Abstract
Premise of the study
Bio-ontologies are essential tools for accessing and analyzing the rapidly growing pool of plant genomic and phenomic data. Ontologies provide structured vocabularies to support consistent aggregation of data and a semantic framework for automated analyses and reasoning. They are a key component of the semantic web.
Methods
This paper provides background on what bio-ontologies are, why they are relevant to botany, and the principles of ontology development. It includes an overview of ontologies and related resources that are relevant to plant science, with a detailed description of the Plant Ontology (PO). We discuss the challenges of building an ontology that covers all green plants (Viridiplantae).
Key results
Ontologies can advance plant science in four keys areas: (1) comparative genetics, genomics, phenomics, and development; (2) taxonomy and systematics; (3) semantic applications; and (4) education.
Conclusions
Bio-ontologies offer a flexible framework for comparative plant biology, based on common botanical understanding. As genomic and phenomic data become available for more species, we anticipate that the annotation of data with ontology terms will become less centralized, while at the same time, the need for cross-species queries will become more common, causing more researchers in plant science to turn to ontologies.
Keywords: bio-ontologies, genome annotation, OBO Foundry, phenomics, plant anatomy, plant genomics, Plant Ontology, plant systematics, semantic web
Data overload is an issue for nearly every branch of plant science. Complete genomes exist for 25 plant species, with more in progress (Joint Genome Institute, 2012), and new high throughput gene expression, proteomics, and phenomics data sets are being generated continuously. Character matrices for systematic studies (both molecular and morphological) are growing larger and more complex. All this information creates exciting new research possibilities, but it comes with the challenge of accessing and integrating data from disparate sources. As plant science research spanning several subdisciplines becomes more integrative and automated, tools that allow both scientists and computers to communicate more effectively with one another are imperative. Ontologies provide such tools, by standardizing terminology, supporting data aggregation and retrieval, and creating a framework for computerized reasoning. While biological ontologies (bio-ontologies) have become indispensable tools for organizing and accessing genomic data from model species, their application in other areas of plant science remains largely in its infancy (Berardini et al., 2004; Yamazaki and Jaiswal, 2005).
An ontology is a way to represent knowledge, by describing the types or classes of entities within a given domain and the relationships among them. By providing standardized definitions for the terms used by scientists to represent these classes, and by defining the logical relationships among these terms, ontologies make information about content explicit for computers, allowing them to discover common meaning in diverse data sets. Thus, ontologies are an important component of many bioinformatics applications (Jensen and Bork, 2010), and they form the foundation of the semantic web (Berners-Lee et al., 2001; Gkoutos, 2006). While ontologies are useful for scientists who want to organize and aggregate knowledge, they are essential for computer applications that need to find, retrieve, and analyze large quantities of data from multiple sources. Bio-ontologies were embraced early on by the medical and model-species genomics communities as a way to effectively access and analyze large quantities of data and to make cross-species comparisons in ways that were relevant to the understanding of human disease (Ashburner et al., 2000; Ashburner and Lewis, 2002). As knowledge of ontologies spreads, ontological applications are being developed for subdisciplines of biology other than genomics (e.g., Madin et al., 2008; Balhoff et al., 2010; Deans et al., 2012). The primary use of ontologies in life sciences is for semantic tagging—associating data with terms in one or more ontologies, including literature annotation (Hill et al., 2008). Tagging allows computers to access and process data based on biological relevance, rather than simple matching of words. This paper provides a background on what bio-ontologies are and why they are relevant to botany and to plant sciences in general. It includes a description of the Plant Ontology and an overview of other relevant ontologies, plus a discussion of the potential uses of ontologies in plant science.
