Summary
Epidemic disease caused by Neisseria meningitidis, the meningococcus, has been recognised for two centuries, but remains incompletely controlled and understood. There have been dramatic reductions in serogroup A and C meningococcal disease following the introduction of protein-polysaccharide conjugate vaccines but there is currently no comprehensive vaccine against serogroup B meningococci. Genetic analyses of meningococcal populations have provided many insights into the biology, evolution, and pathogenesis of this important pathogen. The meningococcus, and its close relative the gonococcus, are the only pathogenic members of the genus Neisseria, and the invasive propensity of meningococci varies widely, with around a dozen ‘hyper invasive lineages’ responsible for most disease. Despite this, attempts to identify a ‘pathogenome’, a subset of genes associated with the invasive phenotypes have failed; however, genome-wide studies of representative meningococcal isolates using high throughput sequencing are beginning to provide detail on the relationship of invasive phenotype and genotype in this fascinating organism and how this relationship has evolved.
Keywords: Neisseria meningitidis, population genomics, pathogenomics, molecular evolution, molecular epidemiology
1. A short history of the meningococcus and meningococcal disease
Historical evidence suggests that epidemic meningococcal disease emerged at the beginning of the nineteenth century, with the first likely outbreaks reported only two years apart but in different continents: in Geneva, Switzerland in 1805 [1] and in Medfield, MA, USA a year later [2]. Throughout the succeeding decades, the disease became widely recognized throughout the United States and Europe with a number of large-scale outbreaks. Epidemic meningococcal disease was not described in Africa until 1905, but major epidemics occurred in the meningitis belt for much of the 20th century [3]. The causative organism, now known as Neisseria meningitidis or the meningococcus, was isolated from human cerebrospinal fluid in 1887 [4], and by 1890 it was acknowledged that the organism could be found in an asymptomatic carrier state, in addition to being isolated from cases of invasive disease [5]. Fulminant meningococcal septicaemia, or Waterhouse–Friderichsen syndrome, the most clinically serious manifestation of the disease, was recognised in the early 20th century [6] and, as the century progressed, serological analyses were increasingly used to identify distinct types of meningococcal isolates, eventually recognised as capsular serogroups, of which serogroups A, B, C, W, and Y are most commonly associated with disease [7]. The incidence of meningococcal disease is influenced by the virulence potential of circulating meningococci, which depends at least in part on host and environmental factors, and by host susceptibility to disease [8]. At the time of writing, the disease had an annual incidence rate that ranged from 1 to 1,000 cases per 100,000 individuals in different parts of the world.
The advent of penicillin, and a variety of other antimicrobial agents in the mid 20th century, made a major impact on mortality as invasive meningococci are very susceptible to treatment with systemic antimicrobials. Indeed as long as an appropriate antimicrobial is administered promptly, prognosis is extremely good [9]; however, the principal problem with the treatment of meningococcal disease is that it can develop extraordinarily quickly, with an apparently healthy individual becoming a medical emergency within a matter of hours [8]. This problem is exacerbated by the non-specific nature of early symptoms. Therefore prophylaxis, especially the development and implementation of vaccines, is the best public health intervention against this disease.
The first effective vaccines against this disease, the ‘plain’ polysaccharide vaccines, were developed in the 1960s [10] but were poorly immunogenic and ineffective in interrupting person-to-person transmission over more than a few weeks [11, 12]. At the end of the 20th and beginning of the 21st century, these were replaced with protein conjugate polysaccharide vaccines [13], which were much more immunogenic and capable of interrupting transmission [14], generating herd immunity [15]. Whilst these vaccines proved to be extremely effective against meningococci of serogroups A, C, W and Y, the similarity of the B polysaccharide to the human antigen NCAM (neural cell adhesion molecule) has precluded the development of a serogroup B vaccine [16, 17]. As serogroup B meningococci are the most prevalent causes of disease in many countries [18], the development of a ‘group B’ substitute vaccine remains a major research priority [14]; however, the high antigenic and genetic variability of the meningococcus have proved major obstacles in the development of such a vaccine.
Initial studies of meningococcal variation employed serological analyses, i.e. the comparison of the reactivity of sera raised against a particular meningococcus with other meningococci. This led to the identification of capsular serogroups, which are the consequence of the expression of one of 11 immunochemically distinct capsular polysaccharides [19]. Later studies identified additional antigenic variation in the outer membrane proteins (OMPs) and short-chain lipopolysaccharides (often referred to as lipooligosaccharides, LOSs) of this Gram negative organism, leading to the development of meningococcal serotypes and serosubtypes (OMPS PorB and PorA respectively) and immunotypes (LOS) [20]. Since the widespread availability of nucleotide sequencing, sequence-based typing has become the method of choice for characterising the highly variable surface proteins of this organism [21].
The meningococcus has been in the forefront of work on the population biology of bacteria in general and pathogens in particular, being among the first organisms to be investigated by multilocus enzyme electrophoresis (MLEE) [22] and the first to be examined by multilocus sequence typing (MLST) [23]. Despite intensive research effort over several decades, which has led to a detailed appreciation of many aspects of the genetics of this organism, much remains to be learned concerning its biology and, particularly, why some meningococci are appreciably more invasive than others. Much has been learned, however, by placing this organism in the context of its close relatives, which in general do not cause a pathology similar to the meningococcus and comparative studies have provided valuable insights into meningococcal biology and continue to represent a promising approach for improving our understanding of the pathogenicity of this bacterium in the genomic era [24].
2. The Genus Neisseria
The genus Neisseria comprises Gram-negative, oxidase-positive, aerobic β-Proteobacteria that inhabit human and animal mucosal and dental surfaces [25]. Apart from the meningococcus only one other species, Neisseria gonorrhoeae the gonococcus, is regarded as pathogenic, the other members of the genus being apparently harmless commensal inhabitants of the microbiota that remain relatively stable colonisers over time, despite regular minor perturbations to the oral environment [26].
Unlike its relatives, which inhabit the human oropharynx, the gonococcus colonises the mucosal surface of the urogenital tract and is considered to be an obligate mucosal pathogen that occasionally causes disseminated infection [27]; although it can on rare occasions also inhabit other mucosal sites. Gonococci are most commonly found to infect young adults and do not always cause symptoms, especially in women. The gonococcus is the least diverse of the Neisseria and the present day gonococcal population is likely to be descended, relatively recently, from a single clone, which adapted to the change in niche [28]. Unusually for a ‘single clone pathogen’, that is a pathogen which has arisen by the invasion of a single clone into a pathogenic niche accompanied by genetic isolation [29], the gonococcus has continued to recombine within its own population, whilst being reproductively isolated from other Neisseria, and it has exhibited a remarkable ability to acquire novel genetic elements encoding phenotypes such as antimicrobial resistance determinants [28, 30]. A distinguishing feature of the gonococcus relative to the meningococcus is that, despite the absence of a polysaccharide capsule, it is nonetheless virulent and causes symptomatic disease quite unlike invasive meningococcal strains.
