Abstract
A genetic bottleneck explains the marked changes in mitochondrial DNA (mtDNA) heteroplasmy observed during the transmission of pathogenic mutations, but the precise timing remains controversial, and it is not clear whether selection plays a role. These issues are critically important for the genetic counseling of prospective mothers, and developing treatments aimed at disease prevention. By studying mice transmitting a heteroplasmic single base-pair deletion in the mitochondrial tRNAMet gene, we show that mammalian mtDNA heteroplasmy levels are principally determined prenatally within the developing female germ line. Although we saw no evidence of mtDNA selection prenatally, skewed heteroplasmy levels were observed in the offspring of the next generation, consistent with purifying selection. High percentage levels of the tRNAMet mutation were linked to a compensatory increase in overall mitochondrial RNAs, ameliorating the biochemical phenotype, and explaining why fecundity is not compromised.
First proposed 30 years ago, the mitochondrial DNA (mtDNA) genetic bottleneck hypothesis provides an explanation for the rapid shifts in mtDNA heteroplasmy, the coexistence of different mtDNA genotypes within the same cell, that occur during the maternal transmission of pathogenic mutations1,2. Based on this hypothesis, only a limited number of identical segregating units populate the next generation, causing a statistical sampling effect, and explaining why different siblings have markedly different heteroplasmy levels. Recently developed cell-labeling and molecular genetic techniques have made it possible to test this hypothesis directly and to determine the underlying biological mechanism of the bottleneck. The initial report of relatively high amounts of mtDNA within the early female germ line3 was challenged by two independent groups reporting a reduction in the intracellular mtDNA content shortly after germ cell specification4,5. In silico modeling implied that the observed number of randomly segregating single mtDNA molecules could account for the variation in heteroplasmy levels seen in the offspring of heteroplasmic mice4. This was not supported by limited observations in a heteroplasmic mouse lineage5, pointing rather towards a post-natal bottleneck through the replication of a sub-population of mitochondrial genomes within the germ line cells. However, concerns about the reliability of measuring heteroplasmy variance from small sampling sizes questioned the strength of these conclusions6, and the precise timing and mechanism of the mitochondrial genetic bottleneck remains uncertain7.
Initial studies of human pedigrees suggested that mothers were equally likely to pass on either higher or lower levels of mtDNA heteroplasmy to their offspring, even for highly deleterious pathogenic mutations8. However, subsequent population-based studies implied selection against deleterious alleles in carefully ascertained human pedigrees9; raising the possibility that selection was occurring during female germ line development. It is notoriously difficult to exclude the possibility of ascertainment bias when studying human pedigrees transmitting mtDNA heteroplasmy8, and the lack of animal models transmitting apparent pathogenic mtDNA mutations has severely limited progress. To address these issues we generated mice with putatively pathogenic mtDNA mutations with the aim of studying mutations that more closely resemble the situation observed in families segregating mtDNA disease.
By backcrossing female mice carrying a mutant mtDNA polymerase (D257A PolGA exo- or mtDNA mutator mice10) to wild-type males, we generated maternal lines transmitting a range of different mtDNA mutations11. We observed that mutations in protein coding genes were subject to a filter of purifying selection and preferentially eliminated within a few generations11. Yet, mitochondrial tRNA genes evaded the selection, despite the fact that many of these mutations are potentially pathogenic in mice, consistent with tRNA mutations being the most common pathogenic substitutions in humans with mtDNA disease12.
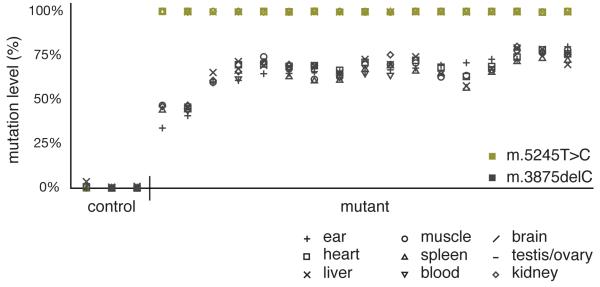
Sequencing of the entire mitochondrial genome (Supplementary Table S1) of one mouse lineage revealed two novel mutations: m.5245T>C in the tRNACys gene and m.3875delC in the tRNAMet gene (Supplementary Fig.1). Initially heteroplasmic, m.5245T>C rapidly segregated to homoplasmy in all organs tested (Supplementary Table S2) with no obvious adverse effects on the organism. The equivalent mutation has been described in humans with mitochondrial encephalopathy (m.5814T>C)13,14. In contrast, m.3875delC remained heteroplasmic and there was no change in heteroplasmy level from birth to adulthood in a variety of tissues relative to levels in ear or tail clips (Supplementary Table S3). The percentage level of the m.3875delC tRNAMet mutation did not vary significantly between different tissues and organs (Fig.1), indicating that the mutation load measured in tail and ear samples were representative of the whole organism and that no tissue-specific selection occurred. However, we were unable to generate offspring with mice harboring >86% mutation levels through selective breeding (Fig. 2a) implicating a selective effect specifically during transmission.
