Abstract
Chitin synthases are critical enzymes for synthesis of chitin and thus for subsequent growth and development in insects. We identified the cDNA of chitin synthase gene (CHS) in Aphis glycines, the soybean aphid, which is a serious pest of soybean. The full-length cDNA of CHS in A. glycines (AyCHS) was 5802 bp long with an open reading frame of 4704 bp that encoded for a 1567 amino acid residues protein. The predicted AyCHS protein had a molecular mass of 180.05 kDa and its amino acid sequence contained all the signature motifs (EDR, QRRRW and TWGTR) of chitin synthases. The quantitative real-time PCR (qPCR) analysis revealed that AyCHS was expressed in all major tissues (gut, fat body and integument); however, it had the highest expression in integument (~3.5 fold compared to gut). Interestingly, the expression of AyCHS in developing embryos was nearly 7 fold higher compared to adult integument, which probably is a reflection of embryonic molts in hemimetabolus insects. Expression analysis in different developmental stages of A. glycines revealed a consistent AyCHS expression in all stages. Further, through leaf dip bioassay, we tested the effect of diflubenzuron (DFB, Dimilin ®), a chitin-synthesis inhibitor, on A. glycines' survival, fecundity and body weight. When fed with soybean leaves previously dipped in 50 ppm DFB solution, A. glycines nymphs suffered significantly higher mortality compared to control. A. glycines nymphs feeding on diflubenzuron treated leaves showed a slightly enhanced expression (1.67 fold) of AyCHS compared to nymphs on untreated leaves. We discussed the potential applications of the current study to develop novel management strategies using chitin-synthesis inhibitors and using RNAi by knocking down AyCHS expression.
Keywords: Chitin synthase, Aphis glycines, Integument, Embryo, Diflubenzuron.
Introduction
The cuticle in exoskeleton of insects has an enormous contribution towards their success in the environment as it provides strength, protection against enemies, and prevents water loss 1. The bulk of insect cuticle is made up of chitin. In addition to the exoskeleton, chitin is present in cuticular lining of foregut, hindgut, and tracheae. Chitin is also a constituent of peritrophic membrane (PM) in the insect midgut. Chemically, chitin is a linear polymer of N-acetylglucosamine residues that are linked together by β-1, 4 glycosidic bonds 2. The terminal step in chitin-synthesis pathway is catalyzed by an enzyme named chitin synthase (CHS). CHS belongs to a family of enzymes called as glycosyltransferases that transfer sugar moieties from activated sugar donors to specific acceptors resulting in a glycosidic bond 3. Thus, CHS transfers the sugar moiety of UDP-N-acetylglucosamine to the non-reducing end of developing chitin polymer. However, the catalytic mechanism of CHS is still unclear.
In insects, two chitin synthases occur which are encoded by two different genes: CHS1 and CHS2 4. CHS1 is mainly expressed in the exoskeleton structures and encodes for an enzyme that catalyzes the production of chitin utilized in cuticle and tracheae 3. In addition to the well perceived role of CHS during molting in immature stages of insects, recent studies suggested an expanded role for this particular gene during insect development 5. CHS2 is expressed in the midgut and encodes for an enzyme that is responsible for production of chitin required in PM present in the midgut of insects 6. Both CHS1 and CHS2 are closely related as both were probably derived from a gene duplication event, however, these can be easily separated phylogenetically 3.The first chitin synthase encoding gene in insects was cloned in the sheep blowfly (Lucilia cuprina) 7. Since then, both CHS1 and CHS2 have been cloned and characterized in many insects, including African malaria mosquito (Anopheles gambiae) 8, yellow fever mosquito (Aedes aegypti) 9, fruit fly (Drosophila melanogaster) 10, tobacco hornworm (Manduca sexta) 11-12, red flour beetle (Tribolium castaneum) 13, and a malaria mosquito (Anopheles quadrimaculatus) 14 (Supplementary Material: Table S1). In all but one of these studies, chitin synthases were characterized from holometabolous insects (those with complete metamorphoses).
Chitin synthase protein is composed of 3 domains namely A, B, and C 15. The A domain, located at N-terminal contains 7-10 transmembrane regions and shows little sequence conservation in different organisms 4. The B domain is the catalytic domain and is directed towards the cytoplasm 3. There are several signature motifs present in the B domain that are vital for catalytic function of this enzyme. The different regions of the catalytic domain are highly conserved in different organisms. The domain C is located near C-terminal, and has 3-5 transmembrane regions. These transmembrane regions are highly conserved in terms of their sequence, location and spacing 15.
As insect growth is central to the molting process that involves the degradation and subsequent replacement of cuticle, the inhibition of chitin synthesis presents an attractive opportunity for insect control 3. Insecticides based on chitin synthesis inhibition are safe to humans as the chitin synthesis pathway is absent in vertebrates. Diflubenzuron (DFB), a member of the class Benzylphenolureas, was the first commercial insecticide that acts by inhibiting chitin synthesis in insects 16. Following the treatment with DFB, reduced chitin content has been reported in A. quadrimaculatus 14, D. melanogaster 17 and T. casteneum 18. However, the exact mode of action of DFB still remains unclear. In a recent study to characterize the effects of DFB in insects, genomic tiling array of the model insect T. casteneum was used 18. Interestingly, only 6% of genes in T. casteneum showed differential expression in treated insects. Furthermore, none of genes involved in chitin metabolism including the gene encoding for chitin synthase were affected by DFB treatment. Thus, reduction in chitin content caused by DFB treatment could be due to events that occur downstream of transcription of chitin metabolism genes 18.
Despite that many non-holometabolous insects are also serious agricultural pests, research on CHS and its potential for their control has been limited. The soybean aphid, Aphis glycines Matsumura, is a major pest of soybean throughout soybean-growing regions of the U.S. 19-20. A. glycines has caused wide spread losses (as high as 40%) of soybean yield in the North-central states where 80% of U.S. soybean crops are grown. In order to control the damage by A. glycines, soybean producers have adopted regular scouting and insecticidal sprays as part of management practices, which eventually have led to a significant economic impact on the soybean production systems 20-21. Host plant resistance (HPR), another potential strategy to manage A. glycines, has been hindered by the development of biotypes i.e. insect populations that are virulent to previously known resistant sources 22-23. Due to high cost associated with chemical control and the complications of HPR in the presence of aphid biotypes, novel strategies to manage A. glycines are necessary. Development of new management strategies necessitates exploration of the molecular physiology of A. glycines, which have been severely lacking due to the recent North American invasion (first found in year 2000) and subsequent rapid emergence as a serious threat.