In addition to the two pathogens, the other member of the genus that has been more extensively investigated is Neisseria lactamica, principally because of its lack of association with invasive disease, despite being found at high rates of carriage in infants and young children [31]. Acquisition of N. lactamica is very rapid in the first few years of life and this organism establishes a long-term relationship with its host, with carriage rates declining gradually as the age of the host population rises [32]. In European populations, carriage of N. lactamica is low in adolescents and young adults [33], while the carriage of the meningococcus increases to very high levels at this age [34], especially in closed or semi-closed environments with populations in close association such as military recruits and university dormitories [35-37]. This has led to the suggestions, on more than one occasion: (i) that carriage of N. lactamica is responsible for the increase in immunity against meningococci which is observed during childhood; and, (ii) that deliberate colonisation or immunisation with N. lactamica is a potential approach to prophylaxis against the meningococcus [31, 38].
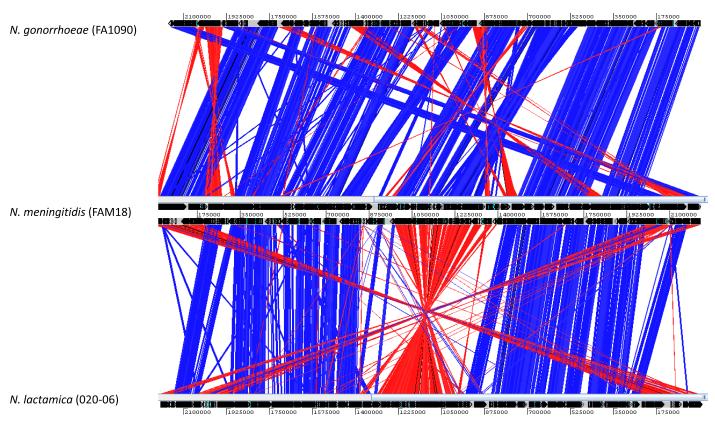
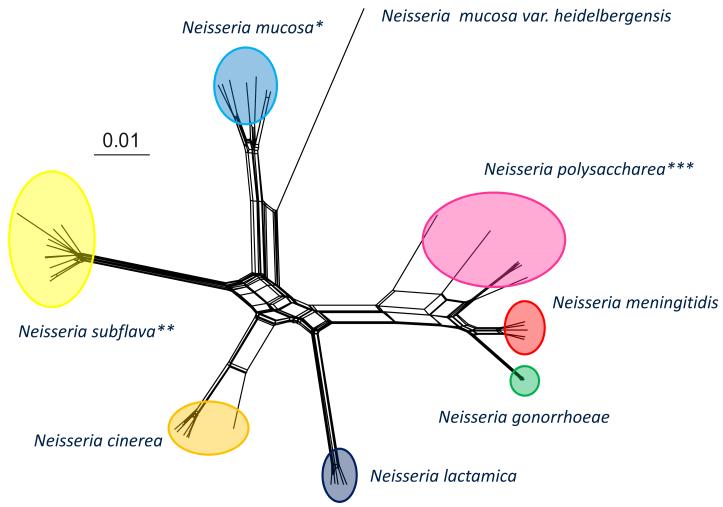
Members of the genus Neisseria are closely related genetically and poorly resolved by techniques for bacterial speciation such as DNA-DNA hybridisation [39] and 16S rRNA sequencing [40], leading to the suggestion that they are not distinct species; however, this suggestion has never found favour among medical microbiologists due to the fact that the defined species nevertheless correspond to particular pathogenic phenotypes. An analysis of three Neisseria genomes using the Artemis Comparison Tool [41] indicates large syntenic regions of similarity; however, further examination of these homologous regions show variability distinct to each species (Fig. 1). Recent genomic analyses show that the microbiologically defined species are, by and large genealogically coherent and distinct groups, which can be reconstructed in a variety of ways including ribosomal MLST, or rMLST (Fig. 2) [42, 43]. The genus provides an excellent model system for studying the genetic traits of closely related organisms that exhibit very different phenotypes, which includes two globally significant diseases [24, 44]. For example why does the meningococcus, normally a generally harmless commensal, occasionally cause disease and what are the differences between it and N. lactamica, which occurs in the same niche? What were the genetic events that led to the gonococcus establishing itself in the urogenital tract and why has this only occurred once [28, 45]? These questions are amenable to population genomic studies which compare the genomes of representative collections of phenotypically defined isolates.
Figure 1.
A comparison of the genome organisation of N. meningitis FAM18 (GenBank accession no. AM421808), N. gonorrhoeae FA1090 (GenBank accession no. NC_002946) and N. lactamica 020-06 (GenBank accession no. FN995097). The red and blue bars indicate regions of similarity with blue bars indicating corresponding regions that are oriented similarly and red bars indicating regions oriented in opposite directions. The coordinates represent the relative nucleotide positions on the genomes.
Figure 2.
Evolutionary relationships among Neisseria isolates based on concatenated sequences of 53 ribosomal protein genes (rMLST). The evolutionary history was inferred using the neighbor-net method. The analysis involved 49 nucleotide sequences consisting of 20927 nucleotides. * This group contained the species Neisseria sicca and Neisseria macacae which are closely related to N. mucosa and are likely to be variants of this species. ** This group also contained the species Neisseria flavescens which is closely related to N. subflava and is likely to be a variant of this species. *** This is a polyphyletic group which suggests that more than one species is present among strains microbiologically defined as N. polysaccharea.
3. Population structure, epidemiology and evolution
The widespread acceptance of seven-locus MLST [46] for the characterisation of Neisseria isolates provided a reproducible and unambiguous means of typing the meningococcus [23], gonococcus [28] and N. lactamica [32], and indeed other species [42]. MLST data confirmed many of the observations made with its predecessor technique MLEE [22, 47], and have enabled studies of the molecular epidemiology [48], evolution [49], and population genetics of these organisms [34]. Other sequence-based identification and typing schemes have been developed, including antigen gene sequence typing (AGST), or ‘fine-typing’, for classification by indexing variation at OMP genes [21] and genes responsible for reduced susceptibility to antimicrobials [50]. These highly reproducible and portable schemes have enabled the assembly of a growing public repository, pubMLST.org/neisseria [51, 52], comprising data from over 17,000 meningococcal isolates from both disease and carriage. At the time of writing this database also contained more than 300 gonococcal, 500 N. lactamica, and over 80 isolates of other Neisseria species.
