Abstract
Translational repression and mRNA degradation are two major mechanisms for post-transcriptional regulation of gene expression. The detailed relationship between these two processes is not yet well established. Zinc-finger antiviral protein (ZAP) inhibits the replication of certain viruses, including human immunodeficiency virus 1, by binding directly to specific viral mRNAs and recruiting cellular mRNA degradation machinery to degrade the target mRNA. Here, we report that ZAP also inhibits the translation of target mRNAs by interfering with the interaction between translational initiation factors eIF4G and eIF4A. Furthermore, we provide evidence that translational repression is required for mRNA degradation and that blocking the degradation of target mRNAs does not affect ZAP-mediated translational repression. We conclude that ZAP can both repress translation and promote degradation of target mRNA, and that translational repression precedes and is required for mRNA degradation.
Keywords: eIF4A, mRNA decay, translational repression, ZAP
Introduction
Translational repression and mRNA decay are two important aspects of post-transcriptional regulation of gene expression in mammalian cells (Garneau et al, 2007; Jackson et al, 2010). Most translational repression occurs at the translational initiation step, which consists of an ordered series of sequential interactions among mRNA, eukaryotic translational initiation factors (eIFs) and ribosome (Jackson et al, 2010). In eukaryotic translational initiation, the m7G cap structure is bound by eIF4E, while the poly(A) tail is bound by poly(A)-binding protein (PABP). Both eIF4E and PABP interact with eIF4G, which may result in a head–tail loop structure that is thought to facilitate translational initiation. To initiate translation, eIF4G recruits the 40S ribosome subunit via the eIF3 complex. The 40S ribosome subunit scans the 5′UTR of the mRNA for the translational start codon. To facilitate this scanning process, eIF4G also recruits RNA helicase eIF4A to disrupt secondary or tertiary structures in the 5′UTR of the mRNA. Once the correct start codon is found, the 60S ribosome subunit joins the 40S ribosome subunit to form an 80S ribosome and translation elongation ensues. Disruption of any of the steps in this process can result in translational repression.
General mRNA decay and most forms of specific mRNA decay begin with removal of the poly(A) tail by various deadenylases, such as the Ccr4–NOT complex, the Pan2/Pan3 complex and poly(A) ribonuclease (PARN) (Garneau et al, 2007). Deadenylated mRNA is degraded by the 3′–5′ exoribonuclease complex exosome (Liu et al, 2006). Deadenylation also promotes decapping of the mRNA by the Dcp1/Dcp2 complex, and removal of the cap structure allows the RNA body to be degraded by the 5′–3′ exoribonuclease XrnI. In mammalian cells, 3′–5′ degradation is considered to be the major pathway for mRNA decay (Couttet et al, 1997; Wang and Kiledjian, 2001; Mukherjee et al, 2002).
There is increasing evidence that mRNA degradation and translational repression are coupled (Coller and Parker, 2005; Wax et al, 2005; Lal et al, 2006; Liao et al, 2007; Isken et al, 2008). However, the precise relationship between translational repression and degradation of mRNA is not well understood. Translational repression of AU-rich element (ARE)-containing mRNAs has been observed in ARE-mediated mRNA decay (Wax et al, 2005; Lal et al, 2006; Liao et al, 2007). Since therein mRNA degradation and translational repression are mediated by separate trans-acting factors, it is difficult to determine the relationship between these two processes. In microRNA-mediated gene silencing, both translational repression and mRNA decay have been observed (Humphreys et al, 2005; Nottrott et al, 2006; Wu et al, 2006; Chendrimada et al, 2007; Kiriakidou et al, 2007; Wakiyama et al, 2007). Recent reports show that translational repression precedes mRNA decay in microRNA-mediated gene silencing (Bazzini et al, 2012; Djuranovic et al, 2012). In nonsense-mediated mRNA decay (NMD), phosphorylation of Upf1 triggers its interaction with eIF3 to block translational initiation and promotes recruitment of mRNA decay machinery (Isken et al, 2008). Escape from translational repression prevents NMD (Isken et al, 2008), supporting the notion that translational repression precedes mRNA decay. It remains to be determined whether the relationship between translational repression and mRNA decay observed with NMD and microRNA-mediated gene silencing can be generalized for other mRNA decay processes.
Zinc-finger antiviral protein (ZAP) is a host factor that specifically inhibits the replication of certain viruses including human immunodeficiency virus 1 (HIV-1), Moloney murine leukaemia virus (MLV) and Sindbis virus (SINV) (Gao et al, 2002; Bick et al, 2003; Zhu et al, 2011). ZAP selectively targets the multiply spliced mRNAs of HIV-1 for degradation and has little effect on unspliced or singly spliced mRNAs (Zhu et al, 2011). ZAP binds directly to its target RNA, recruits PARN to shorten the poly(A) tail and recruits the 3′–5′ exoribonuclease complex exosome to degrade the RNA body (Guo et al, 2004, 2007; Zhu and Gao, 2008; Zhu et al, 2011; Chen et al, 2012). ZAP also recruits the decpapping complex Dcp1a/Dcp2 via its co-factor RNA helicase p72 to initiate mRNA degradation by 5′–3′ exoribonuclease Xrn1 (Zhu et al, 2011).
In the present study, we report that ZAP also represses the translation of target mRNA by disrupting translational initiation via its interaction with eIF4A. We provide evidence that translational repression precedes and is required for mRNA decay in ZAP-mediated inhibition of viral infection.
Results
ZAP inhibits translation of target mRNA
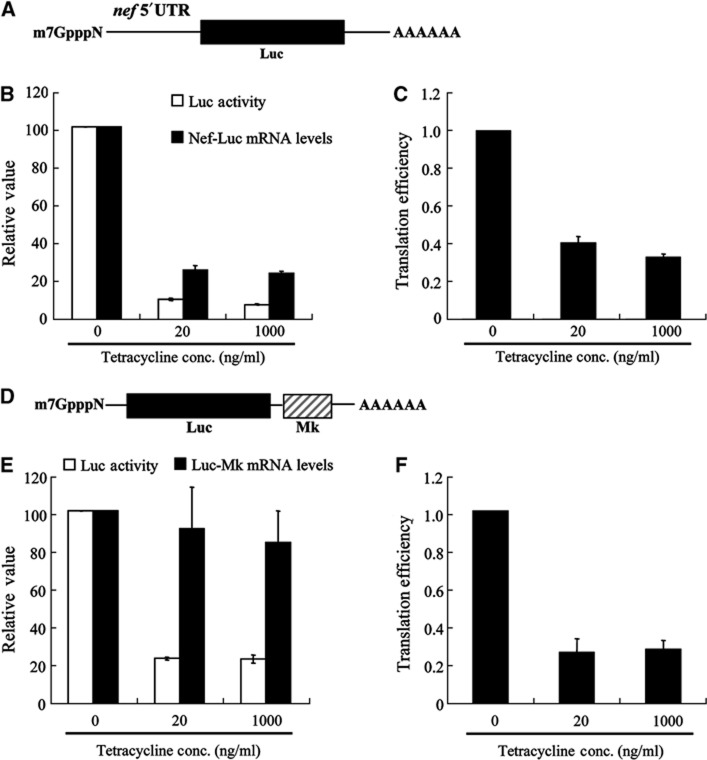
ZAP was expressed in a tetracycline-inducible manner in a stable cell line, 293TREx-ZAP (Zhu et al, 2011; Supplementary Figure S1A). Cells were infected with HIV-1 vector NL4-3-luc, which expresses a multiply spliced Nef-luc mRNA that is targeted by ZAP (Zhu et al, 2011; Figure 1A). ZAP expression was induced with two concentrations of tetracycline. At 20 ng/ml, tetracycline induced ZAP mRNA expression to a level comparable to the endogenous ZAP mRNA level in CD3/CD28-activated human memory CD4 T cells, while at 1000, ng/ml, tetracycline induced about two-fold higher ZAP expression (Supplementary Figure S1B). Luciferase activity and Nef-luc mRNA levels were measured with or without ZAP expression. At both expression levels, ZAP reduced the luciferase activity about 18-fold, but reduced the level of cytoplasmic Nef-luc mRNA only about seven-fold (Figure 1B; Supplementary Table S1 for raw data). This discrepancy suggests that ZAP either promotes the degradation of firefly luciferase protein translated from Nef-luc mRNA or represses the translation of Nef-luc mRNA. To test the first possibility, cells were treated with cycloheximide to block translation and luciferase activity was measured at different time points thereafter. Results show that ZAP did not affect the stability of luciferase (Supplementary Figure S2), suggesting that ZAP may repress the translation of Nef-luc mRNA. A reduction in the relative translation efficiency of Nef-luc mRNA, defined as the protein level divided by the mRNA level, by about 2.5-fold would account for the observed reduction of luciferase protein levels (Figure 1C; Supplementary Table S1 for raw data).
