Abstract
Immunity against infection with Listeria monocytogenes is not achieved from innate immune stimulation by contact with killed but requires viable Listeria gaining access to the cytosol of infected cells. It has remained ill-defined how such immune sensing of live Listeria occurs. Here, we report that efficient cytosolic immune sensing requires access of nucleic acids derived from live Listeria to the cytoplasm of infected cells. We found that Listeria released nucleic acids and that such secreted bacterial RNA/DNA was recognized by the cytosolic sensors RIG-I, MDA5 and STING thereby triggering interferon β production. Secreted Listeria nucleic acids also caused RIG-I-dependent IL-1β-production and inflammasome activation. The signalling molecule CARD9 contributed to IL-1β production in response to secreted nucleic acids. In conclusion, cytosolic recognition of secreted bacterial nucleic acids by RIG-I provides a mechanistic explanation for efficient induction of immunity by live bacteria.
Keywords: inflammasome activation, innate immunity, Listeria infection, nucleic-acid secretion, protective immunity
Introduction
The innate immune system is an evolutionary conserved first-line defense mechanism against invading infectious microorganisms. Recognition of microbial infection is achieved by innate immune sensing receptors that mount an appropriate immune response to eradicate either the infecting microorganism or the infected cells. These sensory receptors are found in all cellular compartments, that is, at cell surface membranes, endolysosomal membranes and in the cytoplasm (Saitoh and Akira, 2010). Membrane-anchored toll-like receptors (TLRs) recognize conserved microbial structural elements or particular nucleic-acid structures and trigger signalling through the adapter molecules MyD88 or TRIF for induction of gene expression eventually resulting in a pro-inflammatory anti-microbial response (Beutler et al, 2006). While the specificity and localization of TLRs support surveillance for infectious microorganisms upon contact or upon endo/phagocytosis, cytosolic immune sensory receptors allow for immune surveillance of those pathogens that gained access to the cytoplasm of the infected cell (Meylan et al, 2006; Franchi et al, 2009). These cytosolic receptors comprise so-called NOD-like receptors (NLRs) that include receptors such as NOD1 and 2 recognizing bacterial peptidoglycans, receptors recognizing microbial RNA such as RIG-I and MDA5 as well as receptors recognizing microbial DNA such as AIM2 (Meylan et al, 2006; Shaw et al, 2008). These cytosolic receptors trigger production of type I interferon (IFN) and can induce formation and activation of inflammasomes, a multi-molecular complex that leads to caspase-1 mediated processing and release of the pro-inflammatory mediators IL-1β and IL-18, that are critical for anti-microbial defense (Mariathasan and Monack, 2007; Schroder and Tschopp, 2010). Recently, access of bacterial nucleic acids into the cytosol of infected cells was shown to be essential for generation of anti-microbial immunity (Sander et al, 2011). The detection of such bacterial nucleic acids within the cytoplasm was linked to viability of infecting bacteria (Sander et al, 2011), indicating that the immune system tailors pathogen-specific immune responses by discriminating between dead and viable bacteria. However, the immune sensory receptors involved in protective cytosolic immune sensing have remained undefined.
Here, we use Listeria monocytogenes, a Gram-positive intracellular bacterium, that infects phagocytic and non-phagocytic cells and serves as a prototypic pathogen to study immunity against intracellular pathogens (Edelson and Unanue, 2000; Pamer, 2004; Hussey et al, 2009), as a model organism to define the parameters critical for the ability of the innate immune system to discriminate between dead and viable pathogens. It is well known that only viable Listeria that gain access to the cytosol but not heat-killed L. monocytogenes (HKLM) or mutant Listeria failing to enter the cytosol generate protective CD8 T-cell immunity (Barry et al, 1992; Lauvau et al, 2001). Apart from its recognition by TLRs, Listeria infection is detected by the cytosolic innate immune receptors NOD1 and NOD2, which induce autophagy and initiate inflammation (Edelson and Unanue, 2000; Yano et al, 2008). Cytosolic Listeria infection is further sensed by NLRP3, NLRC4, NAIP5 and the DNA receptor AIM2, which lead to inflammasome activation and IL-1β release (Corr and O’Neill, 2009; Fernandes-Alnemri et al, 2009; Kim et al, 2010). In addition, second messengers of bacteria, such as c-di-AMP and c-di-GMP, secreted by cytosolic Listeria initiate IFN production through the signalling molecule STING (Kato et al, 2005; Woodward et al, 2010). While recognition of structural bacterial elements by innate immune receptors was clearly demonstrated (Edelson and Unanue, 2000; Yano et al, 2008), it remained poorly understood how Listeria DNA or RNA, which are located within infecting bacteria, could be detected by cytosolic nucleic-acid sensing receptors. Autolysis of bacteria has been suspected to be responsible, but is unlikely to serve as efficient and sensitive detection mechanism (Sauer et al, 2010). Here, we identify the essential role of secreted Listeria nucleic acids in cytosolic immune sensing of viable bacteria through the RNA sensory receptors RIG-I and MDA5 in addition to STING-mediated recognition.
Results
The RNA sensory receptors RIG-I and MDA5 in addition to STING detect cytosolic L. monocytogenes infection in macrophages
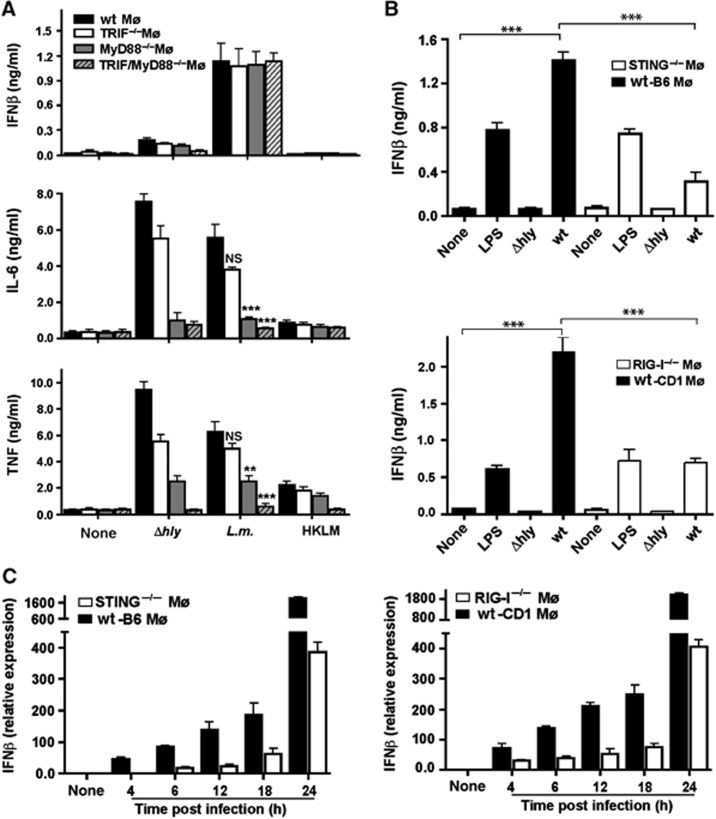
Infection of macrophages with L. monocytogenes is known to induce pro-inflammatory cytokines. Mutant Listeria lacking listerolysin (Δhly), which fail to enter the cytosol after phagocytosis, elicited even more IL-6 and TNF in macrophages than wild-type (wt) Listeria (Figure 1A). Even HKLM elicited some expression of TNF. Consistent with earlier observations (O’Connell et al, 2005), Δhly-dependent IL-6 and TNF expression required TLR signalling because MyD88−/−, TRIF−/− or MyD88−/−/TRIF−/− macrophages failed to produce these cytokines (Figure 1A). However, only viable wt Listeria induced IFN beta (IFNβ). As Δhly and wt Listeria differ with respect to the ability to invade the cytosol, we reasoned that the involvement of cytosolic sensory receptors may explain the differential expression pattern of pro-inflammatory mediators. In line with this hypothesis, we found that MyD88−/−, TRIF−/− or MyD88−/−/TRIF−/− macrophages maintained IFNβ expression after infection with wt Listeria (Figure 1A; Supplementary Figure S1A), indicating that TLR signalling was not essential for IFNβ induction (O’Riordan et al, 2002; Stockinger et al, 2004).