Figure 1.
Levels of heteroplasmy in nine different tissues from three control and 17 mutant adult mice after at least seven generations of backcrossing, showing the homoplasmic m.5245T>C (green) and heteroplasmic m.3875delC (black) mutations. Note that the level of heteroplasmy is similar in all tested tissues.
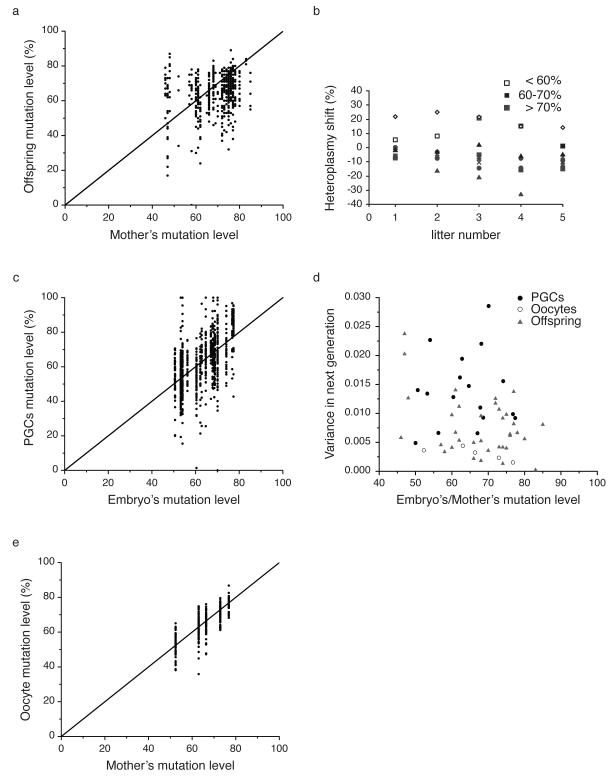
Figure 2.
Measurements of the m.3875delC mutation levels in mothers, primary germ cells (PGCs), oocytes and offspring. (a) Offspring mutation levels (N=533) relative to maternal (N=44) mutation levels for the m.3875delC mutation. (b) Change in average litter mutation load over five generations from 9 individual mothers (symbols). (c) Percentage m.3875delC mutation in 819 PGCs isolated from 18 embryos at 13.5 day-post-conception (dpc) showing no evidence of selection. (d) Normalized variance in heteroplasmy levels in the PGCs, oocytes and offspring. The variance in heteroplasmy is critically dependent on the original heteroplasmy value in the single cell zygote, 3.5 P0. We therefore plotted the variance as a function of the original heteroplasmy value for the different data sets. (e) Percentage m.3875delC mutation in 340 oocytes isolated from the ovaries of five 3.5-day-old neonate females with varying mutation levels showing no evidence of selection.