The current study was intended to better understand the molecular structure and function of chitin synthase in A. glycines. Further, to explore the potential of chitin-synthesis inhibition as a management tool, we conducted an assay to investigate the effect of DFB on fitness of A. glycines. Specifically, in this study, we report (1) complete cDNA sequence encoding full-length chitin synthase from A. glycines, (2) expression profile of AyCHS in different tissues and developmental stages of A. glycines, (3) effect of DFB on A. glycines survival, fecundity and body weight and (4) effect of DFB on the expression of AyCHS during nymphal development of A. glycines.
Materials and Methods
Sequence retrieval and analysis
To retrieve cDNAs for chitin synthase genes in A. glycines, sequences of T. casteneum CHS (TcCHS1: NP_001034491.1 and TcCHS2: NP_001034492.1) were used as query in a tblastn search of A. glycines transcriptomic database [24, R. Bansal, unpublished data]. We identified one A. glycines cDNA contig displaying significant similarity to the chitin synthases of T. casteneum (TcCHS1: 74%; TcCHS2: 64%). The identity of chitin synthase cDNA of A. glycines was further confirmed by blastx search at NCBI-GenBank. Based on known insect chitin synthases, cDNA and deduced protein sequences of AyCHS appeared to be complete (Note: we have chosen the abbreviation Ay to avoid confusion with the Ag abbreviation used for genes from Anopheles gambiae). The ORF finder tool at the National Center for Biotechnology Information (NCBI) was used to identify the open reading frame (ORF) of AyCHS. The transmembrane helices in the AyCHS protein sequence were predicted at TMHMM Server v. 2.0. The putative catalytic domain of AyCHS protein was predicted using SMART tool 25. The putative N-glycosylation sites were predicted by PROSCAN 26. Multiple alignments of various protein sequences were performed by using ClustalW 27-28. The AyCHS cDNA sequence was deposited in the NCBI GenBank (accession number JQ246352).
Phylogenetic analysis of insect chitin synthases
The phylogenetic analysis was conducted in MEGA5.05 software 29. To infer the evolutionary history, the Neighbor-Joining method (with pairwise deletion) was used. A bootstrap test was conducted (10000 replicates) to calculate the percentages of replicate trees in which sequences clustered together. For phylogenetic analysis, chitin synthases were included from A. glycines (Ay), Acyrthosiphon pisum (Ap), Laodelphax striatellus (Ls), Nilaparvata lugens (Nl), Pediculus humanus (Ph), Locusta migratoria (Lm), Manduca sexta (Ms), Aedes aegypti (Aa), Anopheles gambiae (Ag), Apis mellifera (Am), Drosophila melanogaster (Dm), Tribolium castaneum (Tc), Plutella xylostella (Px), and Caenorhabditis elegans (Ce). The GenBank accession numbers are Ay (JQ246352), Ap (XP_003247517.1), Ls (ADR73029.1), Nl (AEL88648.1), Ph1 (XP_002423597.1), Lm1 (ACY38588.1), Ms1 (AAL38051.2), Ms2 (AAX20091.1), Ag1 (XP_321336.4), Ag2 (XP_321951.1), Dm1 (NP_524233.1), Dm2 (NP_524209.2), Aa1 (XP_001662200.1), Aa2 (XP_001651163.1), Px1 (BAF47974.1), Am1 (XP_395677.4), Am2 (XP_001121152.2), Tc1 (NP_001034491.1), Tc2 (NP_001034492.1), Ce1 (NP_492113.2).
Insect culture
For qPCR and leaf dip bioassays, A. glycines insects were obtained from a laboratory colony, referred as biotype 1 (B1) that originated from insects collected from Urbana (IL, USA; 40ᵒ06'N, 88ᵒ12'W) in 2000 30. At Ohio Agricultural Research and Development Center (OARDC, Wooster, OH), a laboratory population of these insects is maintained on susceptible soybean seedlings [SD01-76R (2)] in a rearing room at 23-25°C and 15:9 (Light:Dark) photoperiod.
Tissue and developmental expression of AyCHS
The AyCHS expression was measured primarily in chitin-containing tissues, specifically the gut, integument, fat body, and embryo (developing inside adults) of A. glycines. To obtain selected tissues samples, A. glycines adults (5 days old) were dissected out in phosphate buffer saline (pH 8) under a dissection microscope. The A. glycines embryos which resemble miniature nymphs and adults were cleanly removed from the abdomen of adult aphids. To determine the expression of AyCHS in different developmental stages, all the 4 nymphal and adult samples (whole body) were collected from insects feeding on susceptible soybean [SD01-76R (2)] plants. Both tissue and whole body samples were processed for total RNA extraction by using TRI reagent (Molecular Research Center Inc, Cincinnati, OH, USA), following the protocol provided by the manufacturer. To remove DNA contamination, total RNA samples were treated with TURBO™ DNase (Applied Biosystems/Ambion, Austin, TX, USA). Using iScriptTM cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA), first strand cDNA was prepared with 150 ng and 500 ng RNA (DNA free) from tissue and developmental stages samples, respectively. qPCR was employed to determine the expression of AyCHS in various tissues and developmental stages of A. glycines. The reactions were performed with iQ SYBR green super mix on a CFX-96 thermocycler system (Bio-Rad, Hercules, CA, USA) 31. AyCHS gene-specific primers [forward: AAATATACGCCAAAGTCTT, reverse: GGATAGCAAGGTTATTCAT] were designed using Beacon Designer version 7.0 (Palo Alto, CA, USA). PCR amplification with primers resulted in a 111 bp fragment within the coding region of AyCHS. Due to consistent expression, A. glycines specific EF1α [forward: CTACTGCTACGCCTATTC and reverse: GGTGTCATCAAGAGTGTAA] was used as internal control 32. Prior to PCR, cDNA preparations from developmental stages were diluted 1.5X with nuclease free water. Each reaction was performed with 1 μl of cDNA, 0.5 μM of each primer and 12.5 μl of iQ SYBR green super mix in 25 μl total volume. Each reaction was done in duplicate in a 96-well optical-grade PCR plates, sealed with optical sealing tape (Bio-Rad Laboratories, Hercules, CA). The PCR amplifications were done with the following cycling conditions: one cycle at 95°C (3 min), followed by 35 cycles of denaturation at 95°C (30 seconds), annealing and extension at 55°C for 45 sec. Finally, melt curve analyses were done by slowly heating the PCR mixtures from 55 to 95°C (1°C per cycle of 10 s) with simultaneous measurements of the SYBR Green I signal intensities. A total of three biological replications for tissue and two for developmental stage samples were performed. The data on AyCHS expression at different stages and in different tissues of A. glycines was normalized by subtracting cycle threshold (Ct) values from the corresponding EF1α Ct values. The relative expression level of AyCHS in different tissues and developmental stages was determined by comparative Ct method (2-∆Ct). The significance of differences in the AyCHS expression was determined by t-test.