Figure 1.
ZAP inhibits the translation of target mRNA. (A) Schematic representation of Nef-luc mRNA. (B) 293TREx-ZAP cells were infected with VSV-G pseudotyped NL4-3-luc virus for 3 h, mock-treated (0) or treated with tetracycline at a final concentration of 20 or 1000 μg/ml for additional 48 h to induce ZAP expression. One-tenth of the cells were lysed and luciferase activity was measured (open bars). The luciferase activity in mock-treated cells was set as 100. Cytoplasmic RNA was extracted from the remaining cells and levels of Nef-luc mRNA were measured by real-time PCR (black bars). The levels of Nef-luc mRNA normalized to that of GAPDH mRNA from mock-treated cells were set as 100. (C) Translation efficiency of Nef-luc mRNA was calculated as luciferase activity divided by Nef-luc mRNA level in (B). Translation efficiency of Nef-luc mRNA in mock-treated cells was set as 1. (D) Schematic representation of Luc-Mk mRNA. (E) 293TREx-ZAP cells were transfected with the pCMV-FL-Mk reporter and then mock-treated (0) or treated with tetracycline at a final concentration of 20 or 1000 μg/ml for 48 h to induce ZAP expression. One-tenth of the cells were lysed and luciferase activity was measured (open bars). The luciferase activity in mock-treated cells was set as 100. Cytoplasmic RNA was extracted from the remaining cells and levels of Luc-Mk mRNA were measured by real-time PCR (black bars). The levels of Luc-Mk mRNA normalized to that of GAPDH mRNA from mock-treated cells were set as 100. (F) Translation efficiency of Luc-Mk was calculated as luciferase activity divided by Luc-Mk mRNA level in (E). Translation efficiency of Luc-Mk in mock-treated cells was set as 1. Data presented are mean values±s.d. from three independent experiments.
The ZAP-responsive element (ZRE) in Nef-luc mRNA is in the 5′UTR region (Zhu et al, 2011). To analyse whether the location of ZRE affects ZAP-mediated translational repression, we tested another reporter construct. In addition to HIV-1, ZAP inhibits the replication of SINV (Bick et al, 2003). Luc-MK reporter contains a ZAP-responsive fragment of SINV (named Mk) at the 3′UTR of the reporter mRNA (Figure 1D). Expression of ZAP reduced the luciferase activity from Luc-Mk about five-fold. Unlike Nef-luc mRNA, the Luc-Mk mRNA levels were just slightly reduced (Figure 1E; Supplementary Table S2 for raw data). As a result, the translation efficiency of Luc-Mk was reduced four-fold by ZAP (Figure 1F; Supplementary Table S2 for raw data). To prove that the reduced translation efficiency of Luc-Mk is not a result of ZAP-mediated damage of the mRNA, total RNA was extracted from the cells with or without ZAP expression and subjected to in vitro translation. Comparable luciferase activity was expressed with or without ZAP, indicating that the translatability of the reporter mRNA was not affected by ZAP (Supplementary Figure S3). These results suggest that ZAP represses the translation of ZRE-containing mRNA independent of the location of ZRE or mRNA decay and that ZAP-mediated translational repression of Luc-Mk mRNA is not a result of damage to the mRNA.
ZAP excludes target mRNA from polysome
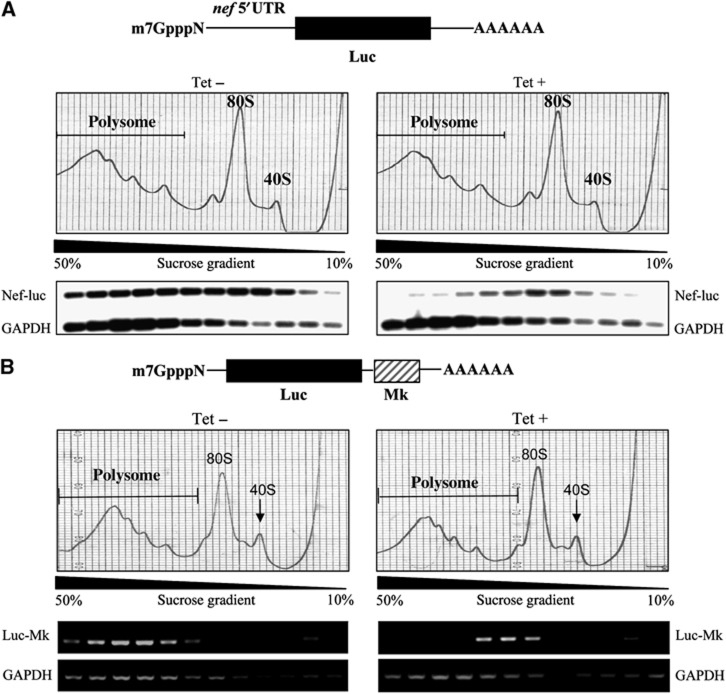
To further substantiate the notion that ZAP represses translation, polysome profile analysis was performed to analyse the translation state of Nef-luc and Luc-Mk mRNAs. ZAP expression did not change the global polysome profile, suggesting that ZAP did not affect global mRNA translation (Figure 2A and B). Without ZAP, more than half of the Nef-luc mRNA was associated with the polysome fractions. However, in the presence of ZAP, most of the Nef-luc mRNA was excluded from polysome fractions, indicating that translation of Nef-luc mRNA was inhibited (Figure 2A). Similar results were also obtained for the Luc-Mk mRNA: expression of ZAP excluded the mRNA out of the polysome fractions (Figure 2B). The distribution of GAPDH mRNA was not affected by ZAP expression (Figure 2). These results confirmed that ZAP specifically inhibits the translation of its target mRNA.
Figure 2.
