight and extending the reads to the appropriate fragment length in the data set. The coverage map was calculated at single base resolution. Enrichment profiles were made from comparing RPM values between sample and IgG control. RPM=(coverage at position)/(total reads in library/10ˆ6). The complete read mapping statistics (including Unique versus Multimapping Reads) are available for each of the data sets used in Supplementary File S2. All figures were generated on the subtelomere reference genome hosted at the Wistar mirror of the UCSC genome browser, http://vader.wistar.upenn.edu.
Plasmids, transfections, and luciferase assay
Luciferase reporter constructs containing 10q subtelomeric fragment (1208–396, 794–396, 1208–768, 1142–768, 926–768, 768–1208, or 768–1142) were generated from 10q bacmid by PCR amplification and cloning into XhoI–HindIII sites of pGL3 basic vector (Stratagene). Reporter plasmid with CTCF site mutations was generated using 10q (1208–396) reporter plasmid as a template by using the QuickChange method (Stratagene, Inc.). The Renilla control vector pGL4.74 was purchased from Promega. Luciferase assay were performed in triplicates by transfecting 0.5 μg of reporter plasmid and 10 ng Renilla control vector into 2 × 105 HCT116 cells with Lipofectamine 2000 reagent (Invitrogen). Transfected cells were collected at 48 h post-transfection and assayed for relative luciferase activity using the Promega dual-luciferase reporter assay system. All data points were averages of relative luciferase activity to Renilla from three independent transfections. Transfections with siRNA were performed by the use of DharmaFECT 1 reagent (Thermo Scientific) according to the manufacturer’s instruction. Briefly, ∼2 × 106 cells were plated in antibiotic-free medium in 10 cm plates 12–16 h prior to transfection. Cells were transfected twice within 24 h with 50 nM final concentration for siRNA, and the transfected cells were collected at 4 days post-transfection for further analysis. siRNAs were purchased from Thermo Scientific with target sequence as followings: siControl (D-001206-03, ATGTATTGGCCTGTATTAG), siCTCF-1 (J-020165-10, GAACAGCCCAUAAACAUAG), and siCTCF-2 (D-020165-19, AAACAUACCGAGAACGAAA).
Lentiviral shRNA infection
pLKO.1 vector-based shRNA constructs for knocking down CTCF and Rad21 were obtained from Open Biosystems. shControl in pLKO.1 vector (target sequence TTATCGCGCATATCACGCG) was designed to poorly target the Escherichia coli DNA polymerase and confirmed not affecting any human gene transcription. Packaging vectors pMDLg/pRRE, RSV-Rev, and CMV-VSVG were used for lentiviral production. Briefly, 8 μg shRNA constructs were cotransfected with 4 μg of each packaging vectors into 3 × 106 low passage 293T cells. Viral supernatants were harvested with 0.45 mm syringe filter at both 48 and 72 h after transfection. About 5 × 106 U2OS cells or HCT116 cells were infected twice with 48 and 72 h lentiviral preparation, respectively in 24 h time period. The infected cells were treated with 1 μg/ml Puromycin 24 h after the second infection for selection, and harvested at 6 days post-infection for further analysis.
RNA preparation and analysis
Total RNA was purified with Trizol reagent (Invitrogen) as per the manufacturer’s instruction. Briefly, each sample was mixed with 1 ml of Trizol followed by adding 200 ml of chloroform. The mix was centrifuged at 12 000 g for 15 min at 4°C, and the aqueous phase were collected and subjected to equal volume of isopropanol precipitation. RNA precipitates were collected by centrifugation at 12 000 g for 10 min, washed with 75% ethanol, air-dried, and resuspended in RNase-free water. These samples were treated with DNase I for 45 min at 37°C, followed by DNase I inactivation in the presence of EDTA at 65°C for 5 min. For Northern blotting, about 7.5–10 μg of total RNA were denatured in sample loading buffer (Ambion) for 15 min at 65°C, separated by 1.2% agarose-formamide gel in 1 × MOPS buffer at 5 V/cm, transferred to GeneScreen Plus blotting membranes (Perkin-Elmer) with 10 × SSC, and UV crosslinked onto membrane at 125 mJ in UV Stratalinker 2400 (Stratagene). Hybridizations were performed using Church buffer (0.5 N Na-phosphate, pH 7.2, 7% SDS, 1 mM EDTA, 1% bovine serum albumin (BSA)) for 16–18 h at 50°C. The membrane was washed twice in 0.2 N Na-phosphate, 2% SDS, 1 mM EDTA at room temperature, once in 0.1 N Na-phosphate, 2% SDS, 1 mM EDTA at 50°C, and analysed by phosphor-imager (Amersham Biosciences). The blots were first hybridized with a 32P-labelled (TAACCC)4 probe, then stripped, and probed with a 32P-labelled 18S probe. When indicated, RNA samples were treated with RNase A (Roche) at a final concentration of 100 μg/ml for 30–60 min at 37°C. Images were processed with a Typhoon 9410 Imager (GE Healthcare) and quantified with ImageQuant 5.2 software (Molecular Dynamics). TERRA RNA levels were calculated as percentage relative to signals from control samples and 18S internal control.