Ontology 101
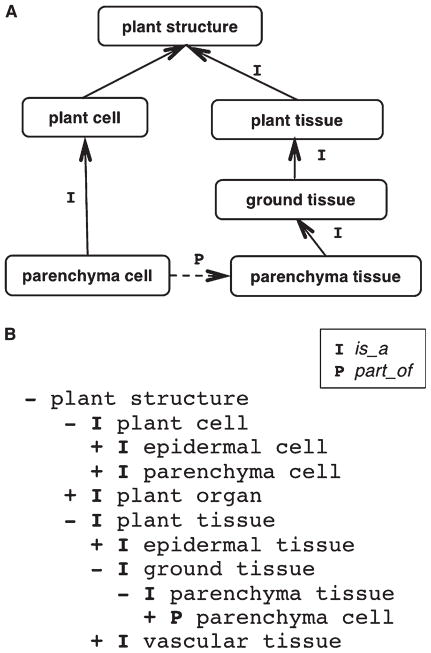
The field of ontology (from the Greek word for the study of being or existence) traditionally falls within the domain of philosophy. The term has been adopted by computer and information scientists, and more recently by biologists, to refer to terminological resources (also called “controlled structured vocabularies”) that are designed to aggregate and classify large quantities of information. When properly constructed, an ontology reflects the consensus understanding of how the reality in a given biological domain is structured, in a way that supports computerized reasoning (Washington and Lewis, 2008). Ontologies model this structure as a collection of types or classes together with certain relationships that hold between them. They do this by constructing the ontology as a graph-theoretic structure consisting of nodes—representing classes—and edges—representing relations (Fig. 1A).
Fig. 1.
(A) A simple ontology in the form of a graph. Boxes represent terms in the ontology and arrows represent relationships. Relationships are read in the direction of the arrow, e.g., “every parenchyma cell is a plant cell” and “every parenchyma cell is part of some parenchyma tissue”. (B) A simple ontology in tree form. Plus signs indicate that a term has additional subclasses not shown in the tree. Relations are read up the tree, from the most indented term to the next highest term of the next highest level, e.g., “every parenchyma cell is a plant cell”.
Each node in the graph is associated with some or all of:
a primary name (typically a scientific term such as parenchyma tissue or parenchyma cell)
synonyms (e.g., equivalent terms used for specific taxa)
equivalent terms in other languages (e.g., “célula parenquimática” and “
 ” as Spanish and Japanese terms, respectively, for parenchyma cell)
” as Spanish and Japanese terms, respectively, for parenchyma cell)a unique alphanumeric identifier forming part of a universal resource identifier (URI)
a definition in English
a corresponding formal definition in some logical language such as the Web Ontology Language (OWL)
citations supporting the definition
comments (e.g., explaining how the definition is to be applied in specific taxa or providing examples).
Throughout this paper, we use italics to mark primary names signifying ontology classes.
From the perspective of philosophical realism, the classes in an ontology are universals (Smith and Ceusters, 2010), each of which is instantiated by spatiotemporal particulars (e.g., the ontology class parenchyma cell is a universal that is instantiated by each of the individual parenchyma cells in the world). The relationships in an ontology define a hierarchy that can also be displayed in a tree-like form (Fig. 1B). The most fundamental relationships represented within an ontology are its SubClassOf (also known as is_a) relationships, specifying the relations between classes and their subclasses, and its part_of relationships.
Computers use the relationships in an ontology, such as is_a and part_of, to support searching and querying. For example, the use of relationships in an ontology allows aggregation of data associated not only with a given class, but also with its subclasses and with classes of its parts. For example, a query for genes expressed in ground tissue using the ontology in Fig. 1 would also identify genes expressed in parenchyma cell and parenchyma tissue, based on their relationships to ground tissue that are captured in the ontology. Using the relationships shown in Fig. 1, software designed to read ontology files could determine that a parenchyma cell is part of some parenchyma tissue, and therefore that any gene expression in a parenchyma cell must also occur in some parenchyma tissue. Likewise, because parenchyma tissue is a subclass of ground tissue, any gene expression in parenchyma tissue must also occur in some ground tissue.