MLST and AGST data have demonstrated that meningococcal populations, whilst highly diverse and dominated by frequent recombination, are nevertheless highly structured into groups of related organisms referred to as clonal complexes [53]. These clonal complexes correspond to clades reconstructed with phylogenetic and genealogical analyses, although with seven and even 20-locus data relationships among these complexes are not apparent [54]. An important insight gained from the comparative study of isolates from invasive disease and asymptomatic carriage [48] is that some of these clonal complexes are more invasive than others: these are referred to as the ‘hyper invasive lineages’, and only a handful of these have caused the majority of reported disease globally in the last half of the 20th century [53]. These lineages exhibit stability in their antigenic and disease phenotype, including their epidemiology, although these properties are different among different hyperinvasive lineages (Table 1). This has established a robust framework for the comparison of Neisseria genomes, both among and within recognised species groups, with the aim of elucidating the relationship between genotype and phenotype, with the ultimate aim of designing improved interventions to reduce or prevent disease.
Table 1.
A summary generated in March 2012 from the pubMLST.org database, showing the top 20 of 51 N. meningitidis clonal complexes, 6 defined N. lactamica clonal complexes, and unspeciated Neisseria strains considered to be either N. meningitidis or N. lactamica on the basis of their MLST profiles. This table represents about 90% of the profiles in the database. Definitive speciation for 60 Neisseria isolates (marked *) is inconclusive. Clonal complexes considered to represent hyperinvasive lineages are highlighted in grey.
| Clonal complex | Neisseria lactamica | Neisseria meningitidis | Neisseria species* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| disease | carriage | undefined | disease | carriage | undefined | disease | carriage | undefined | ||
| hyperinvasive lineages | ST-41/44 complex | 1,474 | 783 | 288 | ||||||
| ST-11 complex | 1,208 | 158 | 126 | |||||||
| ST-5 complex | 623 | 36 | 20 | |||||||
| ST-8 complex | 370 | 12 | 26 | |||||||
| ST-1 complex | 177 | 46 | 11 | |||||||
| ST-4 complex | 18 | 27 | ||||||||
| ST-32 complex | 969 | 254 | 182 | |||||||
| ST-269 complex | 421 | 137 | 82 | |||||||
| ST-23 complex | 192 | 247 | 63 | 1 | ||||||
| ST-22 complex | 155 | 272 | 42 | |||||||
| ST-35 complex | 155 | 243 | 55 | |||||||
| ST-18 complex | 239 | 52 | 36 | |||||||
| ST-213 complex | 114 | 141 | 58 | |||||||
| ST-53 complex | 6 | 290 | 15 | |||||||
| ST-60 complex | 113 | 155 | 29 | 1 | ||||||
| ST-167 complex | 78 | 141 | 20 | |||||||
| ST-198 complex | 8 | 196 | 34 | 1 | ||||||
| ST-254 complex | 49 | 120 | 24 | |||||||
| ST-162 complex | 74 | 78 | 32 | |||||||
| ST-103 complex | 89 | 58 | 31 | |||||||
| ST-624 complex | 50 | 23 | 4 | |||||||
| ST-640 complex | 42 | 5 | ||||||||
| ST-613 complex | 44 | 2 | ||||||||
| ST-1494 complex | 38 | 1 | 8 | |||||||
| ST-595 complex | 32 | 1 | ||||||||
| ST-1540complex | 21 | 1 | ||||||||
| Unassigned STs | 244 | 35 | 1,298 | 1,452 | 355 | 1 | 34 | |||
Horizontal genetic exchange, was once thought to be uncommon in most prokaryotic species, but is now known to be a major factor in the evolution of most bacterial species [55]; indeed, the lack of genetic exchange is now regarded as a special case, i.e. the genetically monomorphic single clone pathogens such as Mycobacterium tuberculosis, Bacillus anthracis, Yersinia pestis, and Salmonella enterica var. Typhi [29]. Work on the Neisseria, and specifically the meningococcus, was influential in establishing this paradigm [56, 57], as they are naturally competent for DNA uptake and horizontal genetic exchange is most frequently mediated by means of transformation followed by integration into the genome, usually by homologous recombination. Although mostly this exchange occurs among meningococci, occasionally this genetic transfer can occur over large phylogenetic distances, an example being the superoxide dismutase gene, sodC, which appears to have been acquired by N. meningitidis via horizontal transfer from Haemophilus influenzae [58].
This type of ‘localised sex’ in bacteria is most often thought of as a means of generating genetic diversity but, more recent evidence suggests that this process is normally conservative, rather than diversifying [59]. The Neisseria have a number of mechanisms that make the transformation process very specific. The first of these is provided by the DNA uptake sequences (DUS) distributed in the Neisseria chromosome, which play a major role in both the DNA uptake and its integration into the chromosome [59]. In addition, Neisseria have a large number of restriction modification systems, which in the meningococci are characteristic of particular clonal complexes [60], suggesting that a given meningococcus is most likely to incorporate DNA from its very closest relatives [61]. This view is supported by the distribution of the DUSs, which are concentrated in conserved rather than variable regions of the chromosome, suggesting that under normal circumstances horizontal genetic exchange is used as a repair mechanism, rather than as a mechanism for diversification in the meningococci [59].
4. Genetic differences among Neisseria isolates with different phenotypes
The existence of genealogically related organisms with distinct properties presents the possibility of conducting ‘association studies’, where the relationships of particular characteristics of organisms with their phenotypes are investigated. As the amount of genetic information increases, so does the opportunity of investigating the relationships of particular genetic variants with specific phenotypes. In the case of the meningococcus, the phenotype of most interest is the propensity to cause disease and the genetic and phenotypic diversity within the genus presents many opportunities to investigate this. Once an association is established, it is then necessary to establish the causality of this association. It is worth reflecting that this general approach is not new, and historically was the way in which the predominant virulence factor of the meningococcus, the polysaccharide capsule, was characterised [19]: virtually all invasive meningococci are capsulate, a property which confers serum resistance. This serum resistance provides a mechanism for the observed association [62]. Whist a necessary property for invasion however, the expression of one of the disease-associated capsules is not sufficient in itself as many capsulate meningococci are nevertheless unlikely to cause disease [48].