Figure 1.
RIG-I and STING detect cytosolic infection with Listeria monocytogenes in macrophages. IFNβ in cell-culture supernatants was determined by ELISA 24 h after transfection or 18 h after infection; IFNβ mRNA was determined by qRT-PCR 6 h after transfection or infection. (A) IFNβ, TNF and IL-6 from cell-culture supernatant of bone marrow-derived macrophages (Mø) generated from TRIF−/−, MyD88−/−, TRIF/MyD88−/− and wild-type (wt) littermate mice. In all, 2 × 105 Mø after infection with 2 × 106 CFU wt Listeria, LLO-deficient Listeria mutant (Δhly) (MOI of 10) or 2 × 108 of HKLM or mock treatment. (B) IFNβ in cell-culture supernatant of RIG-I−/−, STING−/− or wt Mø infected with Listeria. LPS (500 ng/ml) as positive control. (C) Time kinetics of IFNβ mRNA from wt, STING−/− or RIG-I−/− Mø after infection. NS, not significant; **P=0.01, ***P=0.001 (unpaired Student’s t-test). Data are representative of at least three separate experiments (mean and s.d. of triplicates).
A role for STING in the detection of Listeria infection and in particular the recognition of cyclic di-AMP/GMP secreted from Listeria for induction of IFNβ has been demonstrated (Kato et al, 2005; Ishikawa et al, 2009; Woodward et al, 2010). Other cytosolic pattern recognition receptors known to induce IFNβ are the RNA-sensory molecules RIG-I and MDA5. We tested the contribution of these immune sensory receptors in macrophages isolated from STING, RIG-I and MDA5 knockout mice. In principle, all macrophages from these mice were responsive to stimulation by different ligands (Supplementary Figure S1B). To account for any strain differences in responsiveness to stimulation, we always compared macrophages from knockout mice with macrophages from their wt littermates. We found that IFNβ expression in response to Listeria infection was significantly reduced as expected in STING knockout macrophages (Figure 1B; Supplementary Figure S1C) (Kato et al, 2005; Woodward et al, 2010). However, there was a significant reduction in IFNβ production in RIG-I−/− and to a lesser extent in MDA5−/− macrophages (Figure 1B; Supplementary Figure S2A and B). Absence of RIG-I caused a more pronounced reduction in IFNβ than absence of MDA5, suggesting that induction of IFNβ upon sensing of viable Listeria was in addition to STING mediated predominantly through RIG-I. Time-kinetic analysis revealed that there was no IFNβ induction in STING−/− macrophages at 4 h after infection whereas IFNβ mRNA was detectable at this time point in RIG-I−/− macrophages (Figure 1C), indicating that induction of IFN via STING is triggered earlier than RIG-I-mediated IFN induction. As the ligands for STING, that is, c-di-AMP and c-di-GMP, are secreted by cytosolic Listeria (Woodward et al, 2010) we wondered whether RIG-I and MDA5 ligands were also secreted by live Listeria and would thus activate cytosolic immune sensory receptors.
Listeria secretes RNA that triggers IFNβ expression through RIG-I in macrophages
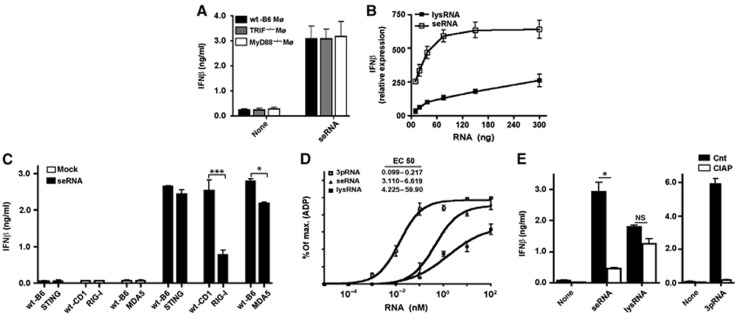
Here, we observed that L. monocytogenes secretes nucleic acids during the exponential growth phase in a cell-free system. DNA (<2 kb) as well as RNA (<2 kb) revealed by DNAse or RNAse treatment prior to nucleic-acid quantification was detected in cell supernatant or bacterial lysates (Supplementary Table S1). There were more of these nucleic acids in cell-culture supernatant after 4 h of culture compared to bacterial lysates from the same culture (Supplementary Table S1). To investigate the importance of such nucleic acids that were secreted from Listeria for induction of IFNβ, we transfected secreted RNA (seRNA) into macrophages. After seRNA transfection, macrophages deficient for the adapter molecules MyD88 and TRIF showed no change in IFNβ induction (Figure 2A; Supplementary Figure S3A). These results suggested that recognition of seRNA occurred mainly by cytosolic sensory receptors. seRNA induced much more IFNβ upon transfection into macrophages than RNA from Listeria lysates (lysRNA) (Figure 2B). Whereas a plateau in IFNβ expression was already reached by 80 ng of seRNA, no such saturation was observed for lysRNA at the concentrations tested here (Figure 2B). We excluded that lysRNA actively suppressed signal transduction by adding increasing high amounts of lysRNA to seRNA in transfection experiments (Supplementary Figure S3B). Assuming a homogenous transfection rate for all macrophages was achieved in our experiments this would mean that ∼10 fg of Listeria RNA per cell was sufficient to trigger IFNβ expression.
Figure 2.
Listeria monocytogenes-secreted RNA triggers type I IFN induction through RIG-I in macrophages. (A) IFNβ in cell-culture supernatants of wt, TRIF−/− or MyD88−/− Mø after transfection with seRNA (10 ng/105 cells) or lysRNA (1 μg/105 cells). (B) Dose kinetic of IFNβ mRNA in Mø after transfection with seRNA or lysRNA. (C) IFNβ in cell-culture supernatant of wt, RIG-I−/−, STING−/− or MDA5−/− Mø after transfection of seRNA (10 ng/105 cells). (D) ATPase assay of purified recombinant human RIG-I protein after incubation with increasing concentration of Listeria RNAs (as described in Materials and methods). EC50 values for the different RNAs were determined by non-linear regression analysis. (E) ELISA of IFNβ in cell-culture supernatant of Mø 24 h after transfection with seRNA (10 ng/105 cells) or lysRNA (1 μg/105 cells) subjected to CIAP treatment. 3pRNA as control. NS, not significant; *P=0.05, ***P=0.001 (unpaired Student’s t-test). Data are representative of at least three separate experiments (mean and s.d. of triplicates).
To investigate which immune sensory receptors were involved in the detection of the secreted Listeria nucleic acids, we analysed the contribution of cytosolic immune sensory receptors. As RIG-I, MDA5 and STING were involved in the sensing of cytosolic Listeria infection, we investigated the contribution of these sensors for the detection of seRNA. Transfection of seRNA into wt, RIG-I−/− or MDA5−/− macrophages revealed the dependence of IFNβ expression on the presence of RIG-I and to a lesser extent on MDA5 (Figure 2C; Supplementary Figure S3C). Transfection of seRNA into STING−/− macrophages, however, resulted in similar IFNβ production compared to wt macrophages (Figure 2C; Supplementary Figure S3C). This demonstrates that STING was not involved in RNA sensing and that no c-di-AMP/GMP was contaminating seRNA. We further excluded a contamination with c-di-AMP/GMP by treating secreted nucleic acids with snake venom phosphodiesterase (SVPD). Furthermore, there was no difference in IFNβ production of RIG-I−/− or MDA5−/− macrophages compared to wt macrophages following exposure to c-di-AMP/GMP (Supplementary Figure S3D) which further supported the notion that no overlap exists between these signalling pathways.