Leaf dip bioassay
DFB toxicity: Two different concentrations (5 ppm and 50 ppm) of DFB were used. Tween-20 @ 0.05% concentration was used as surfactant. For leaf dip bioassay, fresh trifoliate leaves were clipped from soybean plants [SD01-76R (2)]. Detached leaves were dipped into DFB solution for ~20 seconds and were dried for 2 hrs. For control, detached leaves were dipped into water containing only Tween-20. To determine the performance of A. glycines individuals on DFB-treated leaves, a detached leaf assay as described in 33 was followed. Briefly, single detached leaf was placed on a moist filter paper in a petri dish. Freshly hatched nymphs of A. glycines were placed on these leaves using a camel hair brush. Thereafter, these petri dishes were stored in rearing chambers under suitable environmental conditions [23-25°C, 50-70% relative humidity and 15:9 (Light:Dark) photoperiod]. To maintain appropriate moisture for insect growth, 500 µl of water was added to the filter paper every 12 hrs. Both DFB treatments and control experiment were replicated three times, and each replication consisted of 30 individuals. To measure the DFB toxicity, nymphs that were alive were counted daily for 6 days. A. glycines nymphs that turned brown during assay were considered dead. After 6 days, body weight was measured for survivors. To determine the effect of DFB treatment on A. glycines fecundity, nymphs laid by five adults surviving in each treatment were counted every 24 hrs. To avoid error in fecundity data, freshly hatched nymphs were removed after counting. In this way, fecundity data were recorded for three replications. Data on percent mortality of A. glycines nymphs was transformed using arcsine square root transformations. The transformed data was analyzed using one-way ANOVA. To determine the significant differences within different treatments, Tukey's Honestly Significant Difference (HSD) test was performed.
DFB effect on chitin synthase gene expression: To measure the effect of DFB exposure on chitin synthase gene expression, a separate leaf dip bioassay was performed in the same method as described above. The solution containing DFB @ 50 ppm was used for leaf treatment. Insect samples (10 each) were collected at 3 day and 6 day interval from control and treated leaves. Only live insects were collected for gene expression analysis. Insect samples were processed for RNA extraction, DNAse treatment, first strand cDNA synthesis (using 500 ng RNA from each sample) and qPCR analysis as described in the previous section. For every sample, three biological replicates with each having two technical replicates were performed. Fold changes in the expression of AyCHS in treated samples were determined by comparative Ct method (2-∆∆Ct). The significance of differences in the expression of AyCHS was determined by t-test.
Results
AyCHS cDNA and protein sequence in A. glycines
In the cDNA library of A. glycines, we identified a full length 5802 bp long cDNA encoding for a chitin synthase gene (CHS1). AyCHS cDNA was predicted to contain an open reading frame of 4704 bp that encoded for a 1567 amino acid residue protein. The nucleotide and predicted amino acid sequences of the AyCHS are shown in Fig. 1. The sequence included the ATG start codon at positions 129-131, the stop codon TGA at 4830-4832, and one polyadenylation signal, AATAAA, at 5288-5293. Upon blastn search at Genbank, the nucleotide sequence of the ORF region of AyCHS gene showed highest similarity to that of A. pisum (ApCHS, XM_003247469.1, 92% identity), followed by L. migratoria (LmCHS1, GU067731.1, 71% identity), and P. humanus (PhCHS, XM_002423552.1, 69% identity). The predicted molecular mass of putative AyCHS protein was 180.05 kDa with pI of 6.31. Scanning of deduced amino acid sequence of AyCHS at TMHMM Server v. 2.0 predicted 16 hydrophobic, membrane-spanning α-helices with 9 of them in domain A and 7 in domain C. In addition, AyCHS protein was predicted to contain 12 N-glycosylation sites. The putative catalytic domain of AyCHS was 228 amino acid residues long and contained two signature motifs i.e. EDR (863-865) and QRRRW (900-904). Another signature motif TWGTR (1082-1086) that is supposed to play a role in chitin translocation 3 was found in domain C. Multiple sequence alignment of the region encompassing the catalytic domain of chitin synthases in different hemipteran insects shows 90% identity and 97% similarity in amino acid residues (Fig. 2). Full length AyCHS protein had a high level of amino acid sequence identities with different insect chitin synthases found in GenBank. It showed identities of 98%, 73%, 67% and 66% to the chitin synthases of A. pisum (ApCHS, XP_003247517), L. striatellus (LsCHS, ADR73029), Ostrinia furnacalis (OfCHS1, ACB13821) and Spodoptera exigua (SeCHS, AAZ03545), respectively.
Figure 1.
Nucleotide and deduced amino acid sequences of AyCHS cDNA from Aphis glycines. The start codon (ATG), stop codon (TGA) and putative polyadenylation signal (AATAAA) are highlighted in black. The 16 hydrophobic, membrane-spanning α-helices predicted by TMHMM Server v. 2.0 are highlighted in gray. The amino acid sequence of the putative catalytic domain predicted by SMART tool 28 is boxed. The 12 putative N-glycosylation sites predicted by PROSCAN are underlined. The cDNA sequence was deposited in the GenBank with accession number JQ246352.
Figure 2.