We therefore studied the distribution of heteroplasmy levels in 533 offspring born to 44 mothers transmitting the m.3875delC mutation (raw data in Supplementary table S4). Given the known dependence of offspring heteroplasmy levels on maternal heteroplasmy levels, we measured the heteroplasmy distribution in groups of offspring from mothers known to harbor similar mutation levels. The offspring mutation levels were binned into three groups, based on the mothers’ heteroplasmy level: mothers with 40-60% m.3875delC, 61-70% m.3875delC, and >70% m.3875delC. A Kimura distribution most accurately describes mtDNA heteroplasmy, and is in keeping with random genetic drift and no selection15. For each of these three groups, the distribution of offspring mutation level was compared to a Kimura distribution. The 40%-60% group and the >70% group both deviated significantly from the Kimura distribution (P = 0.04 and P = 0.03 respectively, KS test), indicating that a mechanism other than random drift is affecting the segregation of this mutation. However, there was no difference in the frequency of surviving offspring born to mothers with <70% m.3875delC (13 mothers, 192 pups born, 154 surviving) when compared to >70% (11 mothers, 155 pups born, 136 surviving, P=0.11), in spite of a skewed heteroplasmy distribution amongst offspring born to mothers with high heteroplasmy. This suggests that fecundity was not compromised, and that the mothers were not preferentially eating mildly-sick pups harboring high heteroplasmy levels. The significant difference from the expected offspring mutation level distribution predicted from random drift theory, and the obvious lack of offspring with high mutation levels (Fig.2a, b), imply that purifying selection against the m.3875delC tRNAMet mutation occurs prior to the formation of the offspring.
To determine when the selection was occurring, we analyzed cells of the developing female germ line, by crossing mice transmitting the heteroplasmic mtDNA m.3875delC with a germ line-reporter line (Stella-GFP)16. We measured heteroplasmy levels in 819 primary germ cells (PGCs) from 18 embryos at 13.5 day-post-conception (dpc). We observed a wide variation in heteroplasmy levels (Fig.2c) notably with mutation levels extending up to 100% mutant, in stark contrast to the distribution in the offspring where no high mutation level offspring are observed (Fig.2a). The distribution of mutation levels in the PGCs was consistent with a Kimura distribution (KS test: P=0.5 for mothers with 40%-60% mutant, P=0.5 for mothers with 61% to 70% mutant, and P=0.2 for mothers with > 70% mutant), and therefore consistent with random drift determining oocyte m.3875delC heteroplasmy levels before birth. The variance in heteroplasmy in 13.5dpc germ cells was similar to the level found in offspring (Fig.2d). Thus, the variance in heteroplasmy among offspring is determined embryonically, consistent with a prenatal germline genetic bottleneck governing the segregation of mutated mtDNA.
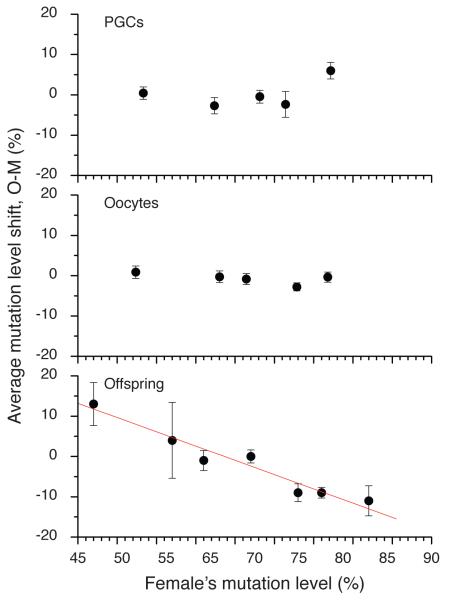
The lack of selection in the germ line at embryonic day 13.5dpc prompted us to investigate the mutation levels in 340 oocytes isolated from the ovaries of five 3.5-day-old neonate females with varying mutation levels (Fig.2e). In all five sets of oocytes, the mutation level distributions were consistent with neutral drift theory (P = 0.31, 0.42, 0.51, 0.08, 0.84, KS test). As a final test for neutral drift, we calculated the difference between the mothers’ mutation level and the mutation levels in the PGCs, oocytes, and offspring (Fig.3). If present, purifying selection would appear in these plots as a significant negative correlation between the mother’s mutation level and the mutation level in the next generation. Consistent with the Kimura distribution tests, both PGCs and oocytes showed no significant correlation, while the offspring showed a very strong and significant (p = 0.002, R = −0.93, linear fit) negative correlation, meaning that the offspring from higher heteroplasmy mothers had lower average heteroplasmy than their corresponding mothers. Thus, all the data and the different analytical approaches showed no evidence for purifying selection at the PGCs and oocyte level, but strong evidence for selection acting on the offspring. Thus, we conclude that selection is likely to have occurred after the oocyte stage. These findings contrast with the conventional view that selection against deleterious mtDNA mutations occurs by oocyte attrition17. However, the absence of oocytes at the high-end of the range (>80% mutant), and the limited data set (n=5) makes it unwise to draw conclusions about the low variance in the oocytes, and we therefore cannot be absolutely certain that selection has not taken place before oocyte formation.