Expression of ZAP excludes target mRNA out of polysome. (A) 293TREx-ZAP cells were infected with VSV-G pseudotyped NL4-3-luc virus for 3 h, and mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet+) for additional 48 h to induce ZAP expression. Cells were then treated with 50 μg/ml cycloheximide to stop global translation. Cell lysates were resolved in a 10–50% continuous sucrose gradient by ultracentrifugation and polysome profiles were obtained by monitoring absorbance at 254 nm. Nef-luc and GAPDH mRNA levels in each fraction were determined by RT–PCR, followed by Southern blotting. (B) 293TREx-ZAP/FL-Mk cells were mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet+) for 48 h to induce ZAP expression, followed by treatment with 50 μg/ml cycloheximide to stop global translation. Cell lysates were resolved in a 10–50% continuous sucrose gradient by ultracentrifugation and polysome profiles were obtained by monitoring the absorbance at 254 nm. Luc-Mk and GAPDH mRNA levels in each fraction were determined by RT–PCR. Figure source data can be found with the Supplementary data.
Downregulation of endogenous ZAP enhances translation of target mRNA
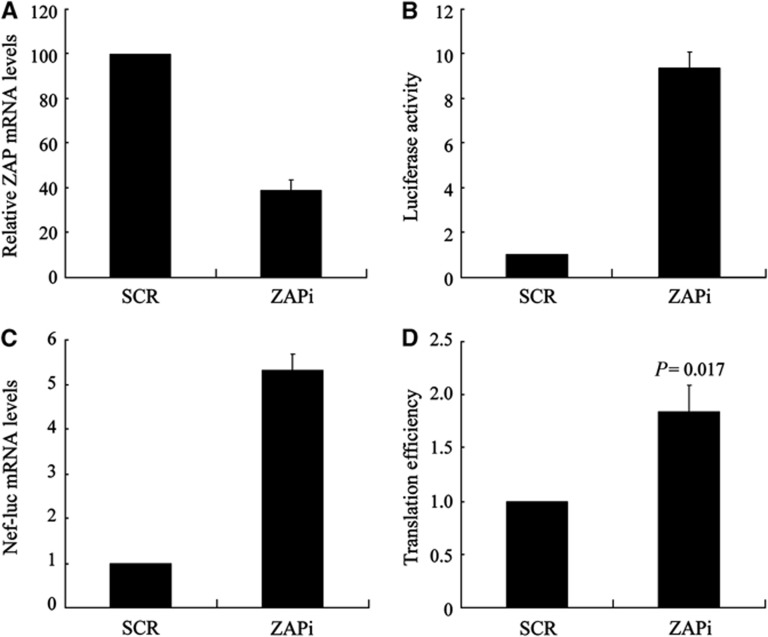
To test whether endogenous ZAP represses the translation of target mRNA, Jurkat cells stably expressing control shRNA (Jurkat-SCR) or an shRNA against ZAP (Jurkat-ZAPi) were generated. The ability of the shRNA to downregulate ZAP expression was confirmed by Western analysis following co-transfection of the shRNA-expressing construct with a plasmid expressing myc-tagged ZAP into HEK293 cells (Supplementary Figure S4). The expression level of ZAP in Jurkat-ZAPi cells was reduced to about 40% of that in control Jurkat-SCR cells (Figure 3A). The cells were infected with VSV-G pseudotyped NL4-3-luc vector. In Jurkat-ZAPi cells, approximately a 10-fold increase in luciferase activity and a five-fold increase in Nef-luc mRNA levels were observed compared with those in the control cells (Figure 3B and C). The translation efficiency of Nef-luc mRNA, calculated by dividing luciferase activity with the mRNA level, was increased about two-fold in Jurkat-ZAPi cells (Figure 3D). These results established that downregulation of endogenous ZAP increased the translation of its target mRNA.
Figure 3.
Downregulation of ZAP enhances the translation of target mRNA. (A) The endogenous ZAP mRNA levels in Jurkat cells stably expressing a control shRNA (SCR) or a shRNA against ZAP (ZAPi) were measured by real-time PCR. The relative ZAP mRNA level in Jurkat-SCR cells was set as 100. (B, C) The cells were infected with VSV-G pseudotyped NL4-3-luc. At 48 h post-infection, 1/10 of the cells were lysed and luciferase activity was measured (B). The luciferase activity in Jurkat-SCR cells was set as 1. Cytoplasmic RNA was extracted from the remaining cells and levels of Nef-luc mRNA were measured by real-time PCR (C). The levels of Nef-luc mRNA normalized to GAPDH mRNA from Jurkat-SCR cells were set as 1. (D) Translation efficiency of Nef-luc mRNA was calculated as luciferase activity in (B) divided by the Nef-luc mRNA level in (C). Translation efficiency of Nef-luc mRNA in Jurkat-SCR cells was set as 1. Data presented are mean values±s.d. of three independent experiments.
HCV IRES escapes ZAP-mediated translational repression
To probe how ZAP inhibits the translation of target mRNA, we modified the NL4-3-luc vector (Supplementary Figure S5). In this reporter (NL4-3-RL-IRES-FL), translation of renilla luciferase (RL) is dependent on the cap structure, and translation of firefly luciferases (FL) is driven by an internal IRES (Figure 4A). Cap-dependent translational initiation requires eIF4E, eIF4G, eIF4A, eIF3, eIF2 and 40S (Jackson et al, 2010). Unlike cap-dependent translation, translational initiation driven by EMCV IRES is independent of eIF4E, but still requires other translational initiation factors (Jang et al, 1988; Kieft, 2008). For HCV IRES, eIF4E, eIF4G and eIF4A are not required for translational initiation (Tsukiyama-Kohara et al, 1992; Kieft, 2008).
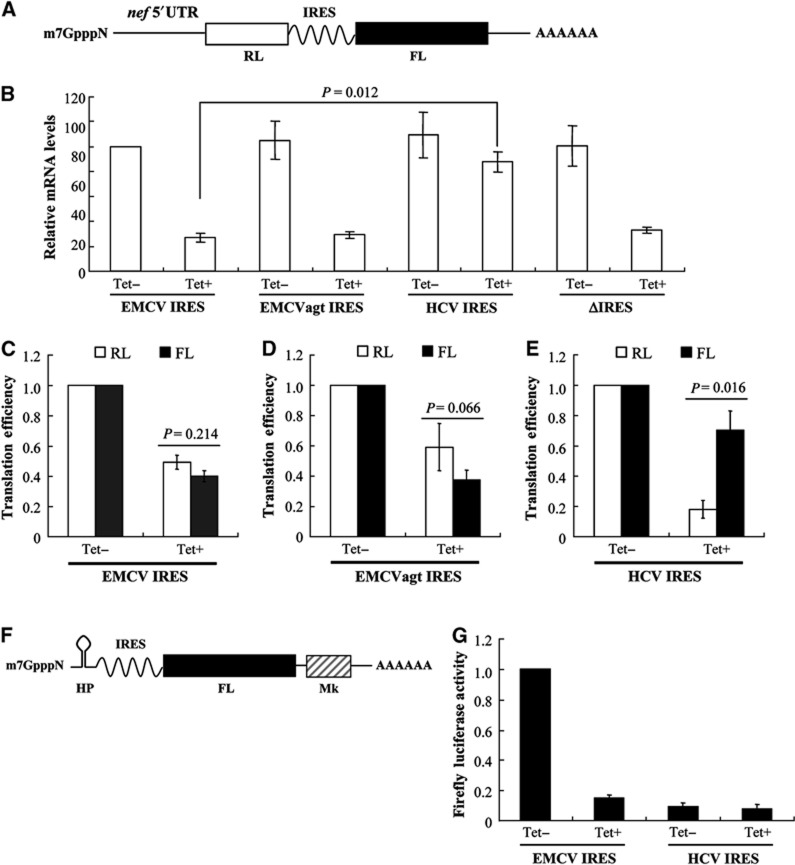
Figure 4.