RT–PCR experiments were performed as described (Deng et al, 2009, 2012). Briefly, 1 μg of RNA was reverse transcribed using (CCCTAA)5 oligonucleotides for TERRA or random decamers for other genes with Super Script III Reverse Transcriptase from Invitrogen. In all, 100 ng of cDNA was then analysed by real-time PCR using a SYBR green probe with ABI Prism 7900 Sequence Detection System (Applied Biosystems) based on the manufacturer’s specified parameters. Relative RT–PCR was determined using ΔΔCT methods relative to control samples and internal control Gapdh. Primer sequences used for real-time PCR are listed in Supplementary Table 1 and the melting curve control for each primer sets is shown in Supplementary Figure S16. Because of the known subterminal sequence organization characteristic of subsets of human subtelomeres (‘subterminal sequence families’; Riethman, 2008a, 2008b) and the known within-family sequence similarities, we anticipated that multiple telomeres with identical predicted TERRA priming sites would be sampled by some TERRA RT–PCR primer sets. The qRT–PCR signal from TERRA relative to the control in a sample should therefore be considered the sum of the signals from TERRA molecules originating from these discrete sites with identical sequence, perhaps contributing to some of the apparent variability in the quantity of TERRA detected by some of the individual TERRA RT–PCR assays. However, comparison of TERRA levels between samples for the same telomere subsets are not affected.
TIF assay
TIF assay was performed as described (Dimitrova and de Lange, 2006) with some modifications. Briefly, cells grown on coverslips were fixed for 15 min in 2% paraformaldehyde at RT, followed by 15 min in 100% methanol at −20°C. After rehydration in PBS for 5 min, cells were incubated for 30–60 min in blocking solution (1 mg/ml BSA, 3% fetal bovine serum, 0.1% Triton X-100, 1 mM EDTA in PBS) before immuno-staining. Primary antibodies were prepared in blocking solution as following dilutions: monoclonal anti-53BP1 (gift of Dr Thanos Halazonetis, 1:40), anti-γH2AX (Millipore, 1:100), and rabbit polyclonal TRF2 (1:1600). For ImmunoFISH, cells were immunostained as described above, and were fixed in 4% paraformaldehyde in PBS for 10 min. Cells were washed in PBS, dehydrated in ethanol series (70, 95, 100%), and air-dried. Coverslips were denatured for 5 min at 80°C in hybridization mix (70% formamide, 10 mM Tris–HCl (pH 7.2), and 0.5% blocking solution (Roche)) containing telomeric PNA-Tamra-(CCCTAA)3 probe. After denaturation, hybridization was continued for 2 h at room temperature in the dark. Coverslips were washed twice for 15 min each with 70% formamide, 10 mM Tris–HCl (pH 7.2), and 0.1% BSA, and followed by three washes for 5 min each with 0.15 M NaCl, 0.1 M Tris–HCl (pH 7.2), and 0.08% Tween-20. Nuclei were counterstained with 0.1 μg/ml DAPI in blocking solution and slides were mounted with VectorShield (Vector Laboratories, Inc.). IF images were taken with a × 100 lens on a Nikon E600 Upright microscope (Nikon Instruments, Inc., Melville, NY) using ImagePro Plus software (Media Cybernetics, Silver Spring, MD) for image processing. Cells with five or more 53BP1 or γH2AX foci colocalizing with TRF2 foci or telomere DNA foci were scored as TIF positive. P-value was calculated by two-tailed Student’s t-test from at least three independent TIF assays.