Figure 3.
The average mutation level shift between the next generation (“O”) and the mothers (“M”) for primary germ cells (PGCs), oocytes and offspring for the data shown in Fig.2. Only the offspring show a significant negative correlation, indicative of purifying selection occurring after oocyte heteroplasmy levels are determined.
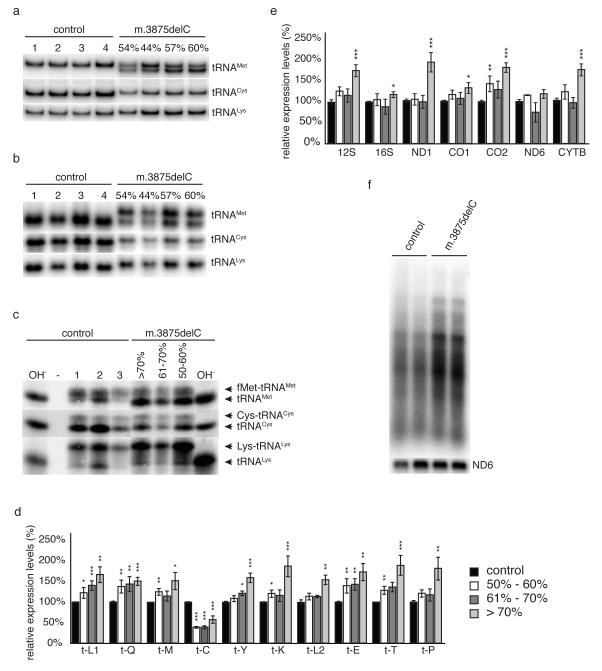
In order to determine the molecular mechanism underpinning the selection, we performed serial cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) histochemistry, respiration measurements, and isolated respiratory chain complex activities in mouse liver and heart homoplasmic for m.5245T>C tRNACys mutation, all showing no defect (not shown). However, given that the heteroplasmic m.3875delC mutation affects the highly conserved tRNAMet anticodon loop, we determined whether the deletion affected tRNA folding and/or wobble base pairing during translation. Separation of total RNA extracts by polyacrylamide gel electrophoresis followed by Northern blot analysis demonstrated that the mutant tRNAMet transcripts were stable, but migrated aberrantly in the gel, consistent with a conformational change in the tRNAMet structure induced by the m.3875delC mutation (Fig.4a&b). In keeping with this, the m.3875delC mutation led to a severe aminoacylation defect in heart and skeletal muscle and, to a lesser extent, in liver (Fig.4c and Supplementary Fig.S2).
Figure 4.
(a-c) Analysis of steady state levels of mitochondrial tRNAs in 16 week old animals with different levels of the m.3875delC mutation of tRNAMet. The percentage of mutated mtDNA refers to the levels in heart. Total RNA was separated by PAGE and blotted and individual tRNAs were detected with radiolabelled probes. (a) Neutral PAGE of total heart RNA demonstrating altered conformation of tRNAMet and decreased levels of tRNACys in mutant animals. (b) Neutral PAGE with non-denatured total heart RNA, demonstrating altered conformation of tRNAMet and tRNACys in mutant animals. (c) Acid-UREA PAGE of total heart RNA extracted under acidic conditions to retain aminoacylation. Base treatment (OH-) of samples prior loading deacylates all tRNAs. (d&e) Relative steady-state levels of mitochondrial (d) tRNAs, (e) mRNAs and rRNAs in hearts from 16 week-old (± 1) mice measured by Northern blot analysis. C57BL/6N (black, n=10-15) m.3875delC mutation at 40-60% (white, n=4-7), 61-70% (dark grey, n=4-7) and >71% (light grey, n=6-11). Levels are normalized to the nuclear encoded 18S rRNA. Error bars show SEM. p<0.05 (*), p<0.01 (**), p<0.001 (***) calculated against C57BL/6N with two-tailed equal variance student t-test. (f) In organello transcription in isolated heart mitochondria from 25 week-old controls and mutant or mice carrying the m.3875delC mutation at >70%.