Despite the increases in the data and technology available to undertake association studies, there has been rather limited success in identifying additional determinants that are consistently associated with invasion. Various studies have employed hybridization approaches with microarrays or filters to compare the genes present among panels of isolates against various reference sequences [63-69]. This approach has the advantage that many isolates can be compared, but has the limitation that the method can only efficiently identify the presence or absence of genes present in the reference isolate or isolates – additional genes present in the tested isolates remain unknown as does the influence of sequence variants within genes that are universally present. Thus in such studies inference is limited to identifying the presence and absence of those members of the accessory genome present in the reference genome(s) in the tested strains and sequence variation in the core genome and shared members of the accessory genome is not detected.
Most CGH studies have concluded that, with the exception of the cps region, which encodes the capsule, the invasive phenotype is polygenic in meningococci, with extensive lateral gene transfer reassorting putative virulence determinates [67]. The meningococcal disease associated (MDA) island which was identified by comparisons of a coherent epidemiological isolate collection [70], contributes to the virulence of certain clonal complexes in teenagers, but interestingly not in infants [71], and appears to be an integrated bacteriophage. Other studies have associated particular iron metabolism genes to be positively or negatively associated with the likelihood of causing invasion and these are associated with particular clonal complexes, but the associations are not absolute [72, 73]. Both human and bacterial factors are likely to be important in determining the outcome of infection and although human genetic association studies are difficult to perform for meningococcal disease, because of the low number of well-characterised patients, variants in human factor H have been implicated in the severity of meningococcal disease [74].
5. Meningococcal genomics and vaccine design
The meningococcus was among the first bacteria to have its complete sequence determined [75], an endeavour which was, to a large extent, motivated by the attempts to develop a ‘serogroup B’ meningococcal vaccine [76]. The first isolate to be genome sequenced was a serogroup B meningococcal variant of the ST-32 complex, the laboratory strain MC58 which was derived from an isolate from a protracted hyperendemic outbreak in Stroud Gloucestershire, United Kingdom [77]. The Stroud outbreak was one of many hyperendemics caused by members of this clonal complex (originally termed the ET-5 complex) which occurred globally from the late 1960s onwards and continuing until the time of writing (2012) [78-84]. The MC58 sequence was followed by a serogroup A (Z2491) [85] and then a serogroup C isolate (FAM18) [86].
The MC58 genome was used as the basis of the ‘reverse vaccinology’ approach which was developed to identify novel vaccine candidates from genome sequences [87]. This approach started with the genome sequence, exploiting the sequences to express the genes identified in the annotation process. These were then used in a mouse antibody screen to identify novel antigens that would be potentially protective and hopefully conserved. Whilst this approach has led to the identification of four proteins which are included in a vaccine undergoing trials, it was necessary to include them with an existing outer membrane vesicle vaccine and most of them are not conserved among meningococci but are highly diverse [88]. One of these components, the factor H binding protein, was also discovered by more conventional biochemical methods [89] and displays appreciable antigenic diversity [90] such that multiple variants are required for vaccine coverage.
In an era of whole genome sequences alternative approaches to developing vaccines can exploit genome data from populations, rather than single genomes, as the starting point of the analysis. Such approaches can take advantage of models of strain structure imposed by immune selection to identify proteins that are under immune selection and to rationally design vaccine cocktails that take advantage of the biology and antigenic variation, rather than trying to circumvent it [91]. The long life spans of most hyperinvasive meningococcal strain types, which are measured in several decades at least, combined with their antigenic stability, suggest that this is a feasible approach.
6. The opportunities and challenges presented by advances in genome sequencing
The scale and efficiency of high throughput ‘next generation’ parallel sequencing methods, combined with their reduced costs, provides the unprecedented prospect of large numbers of whole genome sequences becoming available for the meningococcus in the near future. At least in the short term these will not be fully annotated, closed genome sequences, but rather a number of contiguous sequence assemblies generated from short-read sequencing technologies, such as the Illumina HiSeq 2000 instrument. With sequence data availability increasing more rapidly than conventional analyses can be performed, novel strategies for data analysis, storage, and output are required if full advantage is to be taken of them.
The initial studies using this type of data concentrated on bacteria with relatively limited genetic variation, either the study of relatively ‘young’ single clone pathogens, or closely related members of more diverse species, often initially chosen on the basis of MLST data [92, 93]. For such analyses, it was possible to borrow analysis techniques from studies of human populations, which are not very diverse, for the identification of single nucleotide polymorphisms (SNPs) by mapping against a complete closed reference genome. This approach, however, is not appropriate with more diverse bacteria like the meningococcus where there is high diversity, i.e. extensive sequence polymorphism rather than SNPs, and especially when this diversity is reassorted by recombination. Another limitation of these studies was the requirement of a reference genome, a high-quality, finished genome, closely related to the genomes being analysed and against which SNPs can be called. These approaches are not scalable to a situation where many hundreds or thousands of partial genome sequences are being generated for diverse members of a bacterial species.
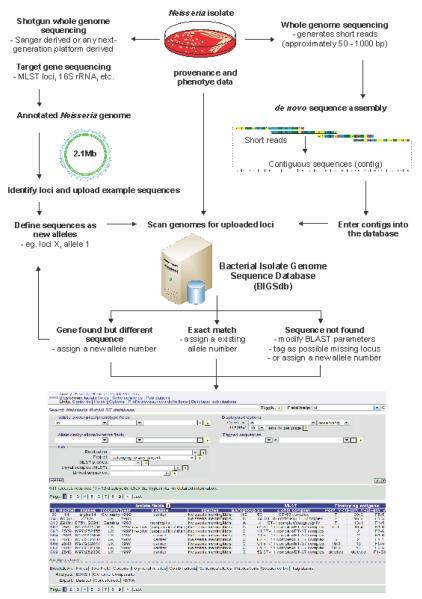
We have recently developed an alternative paradigm for the analysis of whole genome sequence data, which places allelic diversity among bacterial isolates at the centre of multiple genome analysis (Fig. 3). This approach is effectively an expansion of the analysis concept that has been central to the success of MLST [23] and has the advantage that multiple genomes that are highly diverse can be simultaneously analysed. While most MLST schemes index variation at 1-10 ‘housekeeping loci’, usually fragments of genes under stabilising selection [46], there is in principle no limit to the number or type of loci that can be analysed in this way, up to all the loci present in a genome. For the purposes of characterising and cataloguing genetic variation, a locus can be defined as any sequence string, nucleotide or peptide, which can be identified by its sequence, genomic context or a combination of the two. As in MLST, each novel variant of each locus is assigned an allele number and the sequence and the number are stored in a curated data table, providing a comprehensive catalogue of the variation observed to date for that locus. These tables are easily expanded as new variants are detected. The advantage of this approach is that the sequence variation at a given locus can be summarised as a single number, and a genome can be rapidly identified as having a known variant at a particular locus, or a novel variant which can then be added to the curated data set for that locus.