Ontologies are written in specialized languages; the most widely used is the Web Ontology Language or OWL (http://www.w3.org/TR/owl2-overview/; Horridge et al., 2006). OWL is a description logic (DL; Baader, 2003) language with a formally defined semantics. There are many tools and software libraries that support OWL, in particular, a number of computer algorithms called reasoners designed to perform automated classification and consistency checking. Many bio-ontologies, including the PO and the Gene Ontology (GO), have historically been authored using the Open Biomedical Ontologies flat file format (OBOF; http://oboformat.org). The OBOF approximates formally to a subset of OWL. Thus, the OBOF can be mapped to OWL and, subject to certain restrictions, vice versa (Tirmizi et al., 2011). Many bio-ontologies are available for download in both the OBOF and the OWL format. Some ontologies, such as the PO and the GO, can also be viewed using web-based ontology browsers that display not only the ontology, but also associated data that has been annotated using the ontology terms. Appendix S1 (see Supplemental data accompanying online version of this article) provides a screen-shot of the PO view of the term parenchyma cell.
Ontologies go beyond simple terminologies, above all in that they can support automated reasoning by virtue of being written in a formal language like OWL. However, many ontologies, including the PO, provide rich terminological information in addition to formal computable relationships. When discussing bio-ontologies we often advert to this terminological background by referring to the nodes in the ontology as “terms”.
ONTOLOGY RESOURCES FOR PLANT SCIENCE
Overview of ontologies and related resources
Perhaps the best-known bio-ontology is the Gene Ontology (GO), which covers cellular components, biological processes, and molecular functions for all organisms, including plants (Gene Ontology Consortium, 2009). For plant sciences, the main ontology is the Plant Ontology (PO), which covers gross plant anatomy and morphology at the level of the cell and higher (Ilic et al., 2007), as well as plant development stages (Pujar et al., 2006). Plant science researchers can use ontologies such as the PO or GO to associate or “tag” their data with terms for plant anatomy and development, or they can search those ontologies for data that have already been associated with such terms.
The most relevant ontologies for the plant sciences (excluding specialized ontologies used by crop breeders and agronomists), plus resources that aggregate ontologies, are listed in Table 1. These include the Plant Trait Ontology (TO), the Phenotypic Quality Ontology (PATO), and the Protein Ontology (PR), as well as more general ontologies such as the Ontology for Biomedical Investigations (OBI) and the Extensible Observation Ontology (OBOE), both of which can be used to describe experiments or data.
TABLE 1.
Ontologies and other related resources for plant science.
| Resource (abbreviation) | Domain | References |
|---|---|---|
| Plant Ontology (PO) | Plant anatomical entities and plant structure development stages | (Pujar et al., 2006; Ilic et al., 2007) |
| Gene Ontology (GO) | Cellular components, biological processes, and molecular functions | (Gene Ontology Consortium, 2009) http://www.geneontology.org/ |
| Chemical Entities of Biological Interest (ChEBI) | Molecular entities that are natural products or are synthetic products used to intervene in the processes of living organisms | (Degtyarenko et al., 2007; de Matos et al., 2009) http://www.ebi.ac.uk/chebi/ |
| Protein Ontology (PR) | Proteins based on evolutionary relatedness, protein forms produced from a given gene locus, and protein-containing complexes | (Natale et al., 2007; Bult et al., 2011) http://pir.georgetown.edu/pro/ |
| Ontology for Biomedical Investigations (OBI) | Scientific investigations, including the protocols and instrumentation used, the material used, the data generated, and the types of analysis performed | (Brinkman et al., 2010) http://obi-ontology.org |
| Phenotypic Quality Ontology (PATO) | Phenotypic qualities (properties). This ontology can be used in conjunction with other ontologies such as anatomical ontologies to refer to phenotypes. | (Mungall et al., 2010) http://obofoundry.org/wiki/index.php/PATO:Main_Page |
| Plant Trait Ontology (TO) | Phenotypic traits in plants; each trait is a distinguishable feature, characteristic, or quality of a plant | (Jaiswal, 2011) http://www.gramene.org/db/ontology/search?id=TO:0000387 |
| Plant Infectious Disease Ontology (IDOPlant) | Plant infectious diseases, pathogens, and symptoms | (Walls et al., in press) http://purl.obolibrary.org/obo/idoplant.owl. |