Ongoing translation inhibits ZAP-mediated mRNA degradation. (A) Schematic representation of Nef-RL-IRES-FL mRNAs. (B–E) 293TREx-ZAP cells were infected with VSV-G pseudotyped NL4-3-RL-EMCV-FL, NL4-3-RL-EMCVagt-FL NL4-3-RL-HCV-FL or NL4-3-RL-ΔIRES-FL virus for 3 h, and mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet+) for an additional 48 h to induce ZAP expression. (B) Cytoplasmic RNA from 90% of the cells was extracted and mRNA levels were measured by real-time PCR and normalized to that of GAPDH mRNA. Levels of Nef-RL-EMCV-FL mRNA from mock-treated cells (Tet−) were set as 100. (C–E) The RL and FL activities in the remaining 10% cells were measured. Cap-dependent translation efficiency was calculated as the RL activity divided by the mRNA level in (B). IRES-dependent translation efficiency was calculated as the FL activity divided by the mRNA level in (B). The translation efficiency of RL or FL in mock-treated (Tet−) cells was set as 1. Data presented are mean values±s.d. from three independent experiments. (F) Schematic representation of IRES-Luc-Mk mRNA with a hairpin (HP) structure to block cap-mediated translation. (G) 293TREx-ZAP cells were transfected with pHP-EMCV-Luc-Mk or pHP-HCV-Luc-Mk, and then mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet+) for 48 h to induce ZAP expression. Cells were lysed and luciferase activity was measured. The luciferase activity from HP-EMCV-Luc-Mk mRNA in mock-treated cells was set as 1. Data are mean values±s.d. from three independent experiments.
293TREx-ZAP cells were infected with VSV-G pseudotyped NL4-3-RL-IRES-FL vectors, and mRNA levels and translational efficiencies with or without ZAP expression were assayed. For NL4-3-RL-EMCV-FL, ZAP expression reduced the mRNA level by about 70% (Figure 4B; Supplementary Table S3, for raw data). However, for NL4-3-RL-HCV-FL, ZAP expression reduced the mRNA level by only about 25% (Figure 4B; Supplementary Table S3 for raw data). When the IRES was deleted from the reporter (ΔIRES), the mRNA level was reduced by about 80% (Figure 4B; Supplementary Table S3 for raw data), comparable to that of NL4-3-RL-EMCV-FL. The translation efficiency of EMCV IRES is about six-fold higher than that of HCV IRES (Supplementary Figure S6). To address the possibility that the different response of the reporters containing EMCV IRES and HCV IRES was due to different translation efficiencies of the IRES, we mutated the GCGA tetraloop of EMCV IRES to AGTA (EMCVagt) to reduce its translation efficiency to about one-tenth of that of HCV IRES (Supplementary Figure S6). ZAP reduced the mRNA level of the EMCVagt-containing reporter by about 75%, comparable to that of the wild-type EMCV IRES-containing reporter (Figure 4B; Supplementary Table S3 for raw data). Collectively, these results suggest that HCV IRES protects reporter mRNA from degradation to some extent but EMCV IRES fails to do so.
We further compared the effect of ZAP expression on the relative translation efficiency of the reporters. For NL4-3-RL-EMCV-FL and NL4-3-RL-EMCVatg-FL, ZAP repressed cap-dependent translation and IRES-mediated translation comparably (Figure 4C and D; Supplementary Table S3 for raw data). In contrast, for NL4-3-RL-HCV-FL, cap-dependent translation was significantly repressed but HCV IRES-mediated translation was only modestly affected (Figure 4E; Supplementary Table S3 for raw data). These results suggest that HCV IRES-dependent translational initiation escaped ZAP-mediated translational repression. Furthermore, these results also suggest that HCV IRES-mediated escape from translational repression protects the mRNA from degradation since the magnitude of reduction in mRNA levels of HCV IRES-containing reporter was much smaller than that of the control EMCV IRES-containing mRNA (Figure 4B). In line with these results, in a reporter wherein cap-dependent translation is blocked by a hairpin structure (Figure 4F), the translation driven by EMCV IRES was dramatically inhibited by ZAP, while ZAP had only a modest effect on the translation driven by HCV IRES (Figure 4G).
ZAP interacts with eIF4A
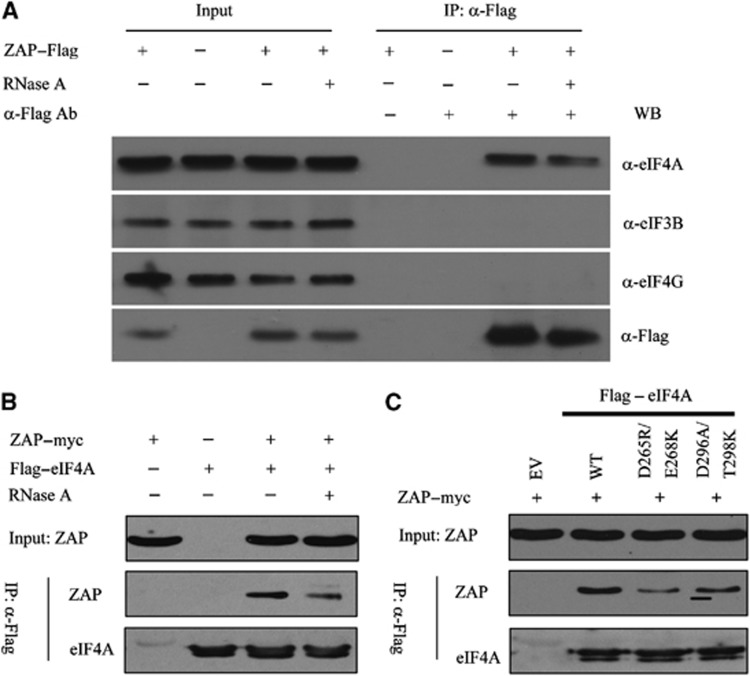
As ZAP inhibited the translation driven by the cap structure and EMCV IRES but not HCV IRES, comparison of the translational initiation factors required by these RNA elements suggests that ZAP may inhibit translational initiation by interfering with interactions involving eIF4G, eIF4A and eIF3. To probe how ZAP interferes with translational initiation, we expressed Flag-tagged ZAP in HEK293T cells and investigated the interactions between ZAP and eIF4A, eIF3 and eIF4G. Co-immunoprecipitation data show that ZAP interacts with endogenous eIF4A (Figure 5A). Moreover, co-immunoprecipitation of ZAP with eIF4A was resistant to RNase A treatment, suggesting that the interaction between ZAP and eIF4A is direct rather than being mediated by nonspecific RNA binding (Figure 5A). In contrast, ZAP did not interact with eIF3 or eIF4G (Figure 5A). The interaction between ZAP and eIF4A was also observed when myc-tagged ZAP and Flag-tagged eIF4A were co-expressed in HEK293T cells, as judged by the co-immunoprecipitation assay (Figure 5B). The direct interaction between ZAP and eIF4A was further confirmed by pull-down assays using purified recombinant ZAP and GST–eIF4A (Supplementary Figure S7).
Figure 5.