Alignment of putative catalytic domain of chitin synthase gene in different hemipteran insects. The conserved and similar amino acid residues are labeled in black and grey backgrounds respectively. Red boxed amino acid residues represent highly conserved regions in glycosyltransferases (family 2) enzymes (based on 3). Sequence alignment was performed using Clustalw2 program 30-31.The following insect chitin synthase sequences were used in the alignment: ApCHS: Acyrthosiphon pisum, XP_003247517.1; AyCHS: Aphis glycines, JQ246352; LsCHS: Laodelphax striatellus, ADR73029.1; NlCHS: Nilaparvata lugens, AEL88648.1.
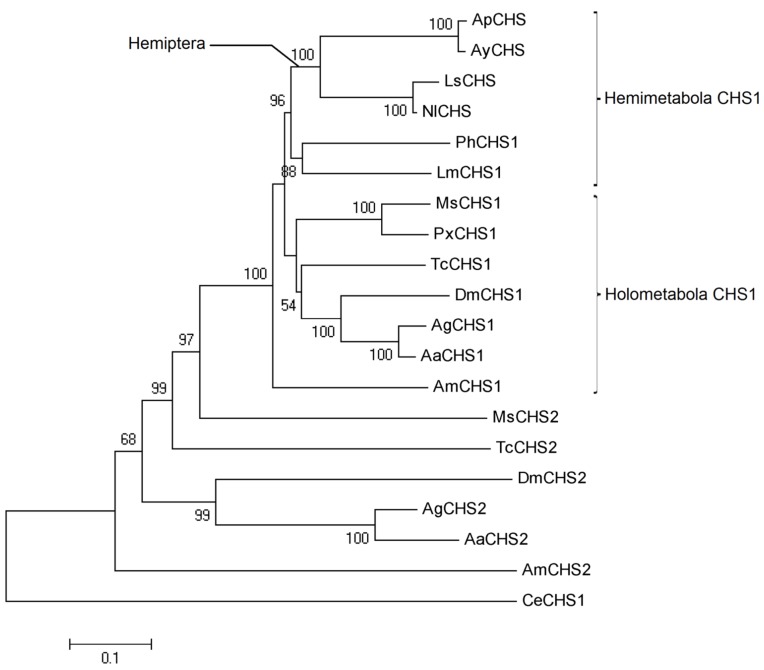
Phylogenetic analysis of insect CHSs
On the basis of amino acid sequence alignment of various insect chitin synthases, a phylogenetic tree was generated using MEGA5.05 (Fig. 3). Clearly, CHS1 and CHS2 from various insect species were placed in two separate clusters. Further, CHS1 from various hemimetabolous and holometabolous insects were grouped into different yet respective clusters. All hemipteran chitin synthases seemed to have a common lineage as high bootstrap value of 88 confirmed their phylogeny. Chitin synthase from A. glycines, AyCHS, was grouped along with that of A. pisum (ApCHS), the only other aphid with known sequence of chitin synthase.
Figure 3.
Phylogeny of insect chitin synthases. A Phylogenetic tree constructed from amino acid sequences of various insect chitin synthases is shown. The percentages of replicate trees in which the sequences clustered together in the bootstrap test (10000 replicates) are shown (only above 50%) next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The scale bar represents 0.1 expected substitutions per amino acid position. The phylogenetic analysis was conducted in MEGA5.05. Chitin synthases were from Aphis glycines (Ay), Acyrthosiphon pisum (Ap), Laodelphax striatellus (Ls), Nilaparvata lugens (Nl), Pediculus humanus (Ph), Locusta migratoria (Lm), Manduca sexta (Ms), Aedes aegypti (Aa), Anopheles gambiae (Ag), Apis mellifera (Am), Drosophila melanogaster (Dm), Tribolium castaneum (Tc), Plutella xylostella (Px), and Caenorhabditis elegans (Ce). The accession numbers for various chitin synthases used in the phylogenetic analysis are provided in the Materials and Methods section.
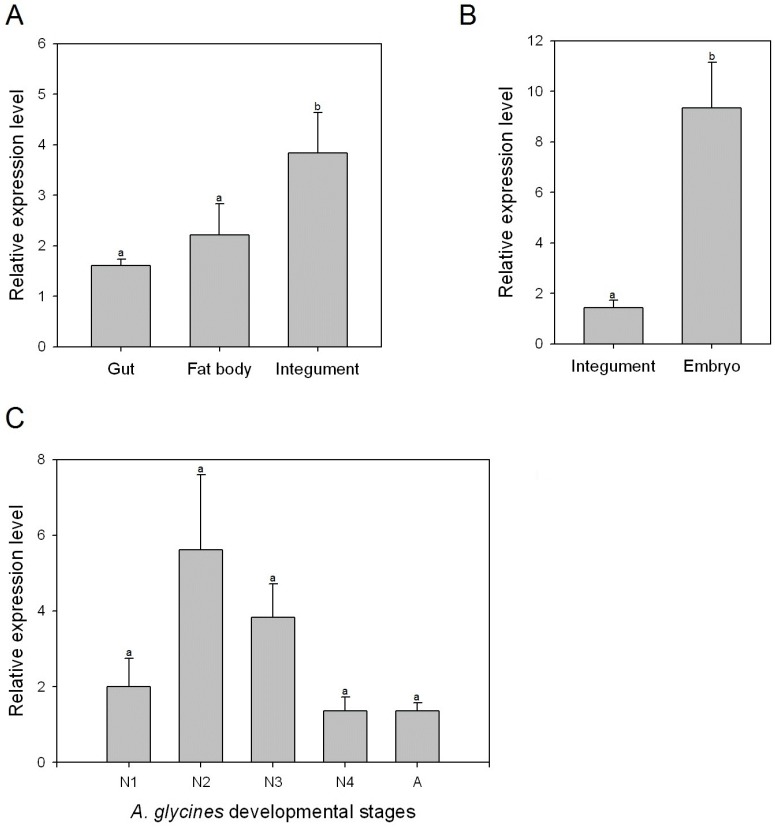
Expression of AyCHS gene in different tissues and developmental stages of A. glycines
Transcript levels of AyCHS in various tissues and developmental stages were determined by qPCR. AyCHS was expressed in all tissues tested (gut, fat body and integument), however, the peak expression was observed in the integument (~3.5 fold compared to gut) (Fig. 4A). Further, the expression of AyCHS in embryos developing inside A. glycines adults was nearly 7 fold higher as compared to that in integument of adults (Fig. 4B). Analysis in different developmental stages of A. glycines revealed that AyCHS is expressed consistently in all stages (Fig. 4C).