| Extensible Observation Ontology (OBOE) | A suite of ontologies for modeling and representing scientific observations | (Madin et al., 2007) https://semtools.ecoinformatics.org/oboe |
| Environment Ontology (EnvO) | Environmental features and habitats | http://environmentontology.org/ |
| NCBI Taxonomy | Biological taxa, based on the classification of the National Center for Biotechnology Information | (Wheeler et al., 2007) http://obofoundry.org/cgi-bin/detail.cgi?id=ncbi_taxonomy |
| BioPortal | Source for finding, searching and querying bio-ontologies | http://bioportal.bioontology.org/ |
| Ontology Lookup Service | Source for finding and searching bio-ontologies | (Côté et al., 2006) http://www.ebi.ac.uk/ontology-lookup/ |
| OntoBee | Source for finding, searching and querying bio-ontologies | (Xiang et al., 2011) http://ontobee.org |
Ontologies can be used on their own or in conjunction with other ontologies. For example, a researcher could combine PO:0020039 (leaf lamina) and PATO: 0001891 (ovate) to describe “ovate leaf lamina shape” (Mungall et al., 2010). Terms from one ontology can also be used to define terms in another ontology, as in the PO definition of plant structure development stage, which refers to GO:0032502 (developmental process). An ontology for plant diseases, linking terms from the PO, the TO, the Infectious Disease Ontology (Cowell and Smith, 2010), and other ontologies, is currently under development and will allow researchers to annotate plant disease data (Walls et al., in press).
Overview of the Plant Ontology
The Plant Ontology project is developing the PO as a general reference ontology for botany and other plant sciences that is designed to establish a semantic framework for queries of gene expression and phenotype data sets across species. The PO was originally designed to cover the model angiosperm species Arabidopsis thaliana, Zea mays, and Oryza sativa, but its scope has been broadened to allow it to keep pace with recent advances in plant science, and particularly with the proliferation of genomic and phenomic data from throughout the plant kingdom. The PO now has terms to cover all green plants (Viridiplantae), from green algae to angiosperms.
The PO contains terms and relations, plus links to data, that cover plant anatomical and morphological entities (such as plant cell or plant organ) and development stages for both plants and plant parts (such as gametophyte development stage or flower development stage). The PO is, accordingly, divided into two main branches: plant anatomical entity and plant structure development stage. As of April 2012, the plant anatomical entity branch of the PO includes 1181 terms for plant morphology and anatomy, including plant structures, plant substances, and plant anatomical spaces or cavities. The subclasses of plant structure represented in the PO include the traditional classifications of plant cell, portion of plant tissue, plant organ, and whole plant, but they also include important categories of structures that have no names in traditional botanical literature, such as collective plant organ structure (a plant structure composed of multiple organs, like a flower or a shoot system), cardinal organ part (a plant structure that is part of a plant organ, such as a lamina or a receptacle), and collective organ part structure (a plant structure composed of parts of multiple plant organs, for example, a pseudostem or a septum). The PO also includes special categories for embryo plant structure and in vitro plant structure.
Most of the common plant structures found in angiosperms and bryophytes are included in the PO, and the ontology has extensive coverage of structures found in lycophytes, pteridophytes, and gymnosperms. Many terms for bryophytes have been added to accommodate annotation of gene expression in the recently sequenced moss Physcomitrella patens (Rensing et al., 2008), with input from the Cosmoss (http://www.cos-moss.org/) and plantco.de (http://plantco.de/) projects. Enrichment for specific structures found in particular angiosperm taxa of economic importance, such as Solanum tuberosum, Z. mays, Musa, and Fabaceae, has been supported by numerous contributors, and new terms are being added continually in reflection of the needs of the users of the PO. Requests for new terms or changes to existing terms can be made by clicking the “Request PO terms” link at the top of Plant Ontology home page (http://plantontology.org). All requests are reviewed by PO curators, who include experts in plant anatomy, morphology, and development, with review of relevant literature and continued feedback from the original submitter. When necessary, experts in particular areas of plant anatomy or development are consulted.