To further characterize the interaction of Listeria RNA with RIG-I, we determined the ability of seRNA and lysRNA to directly activate the recombinant human RIG-I. seRNA bound to and activated recombinant RIG-I more efficiently than lysRNA as determined by ATPase assay (Figure 2D; Supplementary Figure S3E). This remarkable difference in the ability of seRNA and lysRNA to serve as ligand for RIG-I led us to use more lysRNA (100-fold) than seRNA in further experiments. Treatment of seRNA but not lysRNA with calf intestinal alkaline phosphatase (CIAP) led to significantly reduced IFNβ production after transfection into macrophages (Figure 2E; Supplementary Figure S3F), which suggested that seRNA contained more RIG-I ligands than lysRNA. CIAP treatment of Listeria nucleic acids only affected RIG-I-mediated induction of IFNβ mRNA by seRNA (Supplementary Figure S4A), indicating that CIAP treatment did not influence induction of IFNβ mRNA after recognition of these nucleic acids by other immune sensory receptors.
Furthermore, treatment of secreted Listeria nucleic acids with DNAse or RNAse demonstrated the specificity of RIG-I-mediated nucleic-acid recognition (Supplementary Figure S4B). Also, macrophages from NOD1−/− or NOD2−/− mice did not show alterations in IFNβ induction compared to wt macrophages (Supplementary Figure S4C) thereby excluding a contamination of secreted Listeria nucleic acids with ligands for NOD1 or NOD2. These results demonstrate that secreted Listeria RNA served as ligand for RIG-I and that STING did not contribute to IFNβ production under these conditions.
Secreted Listeria DNA induces IFNβ through STING and following recognition through RIG-I via the RNA-polymerase III pathway
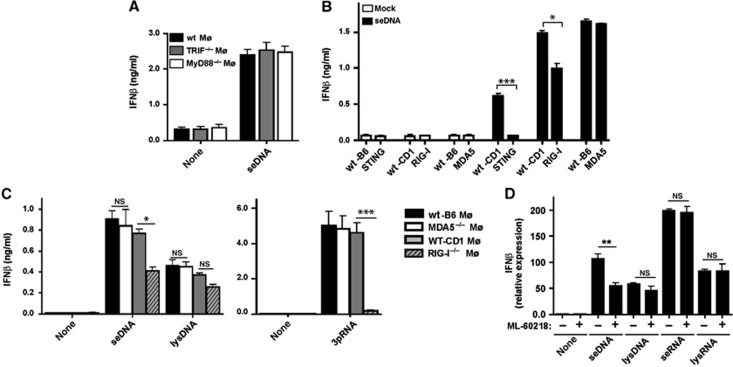
Next, we investigated the importance of secreted Listeria DNA (seDNA) for IFNβ expression. Transfection of seDNA into macrophages revealed that IFNβ induction was independent of MyD88 and TRIF signalling (Figure 3A; Supplementary Figure S5A). These results prompted us to investigate whether seDNA may also be sensed by STING, RIG-I or MDA5. STING is involved in cytosolic sensing of DNA (Ishikawa et al, 2009). STING and RIG-I contributed to the production of IFNβ after transfection with seDNA, because STING−/− or RIG-I−/− macrophages elicited less IFNβ compared to wt macrophages under these conditions (Figure 3B; Supplementary Figure S5B), raising the question how RIG-I was involved in DNA recognition. It has been reported that DNA can activate IFNβ expression via RIG-I in an RNA-polymerase III-dependent fashion (Ablasser et al, 2009; Chiu et al, 2009). Transfection of seDNA (10 ng/105 cells) but not DNA from Listeria lysates (lysDNA) (1 μg/105 cells) into macrophages demonstrated a partial dependence of IFNβ expression on RIG-I but not MDA-5 (Figure 3C; Supplementary Figure S5C). Pharmacologic blockade of RNA polymerase III (by using ML-60218) selectively reduced IFNβ mRNA levels after transfection with seDNA but not with seRNA (Figure 3D), indicating that RIG-I sensing of seDNA was similar to indirect DNA recognition by RIG-I from the Gram-negative bacteria Legionella or viral DNA involving RNA polymerase III (Ablasser et al, 2009; Chiu et al, 2009). Dose-titration kinetics did not show non-specific inhibitory effects of the RNA polymerase III inhibitor on RNA-mediated IFNβ induction in macrophages (Supplementary Figure S5D). Collectively, these data support the notion that seDNA triggered IFNβ production in wt macrophages involving STING as well as RIG-I after transcription of Listeria DNA into RNA by the RNA-polymerase III pathway.
Figure 3.
Secreted Listeria DNA is recognized by RIG-I via the RNA-polymerase III pathway. (A–C) IFNβ in cell-culture supernatants of (A) wt, TRIF−/− or MyD88−/− Mø transfected with seDNA (20 ng/105 cells), (B) wt, STING−/−, RIG-I−/− or MDA5−/− Mø transfected with seDNA (20 ng/105 cells) or (C) wt, MDA5−/− or RIG-I−/− Mø transfected with seDNA (20 ng/105 cells) or lysDNA (2 μg/105 cells). (D) IFNβ mRNA assessed by qRT-PCR in wt macrophages after transfection with seRNA (10 ng/105 cells) or seDNA (20 ng/105 cells), lysRNA (1 μg/105 cells) or lysDNA (2 μg/105 cells) after treatment with the RNA polymerase III inhibitor (ML-60218, 4 μM) for 10 h prior to transfection and during the incubation time. NS, not significant; *P=0.05, **P=0.01, ***P=0.001 (unpaired Student’s t-test). Data are representative of at least three separate experiments (mean and s.d. of triplicates).
Listeria lacking the SecA2 secretion system induce less IFNβ production in macrophages
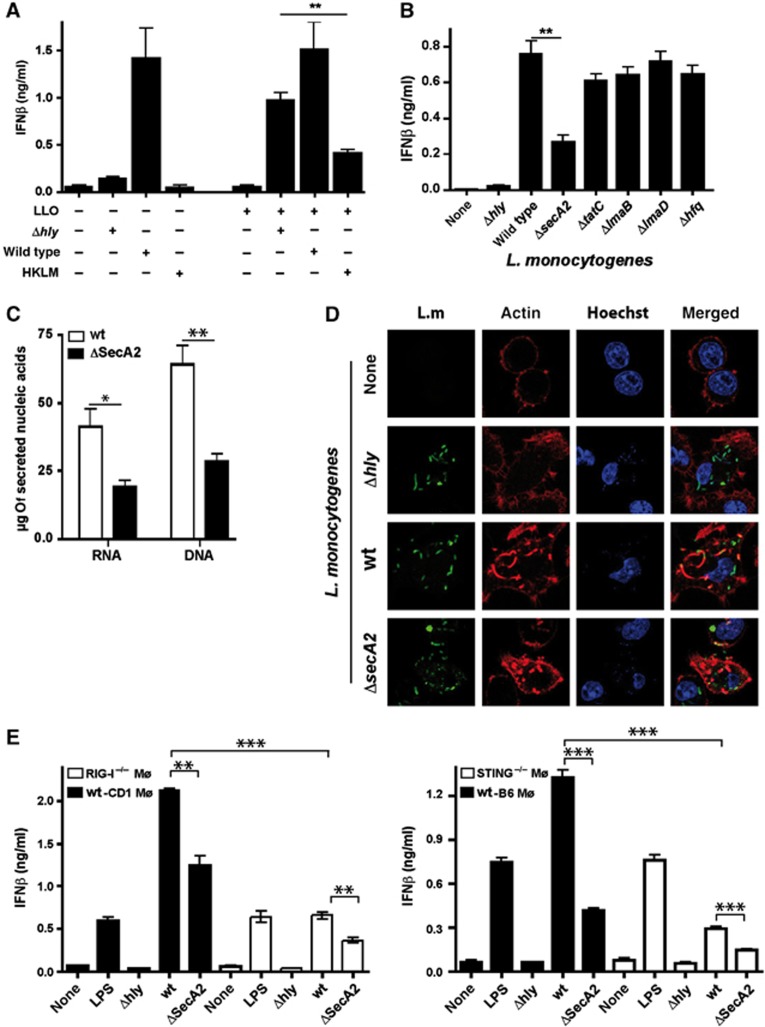
Our data raised the question whether induction of IFNβ by cytosolic Listeria nucleic acids was the result of leakage from degraded bacteria into the cytosol or an active secretion process of viable Listeria. To address this, we incubated macrophages with wt Listeria, Δhly or HKLM in the absence or presence of the pore-forming protein LLO. Clearly, HKLM did not induce IFNβ and even after LLO treatment, which allows phagosomal constituents to enter the cytosol, IFNβ levels did not reach those observed after infection with wt Listeria (Figure 4A; Supplementary Figure S6A). In contrast, Δhly in the presence of LLO elicited macrophage IFNβ expression levels almost similar to that induced by wt Listeria (Figure 4A; Supplementary Figure S6A). This indicated that besides leakage of bacterial debris containing nucleic acids from phagosomal compartments (Fernandes-Alnemri et al, 2010) other mechanisms such as autolysis within the cytosol (Sauer et al, 2010) or active release of nucleic acids from viable bacteria into the cytosol (Sander et al, 2011) were involved in IFNβ production in infected macrophages.