Figure 4.
Relative expression levels of chitin synthase gene (AyCHS) in different tissues and developmental stages of A. glycines as determined by real time PCR. Bars were generated after measuring the relative mRNA level of AyCHS gene in A. glycines. The mean (± S.E) expression level is represented for two biological replicates for developmental profiles and two biological replicates for tissue expression profiles. EF1α gene was used as an internal reference gene. The relative expression was calculated based on the value of the lowest expression which was ascribed an arbitrary value of 1. Different letters on the bars of the histogram indicate significant difference in gene expression compared to the treatment with lowest expression at P value < 0.05 (t-test). Different tissues dissected from A. glycines adults are A. gut, fat body and integument B. integument and embryo. Different developmental stages are C. N1-1st instar nymph, N2-2nd instar nymph, N3- 3rd instar nymph, N4- 4th instar nymph, A-adult.
Effect of DFB on A. glycines survival and AyCHS expression
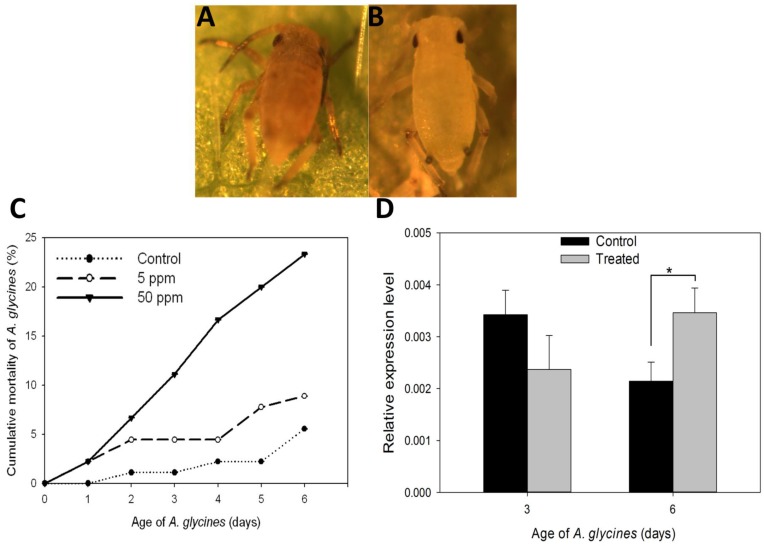
Results on the effect of DFB on A. glycines survival, as determined through leaf dip bioassay, are shown in Fig. 5. Administration of DFB through soybean leaves caused A. glycines' mortality during nymphal molts (F=22.68, P < 0.01) (Fig. 5A). A. glycines nymphs fed with leaves dipped in 50 ppm DFB solution suffered highest mortality at all points of observation except on day 1 (Fig. 5C, Supplementary Material: Table S2). On day 6, cumulative mortality (23.33%) in insects that fed upon leaves dipped in 50 ppm DFB solution was significantly higher as compared to those that fed on control leaves (Tukey's HSD; P < 0.01) and those fed upon leaves dipped in 5 ppm DFB solution (Tukey's HSD; P < 0.01). There were no significant differences in body weight and fecundity of insects that survived the DFB treatment (data not shown).
Figure 5.
Effect on diflubenzuron on A. glycines survival. A. Dead nymphal instar of A. glycines after being fed with soybean leaves that were treated earlier with 50ppm DFB solution. B. Alive and successfully molted nymphal instar of A. glycines after being fed with soybean leaves that were dipped earlier in water (control). C. Mortality of A. glycines nymphs following their feeding upon soybean leaves that were treated earlier with different dosages of DFB. Different data points in the figure represent the percent mortality (± S.E) as calculated from 3 replicates (n=3), each with 30 freshly hatched nymphs. D. Relative expression of the AyCHS in A. glycines feeding upon soybean leaves that were treated earlier with DFB @500ppm (treated) and water (control). Bars were generated after measuring the relative mRNA lelvel of AyCHS in A. glycines nymphs at day 3 and day 6. The mean (± S.D) expression level is represented for three biological replicates (n=3). Asterisk (*) indicates the significant difference at P value < 0.05 (t-test).
On day 3, expression of AyCHS in A. glycines feeding upon treated and control leaves was statistically indistinguishable (t = 2.26, df = 4, P = 0.08) (Fig. 5D). On day 6, expression of AyCHS in A. glycines feeding upon treated leaves increased significantly (by 1.67 fold) compared to those feeding upon control leaves (t = -3.80, df = 4, P < 0.05).
Discussion
Chitin synthase is a key enzyme for the chitin-synthesis pathway in insects. Studies conducted so far on insect chitin synthases have been largely restricted to holometabolous insects. To date, insect chitin synthases have been characterized from at least 15 different holometabolous insects but only 3 hemimetabolous insects 34-36. To better understand the structure and function of chitin synthases, it is important to characterize chitin synthase encoding genes from a diverse group of insects. In this paper, we report, a full-length cDNA encoding chitin synthase from a paraneopteran insect, A. glycines. Paraneoptera is a monophyletic insect group that includes 4 insect orders Hemiptera, Psocoptera, Phthiraptera, and Thysanoptera. Thus, characterization of CHS from A. glycines further enhances the current knowledge and understanding of chitin synthases in insects.