ChIP assays
ChIP assays were performed as described previously (Deng et al, 2009). Quantification of ChIP DNA at subtelomeric regions was determined using real-time PCR and the Absolute Quantification program with ABI 7900 Sequence Detection System (Applied Biosystems). PCR data were normalized to input values that were quantified in parallel for each experiment. Primer sequences used for real-time PCR were designed using Primer Express (Applied Biosystems), and listed in Supplementary Table 2. The melting curve control for each primer sets used in ChIP-qPCR was shown in Supplementary Figure S15. As was described for the ChIP-seq read mappings and the TERRA detection, multiple telomeres having identical sequence will be recognized by some of the ChIP-qPCR primers. The same rational for ChIP-Seq mappings applies to ChIP-qPCR quantitation. Since the sequence organization of distal DNA in each subterminal sequence family is the same and the sequence is unique to subtelomeres, an enrichment in a specific subterminal signal relative to appropriate input controls is a valid measure of the average enrichments for all of the identical mapping sites for the subterminal sequence family.
ChIP DNA at telomeres was quantitated by dot-blotting with probes specific for telomere repeat DNA or Alu repeat. ChIP DNA was denatured, dot-blotted onto GeneScreen Plus blotting membranes (Perkin-Elmer) and crosslinked at 125 mJ. Oligonucleotide probes telomere repeats (4 × TTAGGG or 4 × TAACCC) or Alu repeats (cggagtctcgctctgtcgcccaggctggagtgcagtggcgcga) were labelled with γ-[32P]ATP (3000 Ci/mmol) and T4 nucleotide kinase (New England Biolabs). The membrane was prehybridized in Church hybridization buffer for 2 h at 42°C. A heat-denatured 32P-labelled probe was added and hybridized at 42°C overnight. Membrane was washed three times in 0.04 N Na-phosphate, 1% SDS, 1 mM EDTA at 42°C, developed with a Typhoon 9410 Imager (GE Healthcare) and quantified with ImageQuant 5.2 software (Molecular Dynamics). Antibodies used in ChIP assay include rabbit polyclonal antibodies to TRF1, Rad21, and RNAPII S2 (Abcam), CTCF, histone H3 K4 di- or tri-methylation, histone H3 K9 di- or tri-methylation (Millipore), SMC1 and SMC3 (Bethyl Labs), and RNAPII (Santa Cruz). Rabbit antibodies to TRF1 and TRF2 were generated against recombinant protein and affinity purified.
EMSA
EMSAs by CTCF were performed as described (Chau et al, 2006). Briefly, double-stranded DNA probes covering CTCF sites at XYq, 10q, and 7p subtelomeres were generated by annealing one oligonucleotide to the complementary strand and end-labelling with γ-[32P]ATP (3000 Ci/mmol) and T4 nucleotide kinase (New England Biolabs). In a 20-μl reaction mixture,∼10 000 c.p.m. of 32P-labelled DNA probe (12.5 nM) was added to a reaction mixture containing 0.2 μg poly(dI−dC), 5% glycerol, 0.1 mM ZnSO4, 100 mM KCl, 10 mM Tris–HCl (pH 7.5), 0.1% NP-40, 1 mM β-mercaptoethanol, and purified baculoviral His6-tagged-CTCF (∼0.6–6 μM). Reaction mixtures were incubated for 30 min at 25°C, electrophoresed in a 5% nondenaturing, polyacrylamide gel at 110V, and processed by PhosphorImager and Typhoon 9410 Imager (GE Healthcare).
Fluorescence polarization assay
Fluorescence polarization experiments were used to measure inhibition constant (Ki) of CTCF sites and were performed in 384 well black OptiwellTM plates. All wells were initially filled with 75 μl of assay buffer (100 mM Tris pH 8.0, 50 mM KCl, 0.6 mg/ml BSA, 0.075% Tergitol-type NP-40) for 60 min at room temperature to prevent nonspecific binding. To each well, 2 nM of FAM6-labelled 36 bp dsDNA probe (TCAGAGTGGCGGCCAGCAGGGGGCGCCCTTGCCAGA) with a known dissociation constant of 17 nM for CTCF’s 11 zinc-finger binding domain (CTCF11ZF) was added to increasing concentrations of unlabelled dsDNA probe (0–5 μM). In all, 17 nM CTCF11ZF recombinant protein was then added to a final volume of 30 μl for each well and incubated for 60 min at 4°C. Polarization values in millipolarization units (mP) were measured using an Envision 2104 Multilabel Reader (Perkin-Elmer) at an excitation wavelength at 485 nM and an emission wavelength at 530 nM. Each measurement was completed in triplicate. All experimental data were analysed using Prism 3.0 software and the inhibition constants were determined by fitting to the model logIC50=log(10ˆl[logKi*(1+[Fluo. Probe]/[Fluo. Probe Kd)] where Y=(low binding threshold)+(high binding threshold−low binding threshhold)/(1+10ˆ(X−LogIC50)).