ZAP interacts with eIF4A. (A) ZAP interacts with endogenous eIF4A but not eIF4G or eIF3B. HEK 293T cells were transfected with empty vector (ZAP–) or a plasmid expressing Flag-tagged ZAP. Cells were lysed in the presence (+) or absence (–) of 50 μg/ml RNase A. The lysates were immunoprecipitated with mouse IgG (α-Flag Ab–) or Flag antibody (α-Flag Ab+). eIF4A, eIF4G, eIF3B, and ZAP were detected with antibodies indicated. (B) Plasmids expressing myc-tagged ZAP and Flag-tagged eIF4A were transiently co-transfected into HEK293T cells. Cells were lysed in lysis buffer with (+) or without (−) 50 μg/ml RNase A. Lysates were immunoprecipitated with anti-Flag antibody and detected by western blotting using anti-myc or anti-Flag antibody. (C) Plasmids expressing myc-tagged ZAP and Flag-tagged wild-type or mutant eIF4As were transiently co-transfected into HEK293T cells. Cell lysates were immunoprecipitated with anti-Flag antibody and detected by western blotting using anti-myc or anti-Flag antibody. Figure source data can be found with the Supplementary data.
The finding that ZAP interacted with eIF4A, but not with eIF4G or eIF3 subunit, suggests that ZAP might interfere with the interaction between eIF4A and eIF4G. It has been reported that the D296A/T298K mutations of eIF4A (eIF4A–D296A/T298K) reduce its interaction with eIF4G and that the D265R/E268K mutations (eIF4A–D265R/E268K) abrogate the interaction (Oberer et al, 2005; Supplementary Figure S8). Interestingly, these eIF4A mutants also interacted with ZAP with reduced affinities. Compared with wild-type eIF4A, ZAP’s interaction with eIF4A–D296A/T298K was attenuated, while the interaction between ZAP and eIF4A–D265R/E268K was further reduced (Figure 5C). These results suggest that the binding sites of eIF4A for ZAP and eIF4G partially overlap, supporting the notion that ZAP may compete with eIF4G to bind to eIF4A.
ZAP interferes with the interaction between eIF4A and eIF4G
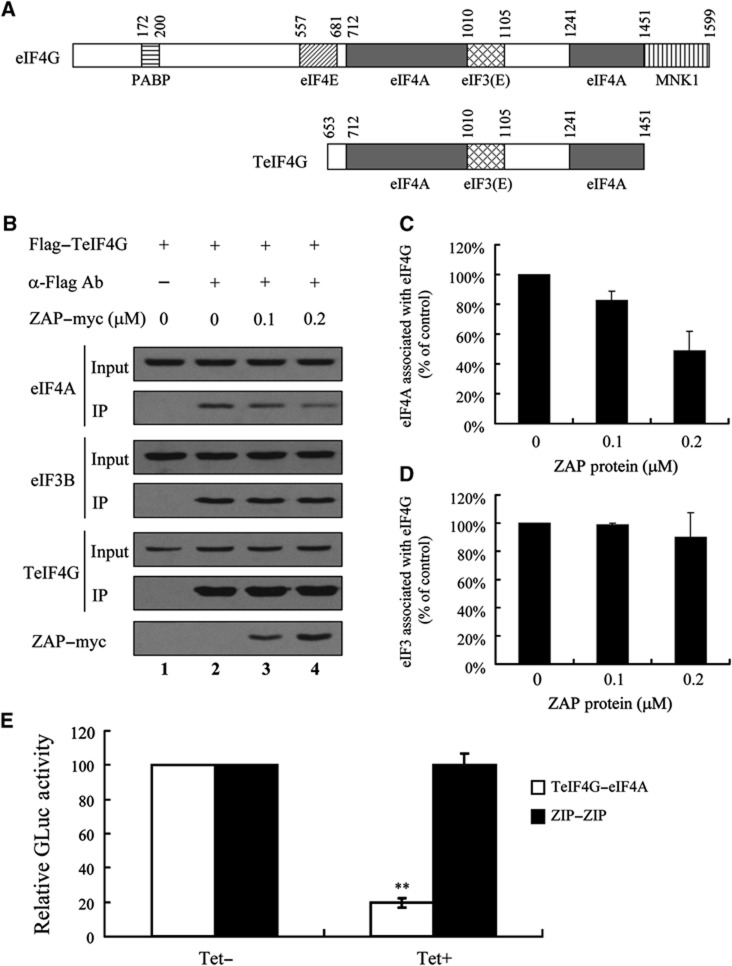
In translational initiation, eIF4G acts as a scaffold protein and interacts with multiple translational initiation factors to facilitate the assembly of the translational initiation complex. eIF4G interacts with the eIF3 complex through the region spanning amino acids 1010–1105 (Korneeva et al, 2000; LeFebvre et al, 2006; Miyakawa et al, 2006). eIF4G interacts with eIF4A through two regions that span amino acids 712–1010 and 1241–1451 (Figure 6A) (Imataka and Sonenberg, 1997; Miyakawa et al, 2006). We used a fragment of eIF4G that spans amino acids 653–1451 (TeIF4G) to recapitulate the interactions of eIF4G with eIF3 and eIF4A (Figure 6A). Co-immunoprecipitation assay data show that both eIF3 and eIF4A associate with TeIF4G (Figure 6B, lane 2).
Figure 6.
ZAP interferes with the interaction between eIF4G and eIF4A. (A) Schematic representation of regions of eIF4G and a fragment of eIF4G (TeIF4G) interacting with indicated translational initiation factors. (B) The plasmid expressing Flag-tagged TeIF4G was transiently transfected into HEK293 cells. At 48 h post-transfection, cells were lysed and the indicated amount of purified myc-tagged ZAP protein (ZAP–myc) was added to the cell lysates. The lysates were immunoprecipitated with mouse IgG (α-Flag–) or with anti-Flag antibody (α-Flag+) and analysed by western blotting to detect endogenous eIF4A, eIF3B, Flag-tagged TeIF4G and myc-tagged ZAP. (C, D) Relative band intensities in (B) representing the amounts of eIF4A (C) and eIF3B (D) were determined using ImageJ software (NCBI). Data presented are mean values±s.e.m. from two independent experiments. (E) 293TREx-ZAP cells were transfected with pZIP-GL1 and pZIP-GL2 (ZIP–ZIP), or p4G-GL1 and p4A-GL2 (TeIF4G–eIF4A) together with pGL3-SV40. At 6 h post-transfection, cells were mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet +) to induce ZAP expression. At 48 h post-transfection, cells were lysed and Gaussia and FL activities were measured. The Gaussia luciferase activity was normalized with that of FL. The Gaussia luciferase activity in mock-treated cells (Tet−) was set as 100. Data presented are mean values±s.d. from three independent experiments. **P⩽0.01. Figure source data can be found with the Supplementary data.
Although the interaction between ZAP and eIF4A suggests that ZAP may interfere with the interaction between eIF4A and eIF4G, ZAP did not affect the translation of GAPDH. We reasoned that ZAP may interfere with the eIF4A–eIF4G interaction in an mRNA-specific manner. Binding of ZAP to its target mRNA is expected to bring it into proximity with eIF4A and eIF4G bound on the target mRNA and thereby increase the local concentration of ZAP. If this is correct, a high concentration of ZAP might be expected to disrupt the interaction between eIF4A and eIF4G. To test this concept, purified recombinant ZAP was added to a lysate of cells expressing TeIF4G (Figure 6B, lanes 3 and 4). Indeed, the interaction between eIF4A and eIF4G was reduced by ZAP in a dose-dependent manner (Figure 6B and C). When 0.1 μM ZAP was added to the cell lysate, the interaction between eIF4A and eIF4G was reduced by about 20%, while 0.2 μM ZAP reduced the interaction between eIF4A and eIF4G by about 50% (Figure 6C). In contrast, the interaction between eIF3 and eIF4G was little affected by ZAP (Figure 6B and D).