Analysis of steady-state levels of mitochondrial transcripts in heart revealed a severe reduction of tRNACys associated with the homoplasmic m.5245T>C mutation (Fig.4d), irrespective of the m.3875delC heteroplasmy level in tRNAMet. However, increasing levels of the m.3875delC mutation were accompanied by a progressive increase in steady-state levels of all other mitochondrial mRNAs and tRNAs apart from ND6 (Fig.4e), likely reflecting a compensatory response to both mtDNA mutations. In keeping with this, we saw up-regulation of de novo transcription (Fig.4f) as well as increased steady-state levels of mitochondrial proteins regulating transcription initiation (POLRMT, TFAM) and transcript stability (LRPPRC). A similar compensatory activation of mitochondrial transcription is well documented in mouse knockout models with defective mitochondrial translation caused by inactivation of nuclear genes regulating mitochondrial ribosomal biogenesis18,19. The compensatory mechanism was not accompanied by an increase in the levels of tRNACys (Fig.4d), presumably because the m.5245T>C mutation reduces tRNA stability. However, the decrease in the levels of tRNACys were not, on their own, sufficient to cause an impairment of mitochondrial function. Increased RNA levels likely explain why mitochondrial respiration and isolated respiratory chain complex assembly and activities showed no obvious difference (Supplementary Fig.S3), and why we did not observe an increase in de novo translation (Supplementary Fig.S4). Interestingly, a similar threshold towards a translational defect has previously been described in a patient with a mutation in tRNALys20.
In conclusion, our observations show that intra-familial differences in heteroplasmy are largely determined during pre-natal oocyte development. In contrast to non-synonymous protein coding mutations in mtDNA, deleterious tRNA mutations are not subject to a purifying selection during this process, resulting in oocyte heteroplasmy levels determined by random genetic drift. This has been observed in human oocytes harboring the most common pathogenic heteroplasmic mtDNA mutation, m.3243A>G21, which causes mitochondrial encephalomyopathy with stroke-like episodes. Despite being predicted to be deleterious, in mice, the m.5245T>C tRNACys and the m.3875delC tRNAMet mutations did not cause a biochemical defect when respiratory chain function was assessed in tissue homogenates or in single muscle fibers, but there was an increase of overall mtRNA expression, which was most likely a compensatory response. This points towards a selective mechanism, which reduces the m.3875delC mutational burden by acting at the cellular or organellar-level in the developing embryo. Our findings corroborate observations in a comparable study in humans transmitting m.3243A>G21, which also showed no evidence of selection against a pathogenic variant during oocyte development.
The compensatory mechanism that we propose may prevent a manifest phenotype in the neonatal period. In keeping with this, we saw no impact on fecundity, although we cannot exclude the possibility that very high levels of m.3875delC might lead to pre-natal or neonatal fatality. If correct, this may also explain why many human mtDNA disorders caused by tRNA mutations do not cause symptoms at, or around birth, but present clinically due to the progressive accumulation of mutation in non-dividing post-mitotic tissues causing neurological, ophthalmological, endocrine and cardiac disease.
METHODS
Breeding
The m.3875delC mouse line was derived by backcrossing a homozygous PolGA mutant female to a C57BL/6N male10, followed by continuous backcrossing of the female germ-line, selecting for PolGA wild-type allele11.
Sequencing and mutation level quantification
Total mtDNA sequencing was performed as previously described11. The m.5245T>C and m.3875delC mutations lead to the loss of an HphI and NlaIV restriction endonuclease restriction sites, respectively. Mutation levels were quantified by last-cycle fluorescent restriction fragment length polymorphism analysis. For all mother/offspring calculations ear-clip or tail-snip samples were obtained at weaning. Tissue samples were taken at various ages. Tissue, ear-clip and tail-snip DNA was isolated either by standard Proteinase K, phenol/chloroform extraction followed by salt precipitation, or by boiling the samples for 60min in 75μl of 25mM NaOH, 0.2mM EDTA and neutralizing with an equal volume of 40mM Tris pH7.4. Individual PGCs and primary oocytes were lysed for 16h at 55°C in 20μl 50mM Tris pH8.5, 0.5% Tween 20 and 100μg/ml Proteinase K, followed by 10min 85°C for inactivation. For all reactions 1μl of lysate was used for PCR. Appropriate primers (Supplementary Table S5) were designed to contain 5’ terminal M13 tag sequences and the relevant restriction sites as internal digestion control. The final cycle of a standard 30-35 cycle PCR was spiked with 5μM M13F-hex primer. PCR products were purified using Agencourt Ampure (Beckmann Coultier) and eluted in 30μl dH2O. A 10μl aliquot was digested over night with 1 unit of the appropriate restriction enzyme (NewEngland Biolabs). 1μl of the digests was separated in the presence of a GS500 size standard by fragment analysis on a 3730 DNA analyzer and analyzed using the Genemapper v. 4.0 software (all Applied Biosystems). Peak heights were used for mutation level quantification. Average values of at least three independent experiments were used for mtDNA mutation level quantification of PGCs and offspring. Oocytes were quantified by a singlee experiment.