Figure 4.
SecA2 mutant Listeria (ΔsecA2) induce less IFNβ than wild-type Listeria. (A) IFNβ in cell-culture supernatants of Mø infected with wild-type Listeria, Δhly (MOI 10) or heat-killed Listeria monocytogenes (HKLM, MOI 100) in the absence or presence of the pore-forming protein LLO. (B) IFNβ in cell-culture supernatants of Mø infected with wild-type or Listeria mutants (MOI=10). (C) Secreted acids from wild-type Listeria or ΔsecA2 were determined in the supernatant from the same number of bacteria. (D) Confocal microscopy of actin polymerization in Mø after infection with wild-type Listeria, ΔsecA2 or Δhly mutants. Representative images from three independent experiments. (E) IFNβ in cell-culture supernatants of wt, STING−/− or RIG-I−/− Mø after infection with wild-type Listeria, ΔsecA2 or Δhly mutants (MOI 10). LPS (500 ng/ml) as positive control. NS, not significant; *P=0.05, **P=0.01, ***P=0.001 (unpaired Student’s t-test). Data are representative of at least three separate experiments.
To study whether a bacterial secretion system was involved in nucleic-acid release by L. monocytogenes that lead to recognition of cytosolic infection by innate immune sensory receptors, we examined different bacterial mutants in currently known secretion systems as well as various chaperons known to bind to nucleic acids (Desvaux and Hebraud, 2006), and determined their contribution to IFNβ expression. Only one mutant in the auxiliary secretion system SecA2 (ΔsecA2) but not the others showed a significant reduction in IFNβ production after infection (Figure 4B; Supplementary Figure S6B) that was TLR independent (Supplementary Figure S6C). SecA2 was initially identified as protein secretion system that contributes to bacterial pathogenesis (Lenz et al, 2003). As expected, in ΔsecA2 there was less release of the SecA2-dependent protein p60 into the supernatant whereas the SecA2-independent LLO was equally well secreted by ΔsecA2 and wt Listeria (Supplementary Figure S6D). We observed reduced levels of nucleic acids secreted from ΔsecA2 compared to wt Listeria (Figure 4C), which suggests that SecA2 contributed to nucleic-acid transport. Although we cannot formally exclude reduced autolysis in ΔsecA2 there is little evidence for this, because similar levels of genomic DNA and ribosomal RNA were observed in ΔsecA2 and wt Listeria whereas small size RNA and DNA were clearly reduced in ΔsecA2 (Supplementary Figure S6E). Similar levels of actin polymerization were observed around cytosolic ΔsecA2 (Figure 4D), indicating that reduced IFNβ expression upon infection of macrophages with this mutant was not due to impaired bacterial access to the cytosol. Along this line, we found similar rates of macrophage infection for wt Listeria and ΔsecA2 (Supplementary Figure S7A). Reduced IFNβ expression in ΔsecA2-infected macrophages was neither a result of diminished intracellular bacterial growth (Supplementary Figure S7B) nor of a more rapid escape from the infected cell, because this mutant has a cell–cell spreading defect (Supplementary Figure S7C; Lenz et al, 2003). In summary, these data suggest that the SecA2 secretion system in Listeria in addition to its function in transport of proteins might be involved in translocation of nucleic acids that triggers IFN expression.
As STING, RIG-I and MDA5 contributed to recognition of secreted Listeria nucleic acids following transfection, we evaluated the relevance of SecA2 for IFNβ induction during Listeria infection. Infection with ΔsecA2 led to reduced IFNβ production in both, RIG-I−/− macrophages and STING−/− macrophages, compared to infection with wt Listeria (Figure 4E; Supplementary Figure S7D), which is compatible with a combinatorial effect of signalling through STING upon recognition of secreted Listeria DNA and recognition of secreted Listeria RNA by RIG-I. IFNβ induction that was still present in RIG-I−/− macrophages infected with ΔsecA2 may have been caused by recognition of ci-di-AMP/GMP or DNA released into the cytosol upon spontaneous bacterial lysis in the cytosol or nucleic-acid secretion by SecA2-independent mechanisms. There was little contribution of MDA5 under the conditions tested (not shown). These results confirmed the notion that both RIG-I and STING are important for sensing of cytosolic Listeria nucleic acids leading to IFNβ production.
Secreted Listeria nucleic acids trigger inflammasome activation in an RIG-I-dependent manner
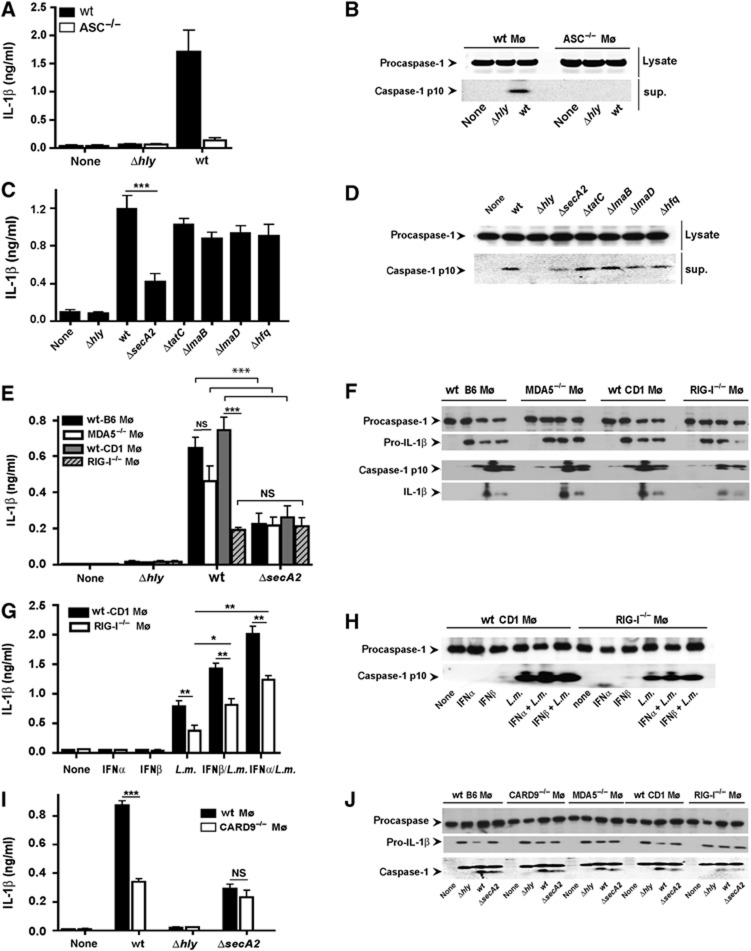
Listeria DNA induces inflammasome activation leading to caspase 1-mediated processing of IL-1β and IL-18, which is relevant for host defense (Fernandes-Alnemri et al, 2009; Kim et al, 2010). Accordingly, ASC-dependent caspase 1 cleavage and release of IL-1β from murine macrophages was observed after entry of Listeria into the cytosol (Figure 5A and B). Similarly to the impaired induction of IFNβ, we observed reduced caspase 1 activation and IL-1β release after infection with ΔsecA2 but not other mutants of Listeria secretion systems (Figure 5C and D), which suggested that cytosolic immune sensory receptors detecting bacterial nucleic acids were involved in inflammasome activation during Listeria infection. There was a reduction in IL-1β release and inflammasome activation after Listeria infection of RIG-I−/− macrophages compared to wt littermate controls, which was not further accentuated upon infection with ΔsecA2 (Figure 5E and F), which indicated that SecA2 may be involved in the cytosolic transport of bacterial nucleic acids that triggered inflammasome activation.