In the transcriptome database of A. glycines, we identified only one transcript encoding an enzyme similar to chitin synthases of T. casteneum. When phylogenetic analysis from putative CHS cDNA and other insect chitin synthases was performed, the cDNA from A. glycines grouped with CHS1 of other insects (Fig. 3). The size of putative CHS cDNA in A. glycines was similar to the CHS1 cDNA in other insects. Taken together, these results suggest that the chitin synthase encoding gene in A. glycines belongs to CHS1 group. From research conducted on insect chitin synthases, it is well established that most insects possess two genes (CHS1 and CHS2) encoding for two distinct chitin synthases 4. CHS1 is mainly expressed in the exoskeleton structures and is important for production of chitin required for cuticle and tracheae. CHS2 is expressed in the midgut and is required for production of chitin in PM of insect midgut. Hemipteran insects, including aphids, are unique in that they lack PM. During the course of evolution, the PM was lost which led to the compartmentalization of the digestive process and ultimately increased digestion of polymers 37-38. Consistent with the lack of a PM, CHS2 seems to be absent in aphids. This is supported by the presence of only one chitin synthase encoding gene (similar to insect CHS1, S. Fig. 1) in A. pisum genome (LOC100162079, Aphidbase). Analyzing the presence or absence of CHS2 in a multitude of aphids and potentially other Hemipterans would be necessary to understand when the loss of CHS2 occurred during the evolution of Aphididae or within the Hemiptera.
Following the absence of PM in aphids, the major chitin containing structure in these insects is integument. Results obtained for the tissue expression studies of AyCHS, for example the highest expression in the integument (Fig. 4A), are in agreement with its perceived role in chitin production in insect exoskeleton 34, 39. Further, chitin is also required for the synthesis of egg shell and embryo cuticle 5. Consistent with the requirement of chitin in embryo cuticle, CHS1 expression occurs in insect embryos 13, 40-41. At the quantitative level, CHS1 expression in embryos is similar to that found in adult insects 13. However, in the current study, we obtained an exceedingly high expression of AyCHS in aphid embryos compared to the adult integument (Fig. 4B). These high transcript levels of AyCHS in A. glycines embryos might probably reflect the uniqueness of embryonic development in hemimetabolous insects. Developing embryos of hemimetabolous insects undergo molting within female reproductive tissues and as a result, three cuticles are formed during embryo development 42. Embryonic molting leads to higher production of chitin during development and thus the higher expression of CHS1 in hemimetabolous insects. The relatively low level of CHS1 in holometabolous embryos is probably due to the absence of embryonic molts within these insects.
Insect CHS1 genes are characterized by the usage of alternate exons leading to the expression of two alternate splice variants 13. These splice variants differ in a short region of 177 nucleotide residues and are differentially expressed in epidermis and tracheae. To detect splice variants of CHS in A. glycines, specific primers 34, 43 were employed to amplify cDNA preparations of different tissues through RT-PCR. These primers were specific for the region corresponding to exon 8 of TcCHS1 13, the well-studied exon for which alternate forms are found in different insects. However, these attempts were not successful as identical AyCHS sequences were recovered from all tissues. Subsequent analysis of genomic fragments encoding chitin synthase in the A. pisum genome (LOC100162079, Aphidbase) (S. Fig. 1) revealed that no alternate form exists for sequence corresponding to exon 8 of TcCHS1 13. It appears that splice variants of CHS1 do not exist in aphids.
As DFB causes a reduction in chitin content, therefore the increased AyCHS expression following the feeding on DFB-treated leaves seems to be plausible as it can lead to an increased enzymatic activity to synthesize chitin. In our study, though the enhanced expression of AyCHS in A. glycines fed with DFB-treated leaves was statistically significant (Fig. 5D), the expression difference (1.67 fold) may not considerable enough to be conclusive. Earlier, it has been reported that CHS expression changes of up to two-fold can occur even among control insect samples 17. Further, results from previous studies do not reveal a consistent pattern on the effect of DFB and other Benzoylphenylureas on insect CHS expression 14, 17, 18, 43. DFB has been found to enhance the CHS1 expression in A.quadrimaculatus 14 but not in D. melanogaster 17 and T. casteneum 18. Thus, additional studies at the transcript and protein levels are required to confirm the biological significance of AyCHS' increased expression as well as the effect of DFB on insect CHS in general.
DFB was the first chitin synthesis inhibitor introduced commercially to control insect pests. It has been recommended for spray in forage crops against the attack of several pests including armyworm, grasshopper, and leafhopper 44. As DFB causes a reduction in chitin content, the insect ultimately dies because of abortive molting 14, 45. We have demonstrated that DFB reduces longevity and causes direct mortality of A. glycines under laboratory conditions (Fig. 5, Supplementary Material: Table S2). To confirm the usefulness and potential of DFB's integration into an IPM strategy to manage this pest, further work should be performed on a larger scale under laboratory and field conditions. Earlier, chloromethiuron, also a chitin synthesis inhibitor, has proved effective in controlling A. glycines under field conditions 46. It was reported that when chloromethiuron was applied at 500 g/ha during the flowering stage, A. glycines populations were controlled without having any adverse effect on natural enemies 46. Given the importance of natural enemies for A. glycines management 19-20, incorporating a chitin-synthesis inhibitor as another tool could be very beneficial for aphid management. Clearly, chitin-synthesis inhibitors including DFB have potential to be a safe and an effective approach to control A. glycines in soybean field.
In addition to chemical applications, chitin synthase genes could be targeted through an RNA-interference (RNAi) based approach 47-48. In this regard, reports on RNAi mediated knockdown of insect CHS genes resulting in lethal phenotype are encouraging 8, 34, 39, 49-51. Even feeding based RNAi that has relevance in field applications has been effective to knockdown CHS 50. The current study provides an ideal platform for future studies to target AyCHS in A. glycines using RNAi. Previous RNAi studies in aphids are promising as A. pisum has shown a robust response to RNAi mediated knockdown 52. In the long term, this approach could prove effective for novel A. glycines management in the field.
Supplementary Material
Table S1. Chitin synthase genes characterized in various insects. Table S2. Effect of DFB at different dosages on the A. glycines' survival. Figure S1. Schematic diagram of the exon-intron organization of the ApCHS gene in A. pisum (LOC100162079, Aphidbase).
Acknowledgments
This research was supported through the North-Central Soybean Research Program and the Ohio Soybean Council (#11-2-360 04).
Abbreviations
- CHS
Chitin synthase
- DFB
Diflubenzuron
- PM
Peritrophic membrane
- RNAi
RNA interference.