Figure 3.
The BIGSdb integrated online database (http://pubmlst.org/neisseria/), which links provenance, phenotype, bibliographic and sequence data for use in applications for epidemiological, evolutionary, and functional studies. Figure is adapted from SK Sheppard, KA Jolley, MCJ Maiden. A Gene-by-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter, Genes 2012, 3(2), 261-277.
For organisms in which recombination is common [49], this analysis approach has the advantage that analyses based on the alleles present, rather than their sequences or individual nucleotides, inherently corrects for bias introduced by nucleotide-based phylogenetic or genealogical analyses by horizontal genetic exchange; indeed, this was why the approach was used for MLST of such organisms [46]. In organisms where recombination is absent or rare, the sequences can be used directly, as the sequence variation conforms to tree-like evolutionary models [94]. A further level of efficiency in cataloguing variation is achieved by grouping alleles, as is done in MLST, to generate STs which effectively summarise variation data at seven loci with a single number. There is no limit to the number of such schemes that can be defined, each of which will group genetic variation in different ways, for example membership of the same macromolecular structure, such as the ribosome [43], or contribution to a particular phenotype, such as antibiotic resistance. Once the genetic variation of a given isolate is catalogued in this way it can readily be associated with provenance and phenotype data for the isolate, often referred to as metadata.
This approach has been implemented with the BIGSdb [52] software on the PubMLST.org/neisseria website. There are three fundamental levels of interrelated information within the website: (i) a table of isolate provenance and phenotype data; (ii) a ‘sequence bin’ associated with each isolate; and (iii) tables of reference sequences. The isolate information table contains data on the provenance and phenotype of the isolate, together with the any alternative names which have been used and links to relevant records in external databases, such as PubMed. The sequence bin, can contain any type of compiled sequence data, including: (i) single locus sequence data, such as MLST data, 16S rRNA gene or and antimicrobial resistance determinant; (ii) assembled draft genomes, such as those generated from ‘next generation’ short read sequences; or (iii) complete closed genomes sequences. Each of these types of data can reside in a single sequence bin, associated with an experiment type, which indicates the likely quality of the data and can be used to analyses the data preferentially, such that complete closed genome data, where available, will be used in preference to draft genome data.
The tables of reference sequences store any number of references sequences from any number of loci, grouped into any number of schemes, enabling rapid and flexible annotation of the sequences within the sequence bin using well established search algorithms such as BLAST. Once identified the loci are tagged within the sequence bin, for easy future identification and the allelic designations reported back to the isolate information table. This scanning process is automated so that as new genomes are added, they are automatically annotated against the loci defined in the reference tables the reference tables themselves not only define variants and associate them with allele numbers, but can also contain links to other data, including publications, which indicate the function of the loci. These tables also give alternative locus names. To facilitate unambiguous labelling each locus is assigned a unique identifier of the form NEIS0001. Loci are curated, but multiple curators are possible such that individuals expert in a particular locus can have responsibility for that locus. All features of the database are accessible through a web interface, so that the system provides a flexible and backwards compatible means of cataloguing and analysing genome wide information. The PubMLST database employing the BIGSdb software provides a platform upon which population genomic analyses of the Neisseria can be efficiently performed. It contains sequence data generated by single locus, multilocus and whole genome approaches that can be analysed together. A number of data analysis, summary and export tools are built into the system and it is possible to conduct phylogenetic and genealogical analysis on the same data set that is being used for functional studies, enabling the effective fusion of these two powerful but not always well integrated functions. This is especially important in bacteria such as the meningococcus, where population structure has to be taken into account when performing association studies to determine which genes or genetic variants are associated with particular phenotype. The pathogenic phenotype in the meningococcus is polygenic and complex, but the data accumulated to date, which has demonstrated the existence of defined genotypes and their association with particular phenotypes suggests that unravelling the genetic elements associated with these traits will be achievable in the foreseeable future.
Conclusion
Large-scale studies of sequence variation in meningococcal populations have established the central role of horizontal genetic exchange in bacterial evolution. The analyses of these data have demonstrated that, notwithstanding high rates of recombination, meningococcal populations are highly structured [95]. This structure, evident from only a handful of housekeeping genes located at various positions on the chromosome, is associated with particular phenotypic properties, including the expression of particular antigens and the likelihood of causing invasive disease [53]. The current models of bacterial population structures and how they evolve were developed in the 1990s from sequence data [96] and the recent increase in sequencing capacity provides access to the whole genome, enabling these models to be refined, developed, and exploited. It is perhaps worth noting, however, that the genomic revolution has not, at least as yet, led to a fundamental change in these models [97]. A particularly intriguing prospect made possible with whole genome data, is to establish definitively the components of the accessory genome associated with particular clonal complexes and the relationship of this repertoire with sequence variation in the core genome, which has been used to identify clonal complexes. As the data and methods to analyse them expand, we are poised to make exciting novel insights into the biology of this intriguing and dangerous, if accidental [98], pathogen. These insights will lead to the development of improved public health interventions.
Future Perspective
There is little doubt that the number of meningococcal genomes available is going to increase dramatically in the next 5-10 years. At the time of writing it was already more cost effective to obtain whole genome sequences on the Illumina platform than to perform MLST using seven PCR reactions, at least at major genome centres. It is very likely that within ten years whole genome sequencing will become the method of choice for the characterisation of clinical isolates of the meningococcus, and perhaps even the sequencing of clinical specimens such as blood and CSF, from which meningococci cannot be cultured due to prior antibiotic treatment [99]. In addition to resolving clinical questions this will generate an invaluable resource for understanding the genetics of the meningococcus, perhaps in conjunction with the genetics of the host. However, exploitation of this resource will depend on the data being available in a structured form and comparison with equivalent data from meningococcal isolates obtained from asymptomatic carriage and other Neisseria species, as exemplified by the gene-by-gene analysis and annotation approach implemented on PubMLST.org.