Isolation of PGCs and primary oocytes
PGCs (gametogonia) were isolated by mating female mice carrying the m.3875delC and m.5245T>C mutations with Stella-GFP BAC–homozygous C57BL/6N males16. Stella-GFP heterozygous embryos were collected at 13.5 dpc in Dulbecco’s minimal essential medium (DMEM, Invitrogen) supplemented with 7.5% FCS and 10mM HEPES. The gonadal ridges of each embryo were dissected using tungsten needles, pooled and washed in PBS, before adding 0.25% trypsin. After 15min incubation at 37°C trypsin was inactivated with FCS and the PGCs released by physical disruption of the gonadal ridges. Single PGCs sorting was performed as previously described4, using a BD FACSAria II or III sorter (Becton Dickinson). PGCs were unidirectionally sorted into single wells of a 96-well plate. GFP was detected using a 100-mW sapphire laser and GFP-positive PGCs were sorted at 20 p.s.i. using a 100-mm nozzle at a sort rate of 2,000 events per second. Instrument sensitivity was proved stable between sorts by internal QC procedures. Plates were stored at –80°C until required. PGCs were lysed and mtDNA mutation levels quantified in triplicate as described above. Primary oocytes were isolated from 3.5day-old neonates (the morning of birth calculated as day 0.5). The ovaries of the neonates were placed in a drop of warm PBS covered by mineral oil (Sigma), and disrupted using fine watchmaker forceps. Primary oocytes were collected by mouth pipetting and transferred through drops of 0.2% hyaluronidase and/or PBS to remove any cumulus cells. Individual primary oocytes were placed into 0.2ml individual capped PCR (Eppendorf) strips by mouth pipetting and lysed analogous to the PGCs.
RNA extraction and Northern blot analysis
Total RNA was extracted using the Totally RNA extraction kit (Ambion) following the manufacturer’s instructions and quantified using a Qubit fluorometer (Invitrogen). Mitochondrial transcript steady-state levels were determined by Northern blot analysis using 750ng of total RNA, essentially as previously described22. Mitochondrial tRNAs were separated in 10% 0.5×TBE polyacrylamide gels, followed by electrical transfer onto nylon membranes (GE Healthcare). For conformational analysis and aminoacylation status of tRNAs, total RNA was isolated with Trizol® reagent (Invitrogen) and resuspended in 0.3M NaOAC (pH5.0), 1mM EDTA. Conformational changes were determined by neutral 0.5×TBE-PAGE, while aminoacylation was performed by acid-UREA PAGE, using 6.5% (19:1) polyacrylamide, 8M UREA gels in 0.1M NaOAc (pH5.0) and run for 48h at 4°C with circulating buffer and regular buffer changes.
Blue-Native PAGE and in-gel complex I activity
Blue-native PAGE for the identification of respiratory chain complexes was performed as previously described23. In-gel complex I activity was measured by incubating blue-native PAGE gels in 2mM Tris-HCl pH7.4, 0.1 mg/ml Nicotinamide adenine dinucleotide (NADH) (Roche) and 2.5 mg/ml iodonitrozolium at room temperature.
Statistical analysis
Mitochondrial transcript steady-state levels were analyzed with two-tailed equal variance student t-test. Error bars show SEM with p<0.05 (*), p<0.01 (**), p<0.001 (***) calculated against C57BL/6N (control) samples. Distributions of mutation level were compared against a distribution calculated from neutral drift theory, the Kimura distribution15, by a Kolmogorov-Smirnov (KS) test comparing the cumulative probability distributions.