References
- 1.Chapman RF. The Insects: Structure and Function. Cambridge, MA: Harvard Univ Press; 1998. [Google Scholar]
- 2.Kramer KJ, Muthukrishnan S. Chitin metabolism in insects. In: Gilbert L.I, latrou K, Gill S, editors. Comprehensive Molecular Insect Science; Vol. 4, Biochemistry and Molecular Biology, Chapter 3. Oxford, UK: Elsevier Press; 2005. pp. 111–144. [Google Scholar]
- 3.Merzendorfer H. Insect chitin synthases: a review. J. Comp. Physiol. 2006;176:1–15. doi: 10.1007/s00360-005-0005-3. [DOI] [PubMed] [Google Scholar]
- 4.Merzendorfer H, Zimoch L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and Chitinases. J. Exp. Bio. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 5.Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2008;38:959–962. doi: 10.1016/j.ibmb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Zimoch L, Merzendorfer H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002;308:287–297. doi: 10.1007/s00441-002-0546-7. [DOI] [PubMed] [Google Scholar]
- 7.Tellam RL, Vuocolo T, Johnson SE, Jarmey J, Pearson RD. Insect chitin synthase: cDNA sequence, gene organization and expression. Eur. J. Biochem. 2000;267:6025–6042. doi: 10.1046/j.1432-1327.2000.01679.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Zhang J, Park Y, Zhu KY. Idnetification and characterization of two chitin synthase genes in African malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 2012;42:674–682. doi: 10.1016/j.ibmb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim GH, Smartt CT, Kiley LM, Christensen BM. Cloning and characterization of a chitin synthase cDNA from the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2000;30:1213–1222. doi: 10.1016/s0965-1748(00)00100-4. [DOI] [PubMed] [Google Scholar]
- 10.Gagou ME, Kapsetaki M, Turberg A, Kafetzopoulos D. Stage-specific expression of the chitin synthase DmeChSA and DmeChSB genes during the onset of Drosophila metamorphosis. Insect Biochem. Mol. Biol. 2002;32:141–146. doi: 10.1016/s0965-1748(01)00101-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhu YC, Specht CA, Dittmer NT, Muthukrishnan S, Kanost MR, Kramer KJ. Sequence of a cDNA and expression of the gene encoding a putative epidermal chitin synthase of Manduca sexta. Insect Biochem. Mol. Biol. 2002;32:1497–1506. doi: 10.1016/s0965-1748(02)00070-x. [DOI] [PubMed] [Google Scholar]
- 12.Hogenkamp DG, Arakane Y, Zimoch L, Merzendorfer H, Kramer KJ, Beeman RW, Kanost MR, Specht CA, Muthukrishnan S. Chitin synthase genes in Manduca sexta: Characterization of a gut-specific transcript. Insect Biochem. Mol. Biol. 2005;35:529–540. doi: 10.1016/j.ibmb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Arakane Y, Hogenkamp DG, Zhu YC, Kramer KJ, Specht CA, Beeman RW, Kanost MR, Muthukrishnan S. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem. Mol. Biol. 2004;34:291–304. doi: 10.1016/j.ibmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JZ, Zhu KY. Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron. Insect Biochem. Mol. Biol. 2006;36:712–725. doi: 10.1016/j.ibmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Merzendorfer H. The cellular basis of chitin synthesis in fungi and insects: Common principles and differences. European J. Cell Biol. 2011;90:759–769. doi: 10.1016/j.ejcb.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Cohen E. Chitin synthesis and inhibition: a revisit. Pest Manag. Sci. 2001;57:946–950. doi: 10.1002/ps.363. [DOI] [PubMed] [Google Scholar]
- 17.Gangishetti U, Breitenbach S, Zander M, Saheb SK, Müller U, Schwarz H, Moussian B. Effects of benzoylphenylurea on chitin synthesis and orientation in the cuticle of the Drosophila larva. Eur J Cell Biol. 2009;3:167–180. doi: 10.1016/j.ejcb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Merzendorfer H, Kim HS, Chaudhari SS, Kumari M, Specht CA, Butcher S, Brown SJ, Robert Manak J, Beeman RW, Kramer KJ, Muthukrishnan S. Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum. Insect Biochem Mol Biol. 2012;42:264–276. doi: 10.1016/j.ibmb.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragsdale DW, Landis DA, Brodeur J, Heimpel GE, Desneux N. Ecology and management of the soybean aphid in North America. Annu. Rev. Entomol. 2011;56:375–399. doi: 10.1146/annurev-ento-120709-144755. [DOI] [PubMed] [Google Scholar]
- 20.Tilmon KJ, Hodgson EW, O'Neal ME, Ragsdale DW. Biology of the soybean aphid, Aphis glycines (Hemiptera: Aphididae) in the United States. J. Integ. Pest Mngmt. 2011;2(2):2011. [Google Scholar]
- 21.Song F, Swinton SM. Returns to integrated pest management research and outreach for soybean aphid. J. Econ. Entomol. 2009;102:2116–2125. doi: 10.1603/029.102.0615. [DOI] [PubMed] [Google Scholar]
- 22.Kim KS, Hill CB, Hartman GL, Mian MAR, Diers BW. Discovery of soybean aphid biotypes. Crop Sci. 2008;48:923–928. [Google Scholar]
- 23.Hill CB, Crull L, Herman T, Voegtlin DJ, Hartman GL. A new soybean aphid (Hemiptera: Aphididae) biotype identified. J. Econ. Entomol. 2010;103:509–515. doi: 10.1603/ec09179. [DOI] [PubMed] [Google Scholar]
- 24.Bai X, Zhang W, Orantes L, Jun TH, Mittapalli O, Mian MAR, Michel AP. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS One. 2010;5(6):e11370. doi: 10.1371/journal.pone.0011370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25(1):217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr121. doi: 0.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill CB, Li Y, Hartman GL. Resistance to the soybean aphid in soybean germplasm. Crop Sci. 2004;44:98–106. [Google Scholar]