Executive Summary
Epidemic meningococcal disease, which takes to the form of meningitis and septicaemia, was first recognised in the early 19th century and continues to have a global impact on human health.
With the exception of the meningococcus, Neisseria meningitidis, and the gonococcus, Neisseria gonorrhoeae the causative agent of gonorrhoea, the members of the genus Neisseria are usually harmless commensal members of the microbiota of humans and other animals.
Towards the end of the 20th century, the application of genetic methods established that meningococcal populations are very diverse but highly structured, with some meningococcal genotypes much more likely to cause disease than others.
Comparisons of a number of meningococcal and other genomes have failed to reveal a ‘pathogenome’, that is a set of genes that are limited to the more invasive meningococci. Rather, these studies suggest that there are many ways in which a meningococcus can become more invasive.
Population genomics, the application of ‘next generation’ sequencing methods to large numbers of meningococcal isolates, is generating novel insights into the biology of the meningococcus with implications for improved understanding of the biology of this disease and the development of novel methods for its control.
Acknowledgements
We are grateful for the support of the Wellcome Trust. MCJM is a Wellcome Trust Senior Research Fellow. This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford [52]. The development of this site has been funded by the Wellcome Trust and European Union.
References
Reference notations
(*) – papers of interest 28, 53, 68, 94
(**) – papers of considerable interest 43, 59, 60
- 1.Vieusseux G. Memoire sur le maladie qui a régné a Genêve au printemps de 1805. Journal de Médecine,Chirurgie et Pharmacie. 1806;II:163–165. [Google Scholar]
- 2.Danielson L, Mann E. The history of a singular and very mortal disease which lately made its appearance in Medfield. Med Agric Reg. 1806;1:65–69. [Google Scholar]
- 3.Greenwood B. Manson Lecture. Meningococcal meningitis in Africa. Trans. R. Soc. Trop. Med Hyg. 1999;93(4):341–353. doi: 10.1016/s0035-9203(99)90106-2. [DOI] [PubMed] [Google Scholar]
- 4.Weichselbaum A. Uber die aetiologie der akuten meningitis cerebrospinalis. Fort.der.Med. 1887;5:573–575. [Google Scholar]
- 5.Kiefer F. Zur differential diagnose des erregers der epidemischen cerebrospinalmeningitis und der gonorrhoea. Berl Klin Wochenschr. 1896;33:628–630. [Google Scholar]
- 6.Waterhouse R. A case of suprarenal apoplexy. The Lancet. 1911;1:577–578. [Google Scholar]
- 7.Flexner S. The results of the serum treatment in thirteen hundred cases of epidemic meningitis. J. Exp. Med. 1913;17:553–576. doi: 10.1084/jem.17.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. The Lancet. 2007;369(9580):2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright K, Reilly S, White D, Stuart J. Early treatment with parenteral penicillin in meningococcal disease. BMJ. 1992;305:143–147. doi: 10.1136/bmj.305.6846.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotschlich EC, Goldschneider I, Artenstein MS. Human immunity to the meningococcus IV. Immunogenicity of group A and group C meningococcal polysaccharides. J. Exp. Med. 1969;129:1367–1384. doi: 10.1084/jem.129.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artenstein MS, Gold R, Zimmerly JG, Wyle FA, Schneider H, Harkins C. Prevention of meningococcal disease by group C polysaccharide vaccine. N. Engl. J. Med. 1970;282:417–420. doi: 10.1056/NEJM197002192820803. [DOI] [PubMed] [Google Scholar]
- 12.Sivonen A. Effect of Neisseria meningitidis group A polysaccharide vaccine on nasopharyngeal carrier rates. J. Infect. 1981;3(3):266–272. doi: 10.1016/s0163-4453(81)90934-8. [DOI] [PubMed] [Google Scholar]
- 13.Robbins JB, Schneerson R, Szu SC, et al. Prevention of invasive bacterial diseases by immunization with polysaccharide-protein conjugates. Current Topics in Microbiology and Immunology. 1989;146:169–180. doi: 10.1007/978-3-642-74529-4_18. [DOI] [PubMed] [Google Scholar]
- 14.Tan LKK, Carlone GM, Borrow R. CURRENT CONCEPTS Advances in the Development of Vaccines against Neisseria meningitidis. N. Engl. J. Med. 2010;362(16):1511–1520. doi: 10.1056/NEJMra0906357. [DOI] [PubMed] [Google Scholar]
- 15.Maiden MC, Ibarz-Pavon AB, Urwin R, et al. Impact of Meningococcal Serogroup C Conjugate Vaccines on Carriage and Herd Immunity. J. Infect. Dis. 2008;197(5):737–743. doi: 10.1086/527401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. The Lancet. 1983;2(8346):355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 17.Robbins JB, Schneerson R, Xie G, Hanson LA, Miller MA. Capsular polysaccharide vaccine for Group B Neisseria meningitidis, Escherichia coli K1, and Pasteurella haemolytica A2. Proceedings of the National Academy of Sciences U S A. 2011;108(44):17871–17875. doi: 10.1073/pnas.1114489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27(Supplement 2):B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 19.Vedros NA. Development of meningococcal serogroups. In: Vedros NA, editor. Evolution of meningococcal disease. CRC Press Inc.; Boca Raton, FL: 1987. pp. 33–37. [Google Scholar]
- 20.Frasch CE, Zollinger WD, Poolman JT. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev. Infect. Dis. 1985;7:504–510. doi: 10.1093/clinids/7.4.504. [DOI] [PubMed] [Google Scholar]
- 21.Jolley KA, Brehony C, Maiden MC. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol. Rev. 2007;31(1):89–96. doi: 10.1111/j.1574-6976.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- 22.Caugant DA, Bovre K, Gaustad P, et al. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J. Gen. Microbiol. 1986;132:641–652. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 23.Maiden MCJ, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiden MC. Population genomics: diversity and virulence in the Neisseria. Curr. Opin. Microbiol. 2008;11(5):1–5. doi: 10.1016/j.mib.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tonjum T. Genus I. Neisseria. In: Garrity GM, Brenner DJ, Krieg NR, Staley JR, editors. Bergey’s Manual of Systematic Bacteriology. Springer-Verlag; New York: 2005. pp. 777–798. [Google Scholar]
- 26.Marsh PD, Percival RS. The oral microflora--friend or foe? Can we decide? International Dental Journal. 2006;56(4 Suppl 1):233–239. doi: 10.1111/j.1875-595x.2006.tb00107.x. [DOI] [PubMed] [Google Scholar]
- 27.Morse SA, Knapp JS. The Genus Neisseria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. Springer-Verlag; New York: 1992. pp. 2495–2559. [Google Scholar]