Figure 5.
Listeria infection triggers IL-1β release and inflammasome activation in an RIG-I-dependent manner. (A) IL-1β in cell-culture supernatants of wild-type or ASC−/− Mø 18 h after infection with wt Listeria or Δhly. (B) Immunoblot of full-length (pro)caspase-1 in cell lysates or cleaved caspase 1 in the supernatant of wild-type or ASC−/− Mø 6 h after infection with wt Listeria or Δhly. (C) IL-1β in cell-culture supernatants of wild-type Mø 18 h after infection with wt, or different Listeria mutants. (D) Immunoblot of procaspase-1 in cell lysates or cleaved caspase 1 in the supernatant of wild-type Mø 6 h after infection with wt or different Listeria mutants. (E) IL-1β in cell-culture supernatants of wild-type, MDA5−/− or RIG-I−/− Mø 18 h after infection with wt, Δhly or ΔsecA2 Listeria. (F) Immunoblot of procaspase-1 and pro-IL-1β in cell lysates or cleaved caspase 1 and IL-1β in the supernatant of wild-type, MDA5−/− or RIG-I−/− Mø 6 h after infection with wt, Δhly or ΔsecA2 Listeria. (G) Like in (E) but macrophages were pretreated with 500 U/ml of IFNα or IFNβ for 2 h prior to and during infection. (H) Immunoblot 6 h after infection of procaspase-1 in cell lysates or cleaved caspase 1 in the supernatant of wild-type or RIG-I−/− Mø pretreated with IFNα or IFNβ as in (G). (I) IL-1β in cell-culture supernatants of wild-type and CARD9−/− macrophages 18 h after infection with wt Listeria, Δhly and ΔsecA2. (J) Immunoblot of wild-type, CARD9−/−, MDA5−/− or RIG-I−/− macrophages 6 h after infection with wt Listeria, Δhly and ΔsecA2. *P=0.05, **P=0.01, ***P=0.001. Figure source data can be found with the Supplementary data.
To investigate the mechanisms behind reduced inflammasome activation in RIG-I−/− macrophages, we addressed the question whether this was related to the reduced IFNβ expression in RIG-I−/− macrophages (Henry et al, 2007; Fernandes-Alnemri et al, 2010). We supplemented wt or RIG-I−/− macrophages with IFNα or IFNβ and then infected the cells with wt Listeria. IFN pretreatment increased IL-1β release from infected wt macrophages and also increased IL-1β release and inflammasome activation in RIG-I−/− macrophages, but the levels never reached those of wt macrophages also treated with IFN (Figure 5G and H). There were no changes in the levels of AIM2 in wt or RIG-I−/− macrophages even after IFN treatment (Supplementary Figure S8A).
We have recently shown that cytosolic viral RNA triggers IL-1β production in an RIG-I- and CARD9-dependent fashion by selective control of the NF-κB-dependent pro-IL-1β synthesis (Poeck et al, 2010). Here, we provide evidence that the CARD9 pathway also mediates IL-1β activation in response to cytosolic Listeria without affecting caspase-1 cleavage (Figure 5I and J). Listeria-induced caspase 1 cleavage observed here results presumably from RIG-I-dependent ASC engagement leading to inflammasome activation. The lack of an additional reduction in IL-1β release observed in CARD9-deficient macrophages infected with ΔsecA2 (Figure 5I and J) further supports the notion that SecA2 may be involved as a transporter for nucleic acids that are recognized by those cytosolic sensors that signal via CARD9. Pro-IL1β mRNA was reduced in Listeria-infected CARD9−/− macrophages (Supplementary Figure S8B), which may explain the reduced secretion of IL-1β despite unchanged caspase cleavage.
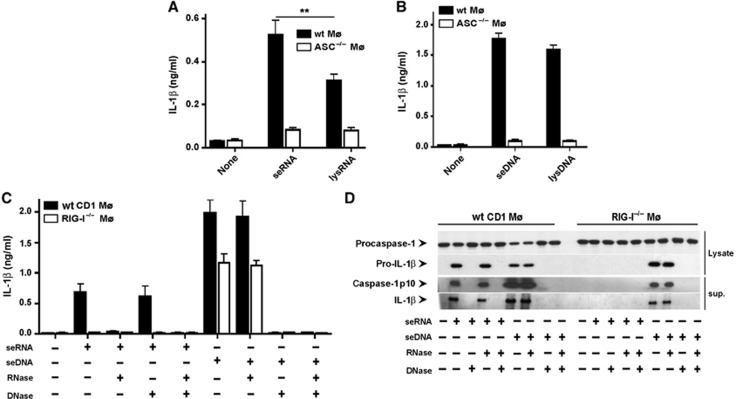
To assess whether the secretion of nucleic acids from Listeria triggered inflammasome activation and release of IL-1β, we transfected seRNA or seDNA into wt or ASC−/− macrophages. Listeria seDNA was more potent than seRNA in releasing IL-1β from macrophages (Figure 6A and B). Both seDNA and seRNA-mediated release of IL-1β required the presence of the inflammasome adapter molecule ASC (Figure 6A and B), indicating inflammasome activation by seDNA as well as by RNA. While transfection of seRNA elicited IL-1β release and inflammasome activation in wt macrophages IL-1β release and inflammasome activation were completely abolished in RIG-I−/− macrophages (Figure 6C and D). As expected for DNA transfection (Fernandes-Alnemri et al, 2010), we observed IL-1β release and inflammasome activation following transfection of seDNA (Figure 6C and D). There was only a modest reduction in IL-1β release and inflammasome activation in transfected RIG-I−/− macrophages, which may be explained by RIG-I-independent pathways of inflammasome activation such as AIM2.
Figure 6.
Secreted Listeria nucleic acids cause IL-1β release and inflammasome activation in an RIG-I-dependent manner. (A, B) IL-1β in cell-culture supernatants of wild-type or ASC−/− Mø 18 h after transfection with (A) seRNA (10 ng/105 cells) or lysRNA (1 μg/105 cells) or (B) seDNA (20 ng/105 cells) or lysDNA (2 μg/105 cells). (C) IL-1β in cell-culture supernatants of wild-type or RIG-I−/− Mø 18 h after transfection of seRNA and seDNA incubated before with RNAse and/or DNAse. (D) Immunoblot of supernatant and cell lysates of cells treated as in (C) 6 h after transfection. **P=0.01. Figure source data can be found with the Supplementary data.