We further employed a sensitive protein–protein interaction assay, the split-Gaussia luciferase interaction system, to analyse whether ZAP affects the interaction between eIF4G and eIF4A (Remy and Michnick, 2006; Law et al, 2010). In this system, Gaussia luciferase is split into an N-terminal half fragment (GL1) and a C-terminal half fragment (GL2), and the luciferase activity is disrupted. When GL1 was fused to TeIF4G and GL2 was fused to eIF4A, the interaction between TeIF4G and eIF4A increased the proximity of GL1 and GL2 and thereby reconstituted Gaussia luciferase activity (Figure 6E). Expression of ZAP significantly reduced Gaussia luciferase activity reconstituted by TeIF4G–eIF4A interaction (Figure 6E). As a control, the GCN4 leucine zipper protein (ZIP)-mediated reconstitution of split-Gaussia luciferase activity was not affected by ZAP (Figure 6E). These results further support the notion that ZAP interferes with the interaction between eIF4G and eIF4A.
Blocking mRNA degradation does not affect ZAP-mediated translational repression
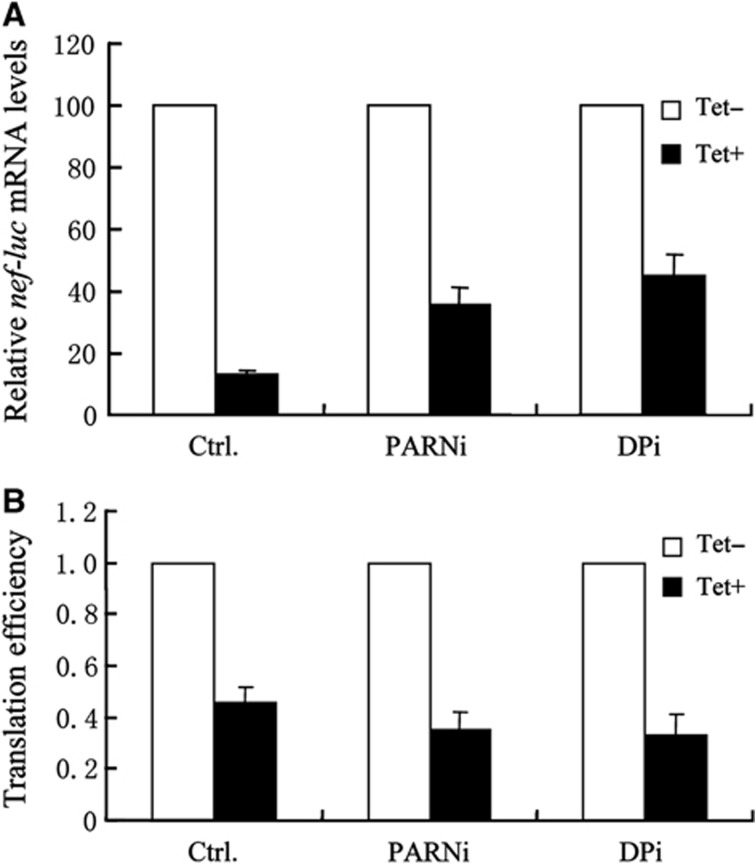
In ZAP-mediated mRNA degradation, deadenylase PARN and decapping enzyme Dcp2 are recruited by ZAP to degrade target mRNA (Zhu et al, 2011). To examine whether ZAP-mediated translational repression is affected by ZAP-mediated mRNA degradation, 293TREx-ZAP cells were transfected with shRNAs to downregulate the expression of PARN and Dcp2, and then challenged with NL4-3-luc vector. Consistent with our previous results, when PARN was downregulated or when PARNA and Dcp2 were downregulated together, ZAP-mediated degradation of Nef-luc mRNA was reduced (Figure 7A). However, downregulation of PARN and Dcp2 modestly enhanced ZAP-mediated translational repression of the mRNA (Figure 7B). These results indicate that ZAP-mediated translational repression is independent of mRNA degradation.
Figure 7.
Blocking mRNA degradation does not affect translational repression. 293TREx-ZAP cells were transfected twice with control shRNA (Ctrl), a shRNA against PARN (PARNi) or shRNAs against PARN and Dcp2 (DPi). Cells were infected with VSV-G pseudotyped NL4-3-luc for 3 h, and mock-treated (Tet−) or treated with 1 μg/ml tetracycline (Tet+) for additional 48 h to induce ZAP expression. (A) Cytoplasmic RNA from 90% of the cells was extracted and the levels of nef-luc mRNA were measured by real-time PCR. Levels of nef-luc mRNA normalized with that of GAPDH mRNA from mock-treated cells were set as 100. (B) FL activity in the remaining 10% cells was measured. Translation efficiency was calculated as FL activity divided by the mRNA level in (A). The translation efficiency of nef-luc mRNA in mock-treated (Tet−) cells was set as 1. Data presented are mean values±s.d. from three independent experiments.
Discussion
We have previously reported that ZAP promotes the degradation of ZRE-containing viral mRNAs (Gao et al, 2002; Guo et al, 2007; Zhu and Gao, 2008; Zhu et al, 2011). In the present study, we report that ZAP also represses translation of target mRNA. mRNA decay itself can result in translational repression; removal of poly(A) tails or the cap structure would disrupt the mRNA cap-eIF4E–eIF4G-PAPB loop and thus repress translation. However, our results indicate that ZAP-mediated translational repression is not a result of mRNA decay. When deadenylation and decapping were inhibited by downregulation of PARN and Dcp2, respectively, ZAP-mediated translational repression was little affected (Figure 7B). Moreover, ZAP affected the stability of Luc-Mk mRNA modestly, but repressed the translation of Luc-Mk mRNA significantly (Figure 1E and F). These results indicated that translational repression is independent of mRNA decay.
ZAP promotes degradation of Nef-Luc mRNA (Figure 1C) but had little effect on Luc-Mk mRNA (Figure 1E). A possible explanation is that the mRNAs were generated differently and the proteins associated with the mRNAs might be different, which are expected to affect the accessibility of cellular mRNA degradation machinery to the mRNA. This possibility is supported by a recent report that suggests that promoters regulate the compositions of mRNP on mRNAs and thus affect the stability of the mRNAs (Bregman et al, 2011). It awaits further investigation to determine under what circumstances ZAP represses the translation and/or promotes the degradation of its target mRNA.
It has been reported that ongoing translation protects mRNA from Upf-1-mediated mRNA decay (Isken et al, 2008; Hogg and Goff, 2010). It is conceivable that translational repression helps to remove protective ribosomes from target mRNA and thus facilitates efficient mRNA degradation. Here, we show that HCV IRES-mediated escape from translational repression protects the target mRNA from ZAP-mediated RNA degradation (Figure 2). Our results provide an additional example supporting the notion that translational repression can facilitate mRNA degradation.