Telomere length assay
Telomere length assay was performed as described previously (Deng et al, 2009). Pulsed-Field gel electrophoresis (PFGE) was performed with 1% agarose gel at 6 V/cm, switch times from 1 to 12 s, 15°C for 12 h using CHEF-DRIII system (Bio-Rad). The blots were first hybridized with either a DIG- or 32P-labelled TTAGGG repeat probe, then stripped with 0.1 × SSC and 0.2 N NaOH at 50°C, and probed with a DIG- or 32P-labelled Alu repeat probe, as indicated. Relative telomere-repeat signals were determined either by DIG detection system (Roche) or by Typhoon 9410 Imager (GE Healthcare).
MNase pattern assay
Telomeric nucleosome patterns were analysed by micrococcal nuclease (MNase) digestion. Briefly, nuclei isolated from siRNA or shRNA transfected cells were treated with increased concentration of MNase for 5 min at 37°C. After digestion, DNA was isolated by phenol/chloroform extraction, fractionated on 1.2% agarose gel, and hybridized with 32P-labelled (TAACCC)4 probe or 32P-labelled Alu probe. The probe specific for XYq subtelomeres was generated by radio-labelling an oligonucleotide (46 mer) spanning XYq CTCF site.
Supplementary Material
Acknowledgments
We thank Pu Wang, Jayaraju Dheekollu, and Andreas Wiedmer in our laboratory for their assistance. We also acknowledge contributions from Ravi Gupta and Fred Keeney at the Wistar Cancer Center Core facilities in Bioinformatics, Genomics, and Microscopy. This work was supported by an American Heart Association grant to ZD, the Philadelphia Health Care Trust, a predoctoral NRSA F31 Diversity award to NS (F31HG006395), a predoctoral training grant to RP (T32GM008216), and NIH grants to PML (RO1CA140652), HR (R21CA143349), and MB (R01HD042026). This work was also supported by the Wistar Cancer Center core Grant (P30 CA10815) and the Commonwealth Universal Research Enhancement Program, PA Department of Health.
Author contributions: ZD, HR, and PL designed the experiments for the project. Bioinformatic analysis of ChIP-Seq data sets was done by NS with the help of SH and PW in the laboratory of HR and RD, respectively; RP in MB’s laboratory performed the experiment and analysis for FP; ZD, ZW, AM, and HSC performed other experiments and generated data for the figures. ZD, HR, and PL analysed and interpreted the data, assembled the figures, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ambrosini A, Paul S, Hu S, Riethman H (2007) Human subtelomeric duplicon structure and organization. Genome Biol 8: R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Brun CM, Azzalin CM (2011) TERRA: long noncoding RNA at eukaryotic telomeres. Prog Mol Subcell Biol 51: 65–94 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J (2008) Telomeres: the silence is broken. Cell Cycle 7: 1161–1165 [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenback P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bah A, Wischnewski H, Shchepachev V, Azzalin CM (2011) The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res 40: 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG (2008) Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S (2011) A role for heterochromatin protein 1gamma at human telomeres. Genes Dev 25: 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S (2009) Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL (2009) MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol 29: 4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Shen Z, Prasanth SG (2011) ‘ORCanization’ on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics 6: 665–670 [DOI] [PubMed] [Google Scholar]
- Chau CM, Zhang XY, McMahon SB, Lieberman PM (2006) Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol 80: 5723–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E (2007) CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27: 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- de Lange T (2004) T-loops and the origin of telomeres. Nat Rev 5: 323–329 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Deng Y, Chan SS, Chang S (2008) Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev 8: 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Campbell AE, Lieberman PM (2010) TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle 9: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wang Z, Xiang C, Molczan A, Baubet V, Conejo-Garcia J, Xu X, Lieberman PM, Dahmane N (2012) Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J Cell Sci advance online publication 28 May 2012 doi:; DOI: 10.1242/jcs.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descipio C, Spinner NB, Kaur M, Yaeger D, Conlin LK, Ambrosini A, Hu S, Shan S, Krantz ID, Riethman H (2008) Fine-mapping subtelomeric deletions and duplications by comparative genomic hybridization in 42 individuals. Am J Med Genet A 146A: 730–739 [DOI] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T (2006) MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev 20: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Jackson K, Weemaes C (2006) Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF). Orphanet J Rare Dis 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Newman T, Linardopoulou E, Trask BJ (2002) Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions. Genome Res 12: 1663–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Forrest AR, Chalk AM, Schroder K, Hayashizaki Y, Carninci P, Hume DA, Grimmond SM (2008) A rescue strategy for multimapping short sequence tags refines surveys of transcriptional activity by CAGE. Genomics 91: 281–288 [DOI] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D (2011) Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol 21: 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati A, Magdinier F, Colasanti V, Bauwens S, Pinte S, Ricordy R, Giraud-Panis MJ, Pusch MC, Savino M, Cacchione S, Gilson E (2012) TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS One 7: e34386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8 416–424 [DOI] [PubMed] [Google Scholar]