Discussion
Innate immune sensing of Listeria can occur at different sites that range from recognition at the cell surface, within phagosomal compartments and in the cytosol. Recent reports indicated that the complexity of innate immune sensing with respect to different receptor systems used in these cellular compartments, that is, TLRs, NLRs, RLRs and inflammasomes, not only serves as a multi-layered redundant detection system but also translates innate immune sensing into graded effector responses of the immune system ranging from cell-autonomous immunity to inflammation and cell-mediated effector responses (Blander and Sander, 2012). This assumption is supported by observations that Listeria unable to enter the cytosol and engage cytosolic immune sensory receptors, such as heat-killed Listeria and some Listeria mutants, provoke inflammation but fail to induce immunity (Lauvau et al, 2001; Hara et al, 2007). It is also unclear, why such immune stimulation requires viable cytosolic bacteria and which cytosolic immune sensory receptors contribute to protective immunity against Listeria infection. Here, we demonstrate that the sensory receptors RIG-I and MDA5 together with STING operate to detect cytosolic live Listeria by recognizing bacterial nucleic acids resulting in expression of IFNβ and inflammasome activation.
Induction of IFN in response to viable cytosolic Listeria has been shown to occur through STING-mediated signalling and direct recognition of the secondary bacterial messenger cyclic di-GMP by STING (Woodward et al, 2010; Burdette et al, 2011). While a role for STING in DNA-mediated recognition of Listeria infection has been suggested (Ishikawa et al, 2009), our results now identify a so far unappreciated role of STING in recognition of Listeria DNA but not RNA secreted via SecA2 leading to production of IFNβ. They also define a novel role for the RNA-specific sensors RIG-I and to a lesser extent MDA5 in detecting secreted Listeria RNA as well as DNA. The contributions of RIG-I, MDA5 and STING were clearly evident using macrophages genetically deficient for either of these receptors. These results also indicate that under the conditions tested here, STING does not contribute to downstream signalling of RIG-I sensing of Listeria RNA. Given its dual function as receptor in c-di-GMP-recognition and as signalling adapter (Burdette et al, 2011), its role in recognizing secreted Listeria DNA remains to be defined.
RNA-specific sensing of cytosolic Listeria infection led to expression of IFNβ, inflammasome activation and IL-1β release. For the first time, we also demonstrate that Listeria infection induces IL-1β production through stimulation of RIG-I and the downstream adapter molecule CARD9. Previously, CARD9-dependent IL-1β production has been shown to be important in combating viral infection (Poeck et al, 2010). Here, we demonstrate the involvement of CARD9 also in IL-1β production in response to bacterial infection. This is consistent with a previous report, which indicated an important role for CARD9 in clearance of Listeria infection in vivo but only observed a link between CARD9 and NOD-mediated recognition of Listeria (Adachi et al, 1998). Our results identify RIG-I and STING-mediated recognition of secreted nucleic acids as a new principle for sensing cytosolic Listeria infection that triggers IFNβ induction, inflammasome activation and IL-1β release. Such sensing of viable Listeria in the cytosol may enhance the ability of the infected cell to detect infection by acting in combination with recognition of Listeria DNA by AIM2 or listeriolysin-mediated inflammasome activation (Fernandes-Alnemri et al, 2009; Kim et al, 2010; Meixenberger et al, 2010).
The secretion of nucleic acids by Listeria, that we report here, reveals a mechanism that is critical to both, the pathogen to increase bacterial virulence and the host to mount anti-bacterial immunity. Presently, SecA2 is known as an auxiliary protein secretion system that is highly conserved and is found in diverse bacteria including the pathogen Mycobacterium tuberculosis as well as Escherichia coli (Cabelli et al, 1988; Lenz et al, 2003; Desvaux and Hebraud, 2006). Proteomic analysis has revealed that several virulence-associated proteins are secreted via the secA2 translocon (Lenz et al, 2003) and a systematic analysis of genes induced in vivo following infection of mice with L. monocytogenes indicate that many of these genes are strongly expressed in vivo (Machata et al, 2005). Protein secretion is of key importance in Listeria virulence and the induced expression of SecA2 following infection indicates that the cargo of this transport machinery is relevant for Listeria survival and pathogenesis, for example, by immune subversion strategies (Lenz et al, 2003). Our data indicate that bacterial proteins transported via SecA2 may also serve as carriers for Listeria RNA and DNA and may thereby function as chaperones to stabilize bacterial nucleic acids.
Recently, extensive changes of small RNA expression in Listeria within infected have emerged as an important feature in regulating infection and intracellular growth (Swanson, 2006; Mraheil et al, 2011). These changes in gene expression likely represent bacterial strategies to adapt to the intracellular environment and to influence the host transcriptome (Lorenz and Wackernagel, 1994; Swanson, 2006; Travassos et al, 2010). Small nucleic acids secreted from infecting Listeria that reach the cytosol might therefore modulate the host transcriptome in favour of bacterial survival. At the same time this very process may represent an ‘Achilles heel’ that can be exploited by the immune system to detect infecting bacteria. As RIG-I and MDA5 are known to recognize non-self small RNAs, the increased expression of Listeria small RNAs may provide more ligands for these RNA-specific sensory receptors. Consistent with this notion, we observed that infection with mutant Listeria lacking SecA2 elicited less RIG-I and MDA5-dependent IFNβ expression and IL-1β release.
Our results support the notion that cytosolic RNA sensing of secreted Listeria RNA allows the infected cell to efficiently discriminate non-infectious material or less virulent bacteria from viable and virulent bacteria within the cytosol. This is corroborated by our findings that even localization of dead bacteria in the cytosol does not elicit strong induction of IFNβ, inflammasome activation and IL-1β release, whereas facilitation of cytosolic invasion of Δhly by exogenous listeriolysin led to strong IFNβ responses. Taken together, recognition through RIG-I allows the infected cell to assemble information on bacterial viability, virulence and localization of bacteria in the cytosol.
It remains an open question whether the cytosolic detection of live Listeria through the immune sensing receptors RIG-I, MDA5 and STING contributes to the development of adaptive immunity. While expression of soluble mediators such as IFNβ or TNF after Listeria infection contributes to but does not suffice for induction of immunity (Lauvau et al, 2001; Auerbuch et al, 2004), inflammasome activation following RIG-I-mediated sensing may trigger development of immunity. Inflammasome activation has been shown to be involved in adaptive immunity against influenza virus infection (Ichinohe et al, 2009), but controversial results are derived from studying the role of inflammasomes in vivo in Listeria infection (Sauer et al, 2011a, b; Warren et al, 2011). The dual role of RIG-I in triggering CARD9-mediated IL-1β expression as well as ASC-mediated caspase activation (Poeck et al, 2010) may also provide a combinatorial signal that is not achieved by AIM2-induced inflammasome activation or NLRC4-inflammasome activation (Rathinam et al, 2010; Sauer et al, 2011a). Future studies will need to address in detail the role of these cytosolic immune sensing receptors in generation of protective immunity against Listeria infection.
Taken together, our results demonstrate that SecA2-mediated secretion of bacterial nucleic acids allows infected macrophages to efficiently detect the presence of viable and virulent Listeria in the cytosol via the immune sensory receptors RIG-I, MDA5 and STING. These results not only reveal a novel cytosolic immune sensing strategy for Listeria infection but also suggest that such immune sensing is linked to recognition of bacterial virulence because SecA2 transports cargo that serves as virulence factors and/or ligands for cytosolic immune sensory receptors. The improved knowledge of the molecular mechanisms determining development of strong immunity during cytosolic bacterial infection will help us to develop rational strategies to overcome persistent intracellular bacterial infection.