The plant structure development stage branch of the PO includes 271 terms that describe stages in the life of a whole plant or plant part during which the structure undergoes developmental processes such as growth, differentiation, or senescence. The PO does not define the developmental processes themselves, which fall within the domain of the Gene Ontology (Ashburner et al., 2000; Gene Ontology Consortium, 2009). Instead, it uses the relevant GO terms to define the stages in the life cycle of a plant or of part of a plant that are delimited by particular developmental landmarks. Terms representing stages are necessary for describing the conditions under which experiments take place, such as when gene expression or physiological parameters are measured.
The branch of the PO devoted to whole plant development stage includes subclasses such as gametophyte stage and sporophyte stage, plus vegetative, reproductive, senescent, and dormant stages for both the gametophyte and the sporophyte. This branch is particularly rich in terms for angiosperms, but it is being revised to include more terms for other groups of plants. Terms for development stages for parts of plants include leaf development stage, which has subclasses such as leaf initiation stage and leaf expansion stage.
Organizing principles of bio-ontologies
Most of the ontologies listed in Table 1 are associated with the Open Biological and Biomedical Ontologies (OBO) Foundry (http://www.obo-foundry.org; Smith et al., 2007). The OBO Foundry is a collaborative initiative with the goal of creating a set of nonoverlapping, interoperable reference ontologies in the biomedical domain. The member and candidate ontologies that comprise the OBO library must follow the ontology development principles agreed upon by the OBO Foundry (http://obofoundry.org/wiki/index.php/Category:Principles). These include principles for ontology management (for example: appoint a person responsible for liaison with the OBO Foundry, provide a tracker for additions and corrections, provide a help desk for inquiries), principles enjoining collaboration with the developers of neighboring ontologies (reuse terms from other ontologies to the maximal possible degree), and also principles pertaining to specific aspects of developing the ontology files (for example, keep careful track of successive versions). The following sections describe some of the important aspects of ontology development within the OBO Foundry framework, using specific examples from the Plant Ontology.
Unique identifiers
Every term in an ontology should have a stable unique identifier (ID) that corresponds to a universal resource identifier (URI; http://tools.ietf.org/html/rfc3986) in OWL. In OBOF, identifiers take the form ID-Space:Local-ID, where ID-Space is an abbreviation for the ontology (e.g., PO) and Local-ID is a number that is unique within that ID-Space. Each of these identifiers maps to a longer persistent URL (http://www.obofoundry.org/id-policy.shtml). For example, PO:0005421 is the unique identifier for the term parenchyma and has the corresponding URI: http://purl.obolibrary.org/obo/PO_0005421. IDs are permanent and unique, that is, once they are publicly released, they can never be used again. If the term represented by an ID needs to be eliminated for some reason, the term and ID are made obsolete by assigning an is_obsolete tag to them, along with meta-data explaining why the term was made obsolete and suggestions for replacement terms. This action prevents the ID from being reused for other terms, thereby preventing conflicts and reasoning errors.
Term names and synonyms
OBO Foundry naming conventions (Schober et al., 2009) specify that primary term names must be singular nouns or noun phrases; their meaning must be clear to human readers and conform to standard usage in biology. In general, a primary name is the most commonly used name, or a close variant thereof. Each primary name is associated in the ontology with other names given as synonyms. The specific needs of describing ontology classes can lead to term names that may appear artificial but are necessary to describe categories of entities that have no common name. For example, the words “whorl” and “rosette” are commonly used to describe collections of leaves, but there is no common name that covers both structures, so the term collective leaf structure was created for the PO. In many OBO Foundry anatomy ontologies, the pre-fix “portion of” is used when naming classes of tissues (e.g., portion of plant tissue in the PO). This naming convention specifies that the ontology is describing an actual material entity that is a concrete portion (or piece or sample) of tissue, rather than an abstract tissue type.
Most bio-ontologies use synonyms of four different scopes: exact, narrow, broad, and related. Exact synonyms are important when the same structure has multiple names, e.g., phellogen is an exact synonym of cork cambium. Narrow synonyms are often used for structures that have different names in different taxa. For example, pod and achene are narrow synonyms of fruit. Broad synonyms are used when the synonym may encompass multiple entities, e.g., adventitious root is a broad synonym of both basal root and shoot-borne root. Related synonyms are used when a word or phrase has been used synonymously with the primary term name in the literature, but the usage is not strictly correct. For example, carpel septum is a related synonym of ovary septum.