- Greenwood J, Cooper JP (2011) Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res 40: 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9: 383–387 [DOI] [PubMed] [Google Scholar]
- Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev 7: 311–322 [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP (2011) Telomeric strategies: means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Lieberman PM (2009) Cell cycle control of Kaposi’s sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. J Virol 83: 6199–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Lieberman PM (2011) Mechanism of glycyrrhizic acid inhibition of Kaposi's sarcoma-associated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J Virol 85: 11159–11169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J (2003) Telomere repeat binding factors: keeping the ends in check. Cancer Lett 194: 189–197 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T (2002) Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449 [DOI] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, Hong MY, Karczewski KJ, Huber W, Weissman SM, Gerstein MB, Korbel JO, Snyder M (2010) Variation in transcription factor binding among humans. Science 328: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, Crawford GE, Iyer VR (2012) Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res 22: 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ (2007) Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet 3: e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ (2005) Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, O’Connor MS, Qin J, Songyang Z (2004) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J (1995) Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol 11: 519–548 [DOI] [PubMed] [Google Scholar]
- Lu F, Tsai K, Chen HS, Wikramasinghe P, Davuluri RV, Showe L, Domsic J, Marmorstein R, Lieberman PM (2012) Identification of host-chromosome binding sites and candidate gene targets for KSHV LANA. J Virol 86: 5752–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Lingner J (2009) TERRA: telomeric repeat-containing RNA. EMBO J 28: 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP (2011) Telomere dysfunction and chromosome instability. Mutat Res 730: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K (2011) Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM (2009) CpG-island promoters drive transcription of human telomeres. RNA 15: 2186–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E (2010) Does CTCF mediate between nuclear organization and gene expression? Bioessays 32: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Rival-Gervier S, Boussouar A, Foerster AM, Rondier D, Sacconi S, Desnuelle C, Gilson E, Magdinier F (2009a) The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet 5: e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Schluth-Bolard C, Gilson E, Magdinier F (2011) D4Z4 as a prototype of CTCF and lamins-dependent insulator in human cells. Nucleus 1: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Schluth-Bolard C, Rival-Gervier S, Boussouar A, Rondier D, Foerster AM, Morere J, Bauwens S, Gazzo S, Callet-Bauchu E, Gilson E, Magdinier F (2009b) Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J 28: 2428–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How Shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S (2004) HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Perrod S, Gasser SM (2003) Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell Mol Life Sci 60: 2303–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A (2012) Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J 31: 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF (2012) Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman H (2008a) Human subtelomeric copy number variations. Cytogenet Genome Res 123: 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman H (2008b) Human telomere structure and biology. Annu Rev Genomics Hum Genet 9: 1–19 [DOI] [PubMed] [Google Scholar]
- Riethman H, Ambrosini A, Castaneda C, Finklestein J, Hu XL, Mudunuri U, Paul S, Wei J (2004) Mapping and initial analysis of human subtelomeric sequence assemblies. Genome Res 14: 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethman H, Ambrosini A, Paul S (2005) Human subtelomere structure and variation. Chromosome Res 13: 505–515 [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A (2008) CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10 228–236 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2009) A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ (2010) Transcript elongation by RNA polymerase II. Annu Rev Biochem 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chaiken MF, Wang F, Price CM (2012) Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res 730: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Sun H, Wu J, Wickramasinghe P, Pal S, Gupta R, Bhattacharyya A, Agosto-Perez FJ, Showe LC, Huang TH, Davuluri RV (2011) Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res 39: 190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, Izumi A, Kobayashi M, Meguro H, Kanki Y, Mimura I, Yamamoto K, Mataki C, Hamakubo T, Shirahige K, Aburatani H et al. (2009) A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA 106: 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Wu P, de Lange T (2008) No overt nucleosome eviction at deprotected telomeres. Mol Cell Biol 28: 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G (2011) Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 31: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Wu Y, Gilson E (2010) Dynamics of telomeric chromatin at the crossroads of aging and cancer. Essays Biochem 48: 147–164 [DOI] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S (2008) Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet 17: 2776–2789 [DOI] [PubMed] [Google Scholar]
- Zakian VA (2012) Telomeres: the beginnings and ends of eukaryotic chromosomes. Exp Cell Res 318: 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.