Materials and methods
Mice and reagents
Mice genetically deficient in MyD88, TRIF, ASC, CARD9, RIG-I, MDA5, NOD1 and NOD2 have been described (Adachi et al, 1998; Mariathasan et al, 2004; Pamer, 2004; Kato et al, 2005, 2006; Gitlin et al, 2006; Gross et al, 2006; Michallet et al, 2008; Sauer et al, 2010). C57BL/6 and CD1 were from Charles River. Mice were 6–12 weeks of age at the onset of experiments and were used according to local guidelines. All mouse strains were bred and maintained in specific pathogen-free conditions according to the FELASA guidelines at the central animal facility at the University Hospital Bonn. RNA polymerase III inhibitor (ML-60218), LPS and SVPD were purchased from Sigma-Aldrich (Germany).
Bacteria, mutants and culture conditions
Chromosomal deletion mutants of hly, secA2, tatC, lmaB, lmaD, hfq and actA2 in the genome of L. monocytogenes were generated as previously described (Chakraborty et al, 1995; Guzman et al, 1995; Schaferkordt and Chakraborty, 1995; Machata et al, 2005). All Listeria strains (EGDe wt and the mutated strains) were grown overnight at 37°C in brain heart infusion (BHI) broth. Next day, cultures were diluted 1:50 with fresh BHI medium. Bacteria were harvested at the logarithmic growth phase (OD600=0.5–1.0).
To determine the CFU of Listeria within infected macrophages, supernatants of infected cells were aspirated then washed twice with PBS. Cells were lysed in 0.01% Triton X in water and serial dilutions were plated on BHI agar to enumerate CFU after growth at 37°C for 24 h.
Cell culture, infection, transfection and stimulation
Bone marrow-derived macrophages were derived from the bone marrow of the mouse strains mentioned above and used after 7 days of culture in RPMI (GIBCO) supplemented with 10% FBS, 100 μM streptomycin, 100 U/ml penicillin (Gibco), 2 mM L-glutamine, and 30% supernatant from L929 cell line.
For Listeria infection, macrophages were plated at a density of 1.5 × 106 cells/ml in antibiotic-free medium overnight prior to infection. Macrophages were infected at an MOI of 10 followed by centrifugation for 10 min at 400 g, 30 min later the medium was replaced with 100 μg/ml Gentamycin-containing medium.
For transfection with Listeria nucleic acids, macrophages were cultured in OptiMEM reduced-serum medium Cells were transfected with seRNA (10 ng/105 cells), lysRNA (1 μg/105 cells), seDNA (20 ng/105 cells), or lysDNA (2 μg/105 cells), cyclic-di-GMP (4 μg/105 cells), 3pRNA (200 ng/105 cells) complexed with 2 μl Lipofectamine 2000 according to manufacturer’s protocol (Invitrogen, Germany).
For the inhibition of RNA-polymerase III, the specific inhibitor ML-60218 (Sigma-Aldrich) was added to macrophages at concentration indicated in the figure legend 10 h prior to, during and after transfection or infection.
Detection of cytokines
Cell-culture supernatants were analysed for cytokine secretion by ELISA (IL-1β, TNF and IL-6 from BD Biosciences, IFN β from R&D Systems) according to manufacturer’s instructions.
Immunoblot
Cell-culture supernatants were harvested and proteins were precipitated by addition of an equal volume of methanol and 0.25 volumes of chloroform; the lower phase was collected after centrifugation for 10 min at 20 000 g and mixed with 500 μl methanol. This mixture was centrifuged for 10 min at 20 000 g and the protein pellet was dried at RT. Protein samples from lysed cells or cell-culture supernatant were resuspended in Laemmli buffer and boiled for 5 min at 99°C. Immunoblotting was performed as previously described (Poeck et al, 2010). Anti-murine caspase-1 p10 (sc-514; Santa Cruz Biotechnology), anti-murine IL-1β (R&D Systems) and anti-AIM2 polyclonal rabbit Ig (Fernandes-Alnemri et al, 2010), anti-β tubulin (Li-Cor, USA).
Isolation of Listeria DNA and RNA
For the isolation of secreted Listeria nucleic acids, bacteria from an overnight culture were diluted 1:50 and expanded for 4 h under exponential growth conditions. Listeria or the cell-culture supernatants were harvested when a density of 5.8 × 108 bacteria/ml was reached.
For isolation of secreted Listeria RNA (seRNA), supernatants were centrifuged at 4000, r.p.m. for 10 min to remove remaining bacteria. Nucleic acids in the supernatant were precipitated with ethanol and incubated with 1% SDS (w/v) and 0.05 mg/ml proteinase K (Merck, Darmstadt, Germany) at 37°C for 60 min to remove proteins. seRNA was then phenolysed with phenol/chloroform, and collected by ethanol precipitation. For separation of small size RNA from the large size ribosomal RNA, we used the miRNAesy kit combined with DNase I treatment (RNase free grade, Promega, Madison, USA), which yields RNA with a molecular size of <1300, bp according to manufacturer’s instructions (Qiagen, Hilden, Germany).
For the isolation of secreted Listeria DNA (seDNA), DNA from the supernatant was first precipitated with ethanol and then small size DNA was enriched by using the QIAGEN Plasmid Midi Kit, which allows the removal of the genomic large DNA. This was followed by purification of small size DNA using the Urine DNA Isolation Micro kit (Norgen Biotec Corp, Canada), which yields DNA with molecular size between 1000 and 50 bp, in combination with RNase treatment (Amersham Bioscience) for 30 min at RT.
Total Listeria RNA was isolated from bacterial lysates using the RNeasy kit according to manufacturer’s instructions (Qiagen) followed by the use of the miRNeasy kit (Qiagen) to isolate RNA from bacterial lysates with a molecular size of <1300, bp (lysRNA). Total DNA was isolated by lysing cells with lysozyme, small size DNA was enriched by using the QIAGEN Plasmid Midi Kit and followed by further purification using Urine DNA Micro kit (Norgen Biotec Corp, Canada) and RNase treatment (Amersham Bioscience) for 30 min at RT yielding DNA from bacterial lysates with a molecular size between 1000 and 50 bp (lysDNA).
All types of Listeria nucleic acids were pretreated with SVPD (7 × 10−5 U/ml for 5 min at 37°C, which did not lead to nucleic-acid degradation (data not shown)) prior to transfection to exclude a contamination of nucleic-acid preparations with cyclic di-AMP or -GMP. Concentration of secreted Listeria nucleic acids was determined with a nanodrop photometer (260/280 nm).
RNA isolation, cDNA synthesis and quantitative real-time PCR
For quantitative real-time PCR (qRT-PCR), cells were lysed in Trizol (Invitrogen). In all, 50–100 ng RNA was reverse transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany). RT-PCR was performed with a LightCyclerTaqman master kit and pre-designed primers and probes for IFNβ, IL-1β, AIM2 and GAPDH gene expression assays (Applied Biosystems, Germany) on a Light Cycler 1.3 instrument (Roche). Analysis was performed using Light-Cycler 4.05 software (Roche) using a calibrator normalized relative quantification based on GAPDH expression.
Immunofluorescence confocal microscopy
Macrophages (5 × 105) were seeded on glass coverslips in 24-well plates overnight in antibiotic-free medium then infected with Listeria (MOI of 10) for 1 h. Macrophages were washed with pre-warmed medium and cultured further in medium containing 50 μg/ml Gentamycin (Sigma). At indicated time points after infection, cells were washed three times with PBS and fixed with 4% paraformaldehyde, permeabilized with 0.1% Saponin. Cells were incubated with anti-Listeria polyclonal antibody (kindly provided by N Papadopoulou), Phalloidin and Hoechst (Molecular Probes) in PBS containing 10% FCS and 0.1% Saponin for 1 h. After three washes with PBS, macrophages were incubated for 1 h at room temperature with FITC-labelled secondary antibody. Confocal microscopy was performed on FV1000 Confocal Microscope (Olympus).