- 31.Bansal R, Hulbert S, Schemerhorn B, Reese JC, Whitworth RJ, Stuart JJ, Chen MS. Hessian fly-associated bacteria: transmission, essentiality, and composition. PLoS One. 2011;6(8):e23170. doi: 10.1371/journal.pone.0023170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bansal R, Mamidala P, Rouf MAR, Mittapalli O, Michel AP. Validation of reference genes for gene expression studies in soybean aphid, Aphis glycines Matsumura. J. Econ. Entomol. 2012;105:1432–1438. doi: 10.1603/EC12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel AP, Mian MAR, Olivas ND, Cañas LC. Detached leaf and whole plant assays for soybean aphid resistance: differential responses among resistance sources and biotypes. J. Econ. Entomol. 2010;103:949–957. doi: 10.1603/ec09337. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Liu X, Zhang J, Li D, Sun Y, Guo Y, Ma E, Zhu KY. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen) Insect Biochem. Mol. Biol. 2010;40:824–833. doi: 10.1016/j.ibmb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Fan H-W, Huang H-J, Xue J, Wu W-J, Bao Y-Y, Xu H-J, Zhu Z-R, Cheng J-A, Zhang C-X. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae) Insect Biochem. Mol. Biol. 2012;42:637–646. doi: 10.1016/j.ibmb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Zhang H, Li S, Zhu KY, Ma E, Zhang J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria. Insect Biochem. Mol. Biol. doi: 10.1016/j.ibmb.2012.09.002. in press. [DOI] [PubMed] [Google Scholar]
- 37.Terra WR, Ferreira C, De Bianchi AG. Distribution of digestive enzymes among the endo- and ectoperitrophic spaces and midgut cells of Rhynchosciara Americana and its physiological significance. J. Insect Physiol. 1979;25:487–494. [Google Scholar]
- 38.Terra WR, Ferreira C. Insect digestive enzymes: properties, compartmentalization and function. Comp. Biochem Physiol. 1994;109B:1–62. [Google Scholar]
- 39.Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect Mol. Biol. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- 40.Moussian B, Schwarz H, Bartoszewski S, Nusslein-Volhard C. Involvement of chitin in exoskeleton morphogenesis in Drosophila melanogaster. J. Morphol. 2005;264:117–130. doi: 10.1002/jmor.10324. [DOI] [PubMed] [Google Scholar]
- 41.Qu M, Yang Q. Physiological significance of alternatively spliced exon combinations of the single-copy gene class A chitin synthase in the insect Ostrinia furnacalis (Lepidoptera) Insect Mol. Biol. 2012;21:395–404. doi: 10.1111/j.1365-2583.2012.01145.x. [DOI] [PubMed] [Google Scholar]
- 42.Chaika SY, Orlova EY. Embryonic molt numbers in hemimetabolous and holometabolous insects. Russian Entomol. J. 2009;18(3):157–164. [Google Scholar]
- 43.Ashfaq M, Sonoda S, Tsumuki H. Developmental and tissue-specific expression of CHS1 from Plutella xylostella and its response to chlorfluazuron. Pestic. Biochem. Physiol. 2007;89:20–30. [Google Scholar]
- 44.North Carolina State University. North Carolina agricultural chemicals manual 2011. College of Agriculture and Life Sciences, North Carolina State University. 2011.
- 45.Mulder R, Gijswijk MT. The laboratory evaluation of two promising new insecticides which interfere with cuticle formation. Pestic. Sci. 1973;4:737–745. [Google Scholar]
- 46.Qi Y, Ma Z, Shen D, Gao X, Wang Q. Effects of insecticides on population of destructive insects and their natural enemies in the soybean field. Plant Prot. 1987;13:4–6. [Google Scholar]
- 47.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J. Control of coleopteran insect pests through RNA interference. Nat. Biotech. 2007;25(11):1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 48.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotech. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 49.Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD, Kanost M, Beeman RW. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix, respectively. Insect Mol. Biol. 2005;14:453–463. doi: 10.1111/j.1365-2583.2005.00576.x. [DOI] [PubMed] [Google Scholar]
- 50.Tian H, Peng H, Yao Q, Chen H, Xie Q, Tang B, Zhang W. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One. 2009;4:e6225. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu M, Yang Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalis- gene organization, expression pattern and physiological significance. Insect Biochem. Mol. Biol. 2011;41:923–931. doi: 10.1016/j.ibmb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Mutti NS, Park YS, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 2006;6:38. doi: 10.1673/031.006.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolognesi R, Arakane Y, Muthukrishnan S, Kramer KJ, Terra WR, Ferreira C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2005;35:1249–1259. doi: 10.1016/j.ibmb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Yang X, Kumar NS, Tang B, Sun X, Qiu X, Hu J, Zhang W. The class A chitin synthase gene of Spodoptera exigua: molecular cloning and expression patterns. Insect Biochem. Mol. Biol. 2007;37:409–417. doi: 10.1016/j.ibmb.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Kumar NS, Tang B, Chen X, Tian H, Zhang W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;149:447–453. doi: 10.1016/j.cbpb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Chang X, Fan D, Piao D. Molecular cloning and sequence analysis of chitin synthase cDNA from Mamestra brassicae (l.) (lepidoptera: noctuidae) cuticle. J. N.E. Agr. Univ. 2010;17(3):12–18. [Google Scholar]
- 57.Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD. RNA interference as a method for target-site screening in the Western Corn Rootworm, Diabrotica Virgifera Virgifera. J. Insect Sci. 2010;10(162):1–16. doi: 10.1673/031.010.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang YR, Chen L, Ruo-Ren W, Jian-Hui Y, Jian-Liang L. Cloning and expression pattern of chitin synthase (CHS) gene in epidermis of Ectropis obliqua Prout. African J. Biotech. 2010;9(33):5297–5308. [Google Scholar]
- 59.Qu M, Liu T, Yang J, Yang Q. The gene, expression pattern and subcellular localization of chitin synthase B from the insect Ostrinia furnacalis. Biochem. Biophys. Res. Commun. 2011;404:302e307. doi: 10.1016/j.bbrc.2010.11.111. [DOI] [PubMed] [Google Scholar]
- 60.Ampasala DR, Zheng SC, Zhang DY, Ladd T, Doucet D, Krell PJ, Retnakaran A, Feng QL. An epidermis-specific chitin synthase cDNA in Choristoneura fumiferana: cloning, characterization, developmental and hormonal regulated expression. Arch. Insect Biochem. Physiol. 2011;76:83e96. doi: 10.1002/arch.20404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Chitin synthase genes characterized in various insects. Table S2. Effect of DFB at different dosages on the A. glycines' survival. Figure S1. Schematic diagram of the exon-intron organization of the ApCHS gene in A. pisum (LOC100162079, Aphidbase).