Mitochondrial preparations
Tissues were collected in ice cold DPBS (Gibco), minced and homogenized with 6 strokes at 600rpm of a Potter S homogeniser (Sartorius) in 10ml ice cold mitochondria isolation buffer (MIB) (310mM sucrose, 20mM Tris-HCl, 1mM EGTA, pH7.2). Samples were enriched for mitochondria by differential centrifugation and re-suspended in an appropriate volume of MIB. Mitochondrial concentration was determined using either the Bradford or Protein DC Lawry assay (BioRad).
Mitochondrial respiratory rates
Mitochondrial oxygen consumption was measured essentially as previously described22, but at 37°C using 65-250μg of crude mitochondria re-suspended in mitochondrial respiratory buffer (120mM sucrose, 50mM KCl, 20mM Tris-HCl, 4mM KH2PO4, 2mM MgCl2, 1mM EGTA, pH7.2) in an oxygraph chamber (OROBOROS). Oxygen consumption was measured using either PGM (10mM pyruvate, 5mM glutamate, 5mM malate) for predominantly complex I respiration, or 10mM succinate and 10mM rotenone for respiration via complex II. Mitochondrial quality of each sample was assessed by measuring the respiratory control rate (RCR), using 1mM ADP (state 3) or 1mM ADP and 2.5μg/ml oligomycin (pseudo state 4). The RCR values were >10 with PGM and >5 with succinate/rotenone. Respiration was uncoupled by successive addition of Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) up to 3μM to reach maximal respiration. Oxygen flux was normalized to mitochondrial protein content as determined by the protein DC kit (BioRad).
Mitochondrial respiratory chain complex activities
15 to 50μg of mitochondria were diluted in phosphate buffer (KH2PO4 50mM, pH7.4), followed by spectrophotometric analysis of isolated respiratory chain complex activities at 37°C, using a HITACHI UV-3600 spectrophotometer. Citrate synthase activity was measured at 412nm (E=13600M−1.cm−1) after the addition of 0.1mM acetyl-CoA, 0.5mM oxaloacetate and 0.1mM 5,5′-dithiobis-2-nitrobenzoic acid (DTNB). Succinate dehydrogenase (SDH) activity was measured at 600nm (E=21000M−1.cm−1) after the addition of 10mM succinate, 35μM dichlorphenolindophenol (DCPIP) and 1mM KCN. NADH dehydrogenase activity was determined at 340nm (E=6220M−1.cm−1) after the addition of 0.25mM NADH, 0.25mM decylubiquinone and 1mM KCN and controlling for rotenone sensitivity. Cytochrome c reductase activity was assessed at 540nm (E=18000M−1.cm−1) in the presence of 1mM cytochrome c, 0.25mM decylubiquinol and 1mM KCN, controlling for antimycin A sensitivity. Cytochrome c oxidase activity was measured by standard N,N,N′,N′-tetramethylphenylene-1,4-diamine (TMPD) ascorbate assays. All chemicals were obtained from Sigma Aldrich.
In organello translation and transcription
De novo translation was analyzed in freshly isolated mitochondria, as previously described19. Mitochondria were incubated at 37°C in mitochondria translation buffer (220 mM mannitol, 70 mM sucrose, 20 mM HEPES, 2 mM EGTA, 0.1% BSA, (pH7.4)) containing 6 μg/ml of all amino acids except methionine. Easy Tag Express35S Protein labeling mix, a mix of [35S]-Methionine and [35S]-Cysteine (Perkin Elmer), was added to a final concentration of 0.35 mCi/ml and after 1h mitochondria were washed in translation buffer and resuspended in a conventional SDS-PAGE loading buffer. Translation products were separated by SDS-PAGE and analyzed by autoradiography. Coomassie-staining of the SDS-PAGE gels was used as loading control.