Flow cytometry
Macrophages were incubated for 20 min on ice with antibodies to CD11b (BD Biosciences) and live/dead-APC-Cy7 cell staining (Invitrogen), then fixed in 4% paraformaldehyde in FACS buffer, and permeabilized in 1 × Perm/Wash (BD Biosciences) for staining with anti-Listeria polyclonal antibody (kindly provided by N Papodopoulou) or control Ig for 20 min on ice. Macrophages were washed and analysed on a FACSCanto flow cytometer (Becton Dickinson) using FlowJo Software.
ATPase assay
The ATPase assay was performed in assay buffer (50 mM KCl, 55 mM HEPES (pH 7.0) 3 mM MgCl2, 0.5 mM DTT, 0.1 mM ATP), as reported previously (Schlee et al, 2009). To calculate EC50, the RNA was titrated in a range from 6 fM to 4 μM. After 30 min of incubation at 37°C, generation of ADP was measured using a very sensitive FRET-based competitive immunoassay (HTRF® Transcreener™ ADP, Cisbio, USA) according to manufacturer’s protocol. FRET was measured using an EnVision® Multilabel Reader (Perkin-Elmer, USA). In this assay, inhibition of FRET correlates with the concentration of ADP generated by ATPase activity of RIG-I. ADP concentrations were calculated from an ADP/ATP titration curve according to manufacturer’s protocol.
Statistical analysis
Two-way analysis of variance followed by the Bonferroni’s post-test was used, unless otherwise stated, with Prism Software. P-values of <0.05 were considered significant and indicated by asterisks: *0.05; **0.01, ***0.001.
Supplementary Material
Acknowledgments
We thank the late J Tschopp (University of Lausanne) for ASC-deficient and IPS-1-deficient mice. We thank R Vance and D Portnoy for providing STING-deficient mice and cyclic-di-AMP; D Busch, München for critical discussion and W Mohamed, Giessen for help in preparing Supplementary figures. We acknowledge excellent technical assistance by K Riethausen and D Bertheloot. We thank the DFG (SFB 670, SFB 704 and SFB TR84), BMBF ERANet Pathogenomices and BONFOR for financial support.
Author contributions: ZA, TC and PK designed experiments and wrote the manuscript; ZA, SR, MAM, SG and FC all performed experiments; TH, MS, K-PH, WB, GH and JR contributed critical reagents, had input into experimental design and contributed to writing of the manuscript. These studies were supported by grants from the BMBF through the ERANET Pathogenomic Network to TH and TC, by NIH U19AI083025 to KPH, and by the SFB 670/SFB 704 to PK.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ablasser A, Poeck H, Anz D, Berger M, Schlee M, Kim S, Bourquin C, Goutagny N, Jiang Z, Fitzgerald KA, Rothenfusser S, Endres S, Hartmann G, Hornung V (2009) Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J Immunol 182: 6824–6833 [DOI] [PubMed] [Google Scholar]
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9: 143–150 [DOI] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA (2004) Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med 200: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry RA, Bouwer HG, Portnoy DA, Hinrichs DJ (1992) Pathogenicity and immunogenicity of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun 60: 1625–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K (2006) Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol 24: 353–389 [DOI] [PubMed] [Google Scholar]
- Blander JM, Sander LE (2012) Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat Rev Immunol 12: 215–225 [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478: 515–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelli RJ, Chen L, Tai PC, Oliver DB (1988) SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell 55: 683–692 [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove CJ, Jockusch BM, Reinhard M, Walter U, Wehland J (1995) A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ (2009) RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138: 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr SC, O’Neill LA (2009) Listeria monocytogenes infection in the face of innate immunity. Cell Microbiol 11: 703–9 [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M (2006) The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol Rev 30: 774–805 [DOI] [PubMed] [Google Scholar]
- Edelson BT, Unanue ER (2000) Immunity to Listeria infection. Curr Opin Immunol 12: 425–431 [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES (2010) The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 11: 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G (2009) The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol 10: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M (2006) Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Nat Acad Sci USA 103: 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J (2006) Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 442: 651–656 [DOI] [PubMed] [Google Scholar]
- Guzman CA, Rohde M, Chakraborty T, Domann E, Hudel M, Wehland J, Timmis KN (1995) Interaction of Listeria monocytogenes with mouse dendritic cells. Infect Immun 63: 3665–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Kawamura I, Nomura T, Tominaga T, Tsuchiya K, Mitsuyama M (2007) Cytolysin-dependent escape of the bacterium from the phagosome is required but not sufficient for induction of the Th1 immune response against Listeria monocytogenes infection: distinct role of Listeriolysin O determined by cytolysin gene replacement. Infect Immun 75: 3791–3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM (2007) Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med 204: 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey S, Travassos LH, Jones NL (2009) Autophagy as an emerging dimension to adaptive and innate immunity. Semin Immunol 21: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A (2009) Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S (2005) Cell type-specific involvement of RIG-I in antiviral response. Immunity 23: 19–28 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V (2010) Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol 40: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvau G, Vijh S, Kong P, Horng T, Kerksiek K, Serbina N, Tuma RA, Pamer EG (2001) Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science 294: 1735–1739 [DOI] [PubMed] [Google Scholar]
- Lenz LL, Mohammadi S, Geissler A, Portnoy DA (2003) SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci USA 100: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MG, Wackernagel W (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58: 563–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machata S, Hain T, Rohde M, Chakraborty T (2005) Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol 187: 8385–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Monack DM (2007) Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol 7: 31–40 [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430: 213–218 [DOI] [PubMed] [Google Scholar]
- Meixenberger K, Pache F, Eitel J, Schmeck B, Hippenstiel S, Slevogt H, N’Guessan P, Witzenrath M, Netea MG, Chakraborty T, Suttorp N, Opitz B (2010) Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J Immunol 184: 922–930 [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M (2006) Intracellular pattern recognition receptors in the host response. Nature 442: 39–44 [DOI] [PubMed] [Google Scholar]
- Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, Kalinke U, Pasparakis M, Tschopp J (2008) TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28: 651–661 [DOI] [PubMed] [Google Scholar]
- Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T (2011) The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39: 4235–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng G (2005) Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinase-binding kinase 1. J Immunol 174: 1602–1607 [DOI] [PubMed] [Google Scholar]
- O’Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA (2002) Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA 99: 13861–13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG (2004) Immune responses to Listeria monocytogenes. Nat Rev Immunol 4: 812–823 [DOI] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J (2010) Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol 11: 63–69 [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA (2010) The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Akira S (2010) Regulation of innate immune responses by autophagy-related proteins. J Cell Biol 189: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM (2011) Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474: 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Pereyre S, Archer KA, Burke TP, Hanson B, Lauer P, Portnoy DA (2011a) Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci USA 108: 12419–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE (2011b) The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA (2010) Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7: 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaferkordt S, Chakraborty T (1995) Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. Biotechniques 19: 720–722724–725 [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J et al. (2009) Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J (2010) The inflammasomes. Cell 140: 821–832 [DOI] [PubMed] [Google Scholar]
- Shaw MH, Reimer T, Kim YG, Nunez G (2008) NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol 20: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T (2004) IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol 173: 7416–7425 [DOI] [PubMed] [Google Scholar]
- Swanson MS (2006) Autophagy: eating for good health. J Immunol 177: 4945–4951 [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ (2010) Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 11: 55–62 [DOI] [PubMed] [Google Scholar]
- Warren SE, Duong H, Mao DP, Armstrong A, Rajan J, Miao EA, Aderem A (2011) Generation of a Listeria vaccine strain by enhanced caspase-1 activation. Eur J Immunol 41: 1934–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA (2010) c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328: 1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S (2008) Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol 9: 908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.