- 28.Bennett JS, Jolley KA, Sparling PF, et al. Species status of Neisseria gonorrhoeae: Evolutionary and epidemiological inferences from MLST. BMC Biology. 2007;5(1):35. doi: 10.1186/1741-7007-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 30.Lewis DA. The Gonococcus fights back: is this time a knock out? Sexually Transmitted Infections. 2010;86(6):415–421. doi: 10.1136/sti.2010.042648. [DOI] [PubMed] [Google Scholar]
- 31.Gold R, Goldschneider I, Lepow ML, Draper TF, Randolph M. Carriage of Neisseria meningitidis and Neisseria lactamica in infants and children. J. Infect. Dis. 1978;137(2):112–121. doi: 10.1093/infdis/137.2.112. [DOI] [PubMed] [Google Scholar]
- 32.Bennett JS, Griffiths DT, Mccarthy ND, et al. Genetic diversity and carriage dynamics of Neisseria lactamica in infants. Infect. Immun. 2005;73(4):2424–2432. doi: 10.1128/IAI.73.4.2424-2432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cartwright KaV, Stuart JM, Jones DM, Noah ND. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol. Infect. 1987;99(3):591–601. doi: 10.1017/s0950268800066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claus H, Maiden MC, Wilson DJ, et al. Genetic analysis of meningococci carried by children and young adults. J. Infect. Dis. 2005;191(8):1263–1271. doi: 10.1086/428590. [DOI] [PubMed] [Google Scholar]
- 35.Caugant DA, Fogg C, Bajunirwe F, et al. Pharyngeal carriage of Neisseria meningitidis in 2-19-year-old individuals in Uganda. Trans. R. Soc. Trop. Med Hyg. 2006;100(12):1159–1163. doi: 10.1016/j.trstmh.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Glitza IC, Ehrhard I, Muller-Pebody B, et al. Longitudinal study of meningococcal carrier rates in teenagers. International Journal of Hygiene and Environmental Health. 2008;211(3-4):263–272. doi: 10.1016/j.ijheh.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Caugant DA, Tzanakaki G, Kriz P. Lessons from meningococcal carriage studies. FEMS Microbiol. Rev. 2007;31(1):52–63. doi: 10.1111/j.1574-6976.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 38.Oliver KJ, Reddin KM, Bracegirdle P, et al. Neisseria lactamica protects against experimental meningococcal infection. Infect. Immun. 2002;70(7):3621–3626. doi: 10.1128/IAI.70.7.3621-3626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guibourdenche M, Popoff MY, Riou JY. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea and “Neisseria polysaccharea”. Annales de l’Institut Pasteur. Microbiologie. 1986;(2):177–185. doi: 10.1016/s0769-2609(86)80106-5. [DOI] [PubMed] [Google Scholar]
- 40.Harmsen D, Singer C, Rothganger J, et al. Diagnostics of neisseriaceae and moraxellaceae by ribosomal DNA sequencing: ribosomal differentiation of medical microorganisms. J. Clin. Microbiol. 2001;39(3):936–942. doi: 10.1128/JCM.39.3.936-942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21(16):3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 42.Bennett JS, Jolley KA, Earle SG, et al. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology. 2012 doi: 10.1099/mic.0.056077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jolley KA, Bliss CM, Bennett JS, et al. Ribosomal Multi-Locus Sequence Typing: universal characterisation of bacteria from domain to strain. Microbiology. 2012;158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falush D. Toward the Use of Genomics to Study Microevolutionary Change in Bacteria. PLoS Genet. 2009;5(10) doi: 10.1371/journal.pgen.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett JS, Bentley SD, Vernikos GS, et al. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics. 2010;11:652. doi: 10.1186/1471-2164-11-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maiden MC. Multilocus Sequence Typing of Bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 47.Caugant DA. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106(5):505–525. [PubMed] [Google Scholar]
- 48.Yazdankhah SP, Kriz P, Tzanakaki G, et al. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 2004;42(11):5146–5153. doi: 10.1128/JCM.42.11.5146-5153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes EC, Urwin R, Maiden MCJ. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 1999;16(6):741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- 50.Taha MK, Vazquez JA, Hong E, et al. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob. Agents Chemother. 2007;51(8):2784–2792. doi: 10.1128/AAC.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jolley KA. Neisseria. MLST website. [Google Scholar]
- 52.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11(1):595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caugant DA, Maiden MC. Meningococcal carriage and disease--population biology and evolution. Vaccine. 2009;27(Suppl 2):B64–70. doi: 10.1016/j.vaccine.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Didelot X, Urwin R, Maiden MC, Falush D. Genealogical typing of Neisseria meningitidis. Microbiology. 2009;155(10):3176–3186. doi: 10.1099/mic.0.031534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323(5915):741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 56.Maynard Smith J, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 57.Maynard Smith J, Smith NH, O’rourke M, Spratt BG. How clonal are bacteria? Proc. Natl. Acad. Sci. USA. 1993;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kroll JS, Wilks KE, Farrant JL, Langford PR. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA. 1998;95(21):12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Treangen TJ, Ambur OH, Tonjum T, Rocha EP. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9(3):R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budroni S, Siena E, Hotopp JCD, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA. 2011;108(11):4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claus H, Stoevesandt J, Frosch M, Vogel U. Genetic isolation of meningococci of the electrophoretic type 37 complex. J. Bacteriol. 2001;183(8):2570–2575. doi: 10.1128/JB.183.8.2570-2575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ram S, Mackinnon FG, Gulati S, et al. The contrasting mechanisms of serum resistance of Neisseria gonorrhoeae and group B Neisseria meningitidis. Mol. Immunol. 1999;36(13-14):915–928. doi: 10.1016/s0161-5890(99)00114-5. [DOI] [PubMed] [Google Scholar]
- 63.Tinsley CR, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA. 1996;93(20):11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect. Immun. 1999;67(11):6119–6129. doi: 10.1128/iai.67.11.6119-6129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stabler RA, Marsden GL, Witney AA, et al. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology. 2005;151(Pt 9):2907–2922. doi: 10.1099/mic.0.28099-0. [DOI] [PubMed] [Google Scholar]
- 66.Hotopp JC, Grifantini R, Kumar N, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152(Pt 12):3733–3749. doi: 10.1099/mic.0.29261-0. [DOI] [PubMed] [Google Scholar]