De novo transcription was performed as previously described22. Mitochondrial pellets were resuspended in mitochondrial transcription buffer (25mM sucrose, 75mM sorbitol, 100mM KCl, 10mM K2HPO4, 50mM EDTA, 5mM MgCl2, 1mM ADP, 10mM glutamate, 2.5mM malate and 10mM Tris–HCl (pH 7.4), with 1mg/ml BSA). Mitochondria containing 500μg of protein, as determined by Bradford method (BioRad), were incubated for 45min in 500μl of transcription buffer containing 30μCi [α-32P]-UTP at 37°C. Mitochondrial RNA was isolated by ToTALLY RNA isolation kit (Ambion), followed by Northern blot analysis.
Supplementary Material
ACKNOWLEDGEMENTS
PFC is a Wellcome Trust Senior Fellow in Clinical Science and an NIHR Senior Investigator who also receives funding from the Wellcome Centre for Mitochondrial Research, the Medical Research Council (UK) Translational Muscle Centre, the UK NIHR Biomedical Research Centre for Ageing and Age-related disease and NIHR Dementia Biomedical Research Unit awards to the Newcastle upon Tyne Foundation Hospitals NHS Trust. NGL is supported by the Swedish Research Council, the Leducq foundation and a European Research Council Advanced Investigator grant. We would like to thank Christoph Göttlinger and Gunter Rappl at the University of Cologne for help with the FACS sorting. T.W. is supported by a long-term HFSP fellowship.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
REFERENCES
- 1.Upholt WB, Dawid IB. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977;11:571–83. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- 2.Olivo PD, Van de Walle MJ, Laipis PJ, Hauswirth WW. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 1983;306:400–2. doi: 10.1038/306400a0. [DOI] [PubMed] [Google Scholar]
- 3.Cao L, et al. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat Genet. 2007;39:386–90. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- 4.Cree LM, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–54. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 5.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–8. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 6.Wonnapinij P, Chinnery PF, Samuels DC. Previous estimates of mitochondrial DNA mutation level variance did not account for sampling error: comparing the mtDNA genetic bottleneck in mice and humans. Am J Hum Genet. 2010;86:540–50. doi: 10.1016/j.ajhg.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels DC, Wonnapinij P, Cree LM, Chinnery PF. Reassessing evidence for a postnatal mitochondrial genetic bottleneck. Nat Genet. 2010;42:471–2. doi: 10.1038/ng0610-471. [DOI] [PubMed] [Google Scholar]
- 8.Chinnery PF, et al. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–5. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 9.Uusimaa J, et al. Prevalence, segregation, and phenotype of the mitochondrial DNA 3243A>G mutation in children. Annals of Neurology. 2007;62:278–87. doi: 10.1002/ana.21196. [DOI] [PubMed] [Google Scholar]
- 10.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 11.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9:657–62. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 13.Manfredi G, et al. Identification of a mutation in the mitochondrial tRNA(Cys) gene associated with mitochondrial encephalopathy. Human Mutation. 1996;7:158–63. doi: 10.1002/(SICI)1098-1004(1996)7:2<158::AID-HUMU12>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Santorelli FM, et al. Mitochondrial tRNA(Cys) gene mutation (A5814G): a second family with mitochondrial encephalopathy. Neuromuscular Disorders. 1997;7:156–9. doi: 10.1016/s0960-8966(97)00444-6. [DOI] [PubMed] [Google Scholar]
- 15.Wonnapinij P, Chinnery PF, Samuels DC. The distribution of mitochondrial DNA heteroplasmy due to random genetic drift. Am J Hum Genet. 2008;83:582–93. doi: 10.1016/j.ajhg.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payer B, et al. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis. 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- 17.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–97. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Camara Y, et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–39. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Boulet L, Karpati G, Shoubridge EA. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF) American Journal of Human Genetics. 1992;51:1187–200. [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. Random genetic drift determines the level of mutant mitochondrial DNA in human primary oocytes. American Journal of Human Genetics. 2001;68:553–536. doi: 10.1086/318190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freyer C, et al. Maintenance of respiratory chain function in mouse hearts with severely impaired mtDNA transcription. Nucleic Acids Res. 2010;38:6577–88. doi: 10.1093/nar/gkq527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–31. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
Reference for Supplementary Table 1
- 24.Hancock DK, Tully LA, Levin BC. A Standard Reference Material to determine the sensitivity of techniques for detecting low-frequency mutations, SNPs, and heteroplasmies in mitochondrial DNA. Genomics. 2005;86:446–461. doi: 10.1016/j.ygeno.2005.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.