We provide evidence that ZAP represses translation by interacting with eIF4A and thus interfering with the interaction between eIF4A and eIF4G, which is required for translational initiation (Figure 6). ZAP-mediated translational repression appears to be ZRE-containing mRNA-specific since the translation of GAPDH mRNA was not affected by ZAP expression (Figure 1C). We hypothesize that ZAP interferes with only the interaction between eIF4A and eIF4G that are associated with the target mRNA, but has no or little effect on the interaction otherwise. Similar mechanisms have been reported with Programmed Cell Death 4, which directly binds to the coding region of c-myb mRNA, and inhibits translational initiation by binding to eIF4A and thus displacing eIF4G and target mRNA from eIF4A (Suzuki et al, 2008).
The 5′UTRs of HIV-1 mRNAs are highly structured, and are thus supposed to impede efficient ribosome scanning and translational initiation (Pelletier and Sonenberg, 1985; Yilmaz et al, 2006). Multiple mechanisms are employed to regulate HIV-1 mRNA translation (Bolinger and Boris-Lawrie, 2009). Notably, the post-transcriptional control element (PCE) that is required for efficient HIV-1 mRNA translation through interaction with RNA helicase A is located in the R-U5 region (Bolinger et al, 2010). The ZRE in HIV-1 nef mRNA was mapped to the 5′UTR (Zhu et al, 2011), which contains the R-U5 region. It remains to be determined whether binding of ZAP to nef mRNA inhibits translation by interfering with the function of the PCE. Nonetheless, binding of ZAP to the 5′UTR is not always required for ZAP to inhibit translational initiation since ZAP also inhibits the translational initiation of a reporter containing a ZRE in the 3′UTR (Figure 2B). It remains to be determined whether ZAP also represses translational initiation of target HIV-1 mRNA by other means.
ZAP interacts with eIF4A, PARN and the exosome even in the absence of viral mRNAs. In a specific process, it would be more efficient for ZAP to recruit only the factors required for translational repression or mRNA degradation. It will be interesting to investigate how translational repression switches to mRNA decay.
Materials and methods
DNA constructs
Plasmid pNL4-3-luc was obtained from Dr Nathaniel Landau through the National Institutes of Health, AIDS Research and Reference Reagent Program (Connor et al, 1995). pCMV-FL-Mk was constructed by replacing the RL coding sequence in phRL-CMV (Clontech) with the Luc-Mk fragment in pGL3-Mk (Guo et al, 2004) using restriction sites NheI and XbaI. Plasmids pSR-SCR, pSR-PARNi, pSR-Dcp2i and pSR-hZAPi, which express control shRNA and shRNAs directed against PARN, Dcp2 and hZAP, respectively, have been described previously (Zhu et al, 2011). Lentivector pNL4-3RL-EMCV-FL, in which RL expression is dependent on the cap structure and FL expression is dependent on the IRES of EMCV was constructed by inserting the RL-EMCV-FL sequence (kindly provided by Drs Peter Sarnow and Roy Parker (Coller and Parker, 2005)) into pNL4-3-luc using NotI and XhaI sites. pNL4-3RL-EMCVagt-FL, in which the tetraloop of EMCV IRES was mutated from GCGA to AGTA, was constructed by overlapping PCR. Lentivector pNL4-3RL-HCV-FL was constructed in a similar manner. pNL4-3-RL-ΔIRES-FL was constructed based on pNL4-3RL-HCV-FL by deleting HCV IRES and the ATG of FL using overlapping PCR. pHP, which is a plasmid containing a hairpin structure sequence, was constructed by inserting the hairpin sequence (ggtttagtgaaccgtcagatcactgcctaggccggagcgcccagatctgggcgctccggcctaggcagg) (Isken et al, 2008) into phRL-CMV (Promega) with PstI and NheI. EMCV-Luc-Mk and HCV-Luc-Mk were digested with XbaI and NotI, and cloned into pHP digested with NheI and NotI to generate pHP-EMCV-Luc-Mk and pHP-HCV-Luc-Mk, respectively.
Human ZAP-expressing plasmid pcDNA4-hZAP2–myc has been previously described (Zhu et al, 2011). pQCXIP-ZAP–Flag, which expresses Flag-tagged hZAP2, was constructed by inserting Flag-tagged ZAP sequence into pQCXIP (Clontech) between NotI and BamHI. pGST–eIF4A was constructed by inserting eIF4A coding sequence into pGEX5X-1 between BamHI and NotI. pCMV-HF-eIF4A, which expresses Flag-tagged eIF4A was constructed by inserting eIF4A coding sequences into pCMV-HF (Guo et al, 2007) using EcoRI and NotI sites. pcDNA3-HA-eIF4G1-1599, which expresses full-length eIF4G, was kindly provided by Dr Nahum Sonenberg (Yanagiya et al, 2009). pCMV-HF-TeIF4G, which expresses a fragment of eIF4G (amino acids 653–1451), was generated by cloning PCR-derived coding sequence into pCMV-HF using EcoRI and NotI sites. Plasmids expressing Flag-tagged eIF4A D265R/E268K and D296A/T298K mutants were generated by overlapping PCR. pZIP-GL1 and pZIP-GL2 were kindly provided by Dr Margaret R MacDonald and described previously (Remy and Michnick, 2006; Law et al, 2010). p4G-GL1 (expressing TeIF4G fused with GL1) and p4A-GL2 (expressing eIF4A fused with GL2) were constructed by replacing the ZIP sequence in pZIP-GL1 and pZIP-GL2 with that encoding TeIF4G and eIF4A, respectively.
Cell culture
All cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Hyclone). 293TREx-ZAP cells, which express human ZAP isoform 2 (hZAP-v2) in a tetracycline-inducible manner, have been described previously (Zhu et al, 2011). 293TREx-ZAP/FL-Mk cell line, in which the Luc-Mk reporter mRNA is stably expressed, was constructed by co-transfecting 293TREx-ZAP cells with pCMV-FL-Mk and pTRE2 (Clontech) at a ratio of 10:1 followed by selection with hygromycin B.
VSV-G pseudotyped lentiviral vectors were generated by co-transfecting pVSV-G with pNL4-3-luc, pNL4-3-RL-EMCV-FL, pNL4-3-RL-HCV-FL or pNL4-3-RL-ΔIRES-FL into HEK293T cells. Virus-containing supernatants were three-fold diluted with medium containing 10 mM HEPES (pH 7.5) and 8 μg/ml polybrene.
Luciferase assay
Luciferase activity was measured using either the Luciferase Assay System for FL or by the Dual-luciferase Reporter Assay System for both RL and FL (Promega).
RNA extraction and reverse transcription
Cytoplasmic RNA was extracted using an RNeasy Mini Kit (QIAGEN). To extract RNA from polysome fractions, 500 μl of each Proteinase K-treated fraction was extracted twice with 500 μl phenol/chloroform/isoamyl alcohol (25:24:1) and once with 500 μl chloroform/isoamyl alcohol (49:1). The RNA was ethanol precipitated. In all, 4 μl of cytoplasmic RNA or all the RNA extracted from polysome fractions was reverse transcribed in a 20-μl reaction (RNA templates, 5 pmol/μl random primer, 0.5 mM of each dNTP, 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM DTT, 1 μl RT).