- 67.Joseph B, Schwarz RF, Linke B, et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS One. 2011;6(4):e18441. doi: 10.1371/journal.pone.0018441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snyder LA, Saunders NJ. The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics. 2006;7:128. doi: 10.1186/1471-2164-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joseph B, Schneiker-Bekel S, Schramm-Gluck A, et al. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J. Bacteriol. 2010;192(20):5363–5377. doi: 10.1128/JB.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bille E, Zahar JR, Perrin A, et al. A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 2005;201(12):1905–1913. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bille E, Ure R, Gray SJ, et al. Association of a bacteriophage with meningococcal disease in young adults. PLoS ONE. 2008;3(12):e3885. doi: 10.1371/journal.pone.0003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tauseef I, Harrison OB, Wooldridge KG, et al. Influence of the combination and phase variation status of the haemoglobin receptors HmbR and HpuAB on meningococcal virulence. Microbiol-Sgm. 2011;157:1446–1456. doi: 10.1099/mic.0.046946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison OB, Evans NJ, Blair JM, et al. Epidemiological evidence for the role of the hemoglobin receptor, HmbR, in meningococcal virulence. The Journal of Infectious Diseases. 2009;200(1):94–98. doi: 10.1086/599377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davila S, Wright VJ, Khor CC, et al. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat. Genet. 2010;42(9):772–U763. doi: 10.1038/ng.640. [DOI] [PubMed] [Google Scholar]
- 75.Tettelin H, Saunders NJ, Heidelberg J, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287(5459):1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 76.Jodar L, Feavers IM, Salisbury D, Granoff DM. Development of vaccines against meningococcal disease. Lancet. 2002;359(9316):1499–1508. doi: 10.1016/S0140-6736(02)08416-7. [DOI] [PubMed] [Google Scholar]
- 77.Cartwright KaV, Stuart JM, Noah ND. An outbreak of meningococcal disease in Gloucestershire. Lancet. 1986;ii:558–561. doi: 10.1016/s0140-6736(86)90124-8. [DOI] [PubMed] [Google Scholar]
- 78.Wylie PaL, Stevens D, Drake W, Stuart J, Cartwright KaV. Epidemiology and clinical management of meningococcal disease in west Gloucestershire: retrospective, population based study. Britisih Medical Journal. 1997;315(7111):774–779. doi: 10.1136/bmj.315.7111.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takahashi H, Kuroki T, Watanabe Y, et al. Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing. J. Med. Microbiol. 2004;53(Pt 7):657–662. doi: 10.1099/jmm.0.45541-0. [DOI] [PubMed] [Google Scholar]
- 80.De Filippis I, Vicente AC. Multilocus sequence typing and repetitive element-based polymerase chain reaction analysis of Neisseria meningitidis isolates in Brazil reveal the emergence of 11 new sequence types genetically related to the ST-32 and ST-41/44 complexes and high prevalence of strains related to hypervirulent lineages. Diagn Microbiol Infect Dis. 2005;53(3):161–167. doi: 10.1016/j.diagmicrobio.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 81.Smith I, Caugant DA, Hoiby EA, Wentzel-Larsen T, Halstensen A. High case-fatality rates of meningococcal disease in Western Norway caused by serogroup C strains belonging to both sequence type (ST)-32 and ST-11 complexes, 1985-2002. Epidemiol Infect. 2006;134(6):1195–1202. doi: 10.1017/S0950268806006248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottfredsson M, Diggle MA, Lawrie DI, et al. Neisseria meningitidis sequence type and risk for death, Iceland. Emerg Infect Dis. 2006;12(7):1066–1073. doi: 10.3201/eid1207.051624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Climent Y, Urwin R, Yero D, et al. The genetic structure of Neisseria meningitidis populations in Cuba before and after the introduction of a serogroup BC vaccine. Infection Genetics and Evolution. 2010;10(4):546–554. doi: 10.1016/j.meegid.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 84.De Filippis I, De Lemos APS, Hostetler JB, et al. Molecular Epidemiology of Neisseria meningitidis Serogroup B in Brazil. PLoS One. 2012;7(3):e33016. doi: 10.1371/journal.pone.0033016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parkhill J, Achtman M, James KD, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404(6777):502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 86.Bentley SD, Vernikos GS, Snyder LA, et al. Meningococcal Genetic Variation Mechanisms Viewed through Comparative Analysis of Serogroup C Strain FAM18. PLoS Genet. 2007;3(2):e23. doi: 10.1371/journal.pgen.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 88.Scarselli M, Arico B, Brunelli B, et al. Rational Design of a Meningococcal Antigen Inducing Broad Protective Immunity. Sci. Transl. Med. 2011;3(91) doi: 10.1126/scitranslmed.3002234. [DOI] [PubMed] [Google Scholar]
- 89.Fletcher LD, Bernfield L, Barniak V, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72(4):2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brehony C, Wilson D, Maiden M. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology. 2009;155:4155–4169. doi: 10.1099/mic.0.027995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buckee CO, Gupta S, Kriz P, Maiden MCJ, Jolley KA. Long-term evolution of antigen repertoires among carried meningococci. Proceedings of the Royal Society B: Biological Sciences. 2010:1635–1641. doi: 10.1098/rspb.2009.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Croucher NJ, Harris SR, Fraser C, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris SR, Feil EJ, Holden MTG, et al. Evolution of MRSA During Hospital Transmission and Intercontinental Spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18(7):315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Buckee CO, Jolley K, Recker M, et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2008;105(39):15082–15087. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spratt BG, Maiden MCJ. Bacterial population genetics, evolution and epidemiology. Proc. R. Soc. Lond. B Biol. Sci. 1999;354(1384):701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Enright MC, Spratt BG. Genomics. The genomic view of bacterial diversification. Science. 2011;331(6016):407–409. doi: 10.1126/science.1201690. [DOI] [PubMed] [Google Scholar]
- 98.Maiden MC. Dynamics of bacterial carriage and disease: lessons from the meningococcus. Adv Exp Med Biol. 2004;549:23–29. doi: 10.1007/978-1-4419-8993-2_5. [DOI] [PubMed] [Google Scholar]
- 99.Ni H, Knight AI, Cartwright K, Palmer WH, Mcfadden J. Polymerase chain reaction for diagnosis of meningococcal meningitis. Lancet. 1992;340:1432–1434. doi: 10.1016/0140-6736(92)92622-m. [DOI] [PubMed] [Google Scholar]