Real-time PCR
The mRNA levels of nef-luc were measured by Taqman real-time PCR in a Rotor-gene 6000 (Corbett Life Science) with the following programme: (1) 50°C 2 min; (2) 95°C 10 min; (3) 95°C 15 s, 60°C 1 min, 40 cycles; (4) 30°C 1 min. TaqMan® GAPDH Control Reagents (Human) (ABI) were used for gapdh mRNA. The Taqman probe and primers for nef-luc, nef-RL-EMCV-FL, nef-RL-EMCVagt-FL and nef-RL-HCV-FL mRNAs were:
qNef probe: 5′FAM-AAGCAACCCACCTCCC-NFQ3′
qNef FP: 5′-ACAGTCAGACTCATCAAGCTTCTCT-3′
qNef RP: 5′-CGGGTCCCCTCGGGATT-3′
The mRNA levels of ZAP and Luc-Mk were measured by SYBR Green real-time PCR in Rotor-gene 6000 (Corbett Life Science) using the following programme: (1) 50°C 2 min, 1 cycle; (2) 95°C 10 min, 1 cycle; (3) 95 °C 15 s–>60 °C 30 s–>72 °C 30 s, 40 cycles; (4) 72°C 10 min, 1 cycle. Sequences of the primers are listed below:
qZAP FP: 5′-CCACATCTTCTAGGGTGGATGA-3′
qZAP RP: 5′-CGTCCAGGTTTTACCAATAAGCA-3′
qLuc-Mk FP: 5′-GTTGGTCGTGAGGCACTGGGTGTCC-3′
qLuc-Mk RP: 5′-GTCCACCTCGATATGTGCATCTGTA-3′
Polysome analysis
NL4-3-luc-infected 293TREx-ZAP cells or 293TREx-ZAP/FL-Mk cells were mock-treated or treated with 1 μg/ml tetracycline to induce ZAP expression. At 36 h post-induction, cycloheximide was added to the medium at a final concentration of 50 μg/ml to stop translation. Cells were lysed with 300 μl Lysis Buffer [300 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.4), 0.5% NP-40 and 100 μg/ml cycloheximide] and centrifuged for 15 min at 4°C. In all, 200 μl of the supernatant was applied to a 10–50% sucrose continuous gradient and centrifuged at 36 000 r.p.m. for 3.5 h at 4°C. Twelve fractions, 1 ml each, were collected on ice. The absorbance at 254 nm was monitored and recorded to indicate the positions of ribosome subunits and polysomes. The RNA in each fraction was extracted and subjected to RT–PCR using specific primers to detect mRNA. Primers used in the polysome analyses are listed below.
Nef-luc FP: 5′-AGGGGCGGCGACTGGAAGAAGCGGAG-3′
Nef-luc RP: 5′-AACTTTACCGACCGCGCCCGGTTTA-3′
FL-Mk FP: 5′-GTTGGTCGTGAGGCACTGGGTGTCC-3′
FL-Mk RP: 5′-GTCCACCTCGATATGTGCATCTGTA-3′
GAPDH FP: 5′-ATGGGGAAGGTGAAGGTCGGAGTCA-3′
GAPDH RP: 5′-TGGAGGCCATGTGGGCCATGAGGTC-3′
Co-immunoprecipitation assay
Cells were lysed in 500 μl co-IP buffer (30 mM HEPES (pH 7.5), 100 mM NaCl, 0.5% NP-40, and a protease inhibitors cocktail). To remove nonspecific RNA-mediated tethering, the lysates were treated with 50 μg/ml RNase A and clarified by centrifugation at 12 000 r.p.m. for 15 min at 4°C. The supernatant was incubated with antibodies and protein G sepharose (Amersham Pharmacia) for 4 h at 4°C. Immunoprecipitates were washed three times with PBS, re-suspended in SDS loading buffer, boiled for 5 min, resolved by SDS–PAGE and detected by western blotting.
Split-Gaussia assay
293TREx-ZAP cells in six-well plates were transfected with 0.45 μg pZIP-GL1 and 0.45 μg pZIP-GL2 or 0.45 μg p4G-GL1 and 0.45 μg p4A-GL2 together with 0.1 μg pGL3-SV40 using lipofectamine 2000. At 6 h post-transfection, cells were mock-treated or treated with 1 μg/ml tetracycline to induce ZAP expression. At 48 h post-transfection, cells were lysed by passive lysis buffer (Promega) and the Gaussia and FL activities were measured by Dual-luciferase Reporter Assay System (Promega).
Statistics
The mean values±s.d. or mean values±s.e. were calculated from at least three independent experiments unless otherwise indicated. The P-values were calculated using the Student’s t-test and P-values of ⩽0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr Peter Sarnow and Dr Roy Parker for providing plasmids containing the RL-EMCV-FL and RL-HCV-FL sequences, Dr Nahum Sonenberg for providing plasmid pcDNA3-HA-eIF4G1-1599 and Dr Margaret R. MacDonald for providing plasmids pZIP-GL1 and pZIP-GL2. We are grateful to Drs Yihui Xu, Xiang Wang and Jia Hao for technical support. This work was supported by grants to Guangxia Gao from the Ministry of Science and Technology (973 Program 2012CB910203), the Ministry of Health (2012ZX10001-006) and the National Science Foundation (81030030 and 81161120414) of China, and by a grant to Stephen Goff from the National Institute of Health (R37CA030488-31S1) of the United States.
Author contributions: YZ and GG designed the experiments; YZ and XW performed the experiments; YZ, XW, SG and GG analysed the data; YZ and GG wrote the manuscript; and SG edited the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bazzini AA, Lee MT, Giraldez AJ (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR (2003) Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77: 11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Boris-Lawrie K (2009) Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K (2010) RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res 38: 1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M (2011) Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y (2012) Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol 19: 430–435 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447: 823–828 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R (2005) General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR (1995) Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206: 935–944 [DOI] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T (1997) Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA 94: 5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Guo X, Goff SP (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297: 1703–1706 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78: 12781–12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ma J, Sun J, Gao G (2007) The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA 104: 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP (2010) Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 17: 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE (2008) Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E (1988) A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS (2008) Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z (2007) An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129: 1141–1151 [DOI] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE (2000) Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 275: 41369–41376 [DOI] [PubMed] [Google Scholar]
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M (2006) Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell 22: 117–128 [DOI] [PubMed] [Google Scholar]
- Law LM, Albin OR, Carroll JW, Jones CT, Rice CM, Macdonald MR (2010) Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. J Virol 84: 4504–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE (2006) Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G (2007) Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol 14: 511–518 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Miyakawa S, Oguro A, Ohtsu T, Imataka H, Sonenberg N, Nakamura Y (2006) RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA 12: 1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD (2006) Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 13: 1108–1114 [DOI] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G (2005) Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19: 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40: 515–526 [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 3: 977–979 [DOI] [PubMed] [Google Scholar]
- Suzuki C, Garces RG, Edmonds KA, Hiller S, Hyberts SG, Marintchev A, Wagner G (2008) PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. Proc Natl Acad Sci USA 105: 3274–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A (1992) Internal ribosome entry site within hepatitis C virus RNA. J Virol 66: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S (2007) Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev 21: 1857–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M (2001) Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762 [DOI] [PubMed] [Google Scholar]
- Wax SD, Nakamura H, Anderson PJ (2005) The tumor necrosis factor-alpha AU-rich element inhibits the stable association of the 40S ribosomal subunit with RNA transcripts. Biochem Biophys Res Commun 333: 1100–1106 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N (2009) Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol 29: 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Bolinger C, Boris-Lawrie K (2006) Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr HIV Res 4: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G (2011) Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA 108: 15834–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Gao G (2008) ZAP-mediated mRNA degradation. RNA Biol 5: 65–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.