Ontologies can also include synonyms in multiple languages. A new and special feature of the PO is the addition of Spanish and Japanese translations for anatomical and morphological terms, with German translations in preparation. Foreign language terms not only make ontologies more accessible to non-English speakers, but also allow text-processing applications to search and annotate foreign literature.
Standardized definitions
A well-developed ontology should include textual definitions with citations for all terms. Whenever possible, definitions should be obtained from published sources such as reference works or journal articles, adapted by the curators to the terminological usage of the host ontology. When a published definition is unavailable, or when published definitions disagree with each other, they may be written by the curators. This is often the case for upper-level ontology terms in multispecies ontologies (such as organ or portion of tissue), because many published definitions are written for specific taxa, while ontological definitions must be appropriate for all species to which a term can apply.
In a well-constructed ontology, terms can have both logical (computable) definitions and textual definitions. Textual definitions specify the necessary and sufficient conditions for the correct application of a term; thus, they tell us exactly what must hold of an entity if it is to be an instance of a given type, or, in other words, if it is to be a member of a given class. Definitions (both textual and logical) should follow the genus-differentia form: an X is a G (the genus) that D, E, F (the differentiae, or characteristics that serve to differentiate the Xs from the other members of G). For example:
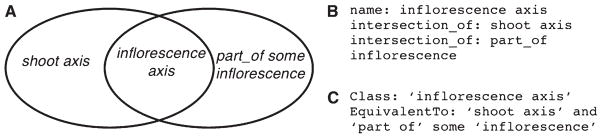
inflorescence axis = def. A shoot axis (genus) that is part of an inflorescence (differentia).
Whereas textual definitions are used by the human users of an ontology, logical definitions are used by computers. They provide the semantic basis to allow the use of tools called reasoners that automatically classify the ontology and detect inconsistencies. This provision is extremely useful for ontologies of moderate to large size that may need to be frequently amended to reflect advances in knowledge. Formally, the computable definition is a statement of equivalence between the class that is being defined and a logical expression involving other simpler classes from the ontology (or from another ontology). For example, in PO, the class inflorescence axis is equivalent to the intersection of the class shoot axis with the class of entities satisfying the condition that they are part of some inflorescence (Fig. 2).
Fig. 2.
(A) Venn diagram to illustrate the logical definition of inflorescence axis as being equivalent to the class intersection of shoot axis and the class of all things that are part of some inflorescence. (B) Syntactic representation in Open Biomedical Ontologies flat file format (OBOF). (C) Equivalent representation in Web Ontology Language (OWL).
Content based on high-level ontologies
To increase their utility and power, ontologies must be interoperable with other ontologies. This interoperability is achieved, in part, by defining terms on the basis of other terms defined in higher-level, more general ontologies—that is, ontologies that are not specific to a narrow domain. For example, the two basic branches of the PO follow the divisions of the Basic Formal Ontology (BFO; Grenon and Smith, 2004) that partition reality into entities that continue to exist through time (continuants such as leaf or stem) and entities in which continuants participate (occurrents such as whole plant developmental stage). Best practice in ontology development involves maximal reuse of terms from existing high-quality ontologies, either by importing such terms using the MIREOT process (Courtot et al., 2011; Xiang et al., 2010) or by using such terms in creating logical definitions. Examples of the latter are the top-level terms in the anatomical entity branch of the PO—plant anatomical entity, plant structure, portion of plant substance, and plant anatomical space—which are defined in terms of the corresponding terms in the Common Anatomy Reference Ontology (CARO; Haendel et al., 2007). Many terms in the PO use terms from the Gene Ontology (GO) in their definitions, and as more logical definitions are added to PO, this reuse of terms from existing ontologies will increase.
Relationships between classes
The power of ontologies to provide a logical framework for data access and analysis lies in the relational graph by which terms are connected. For example, suppose a researcher were to ask a computer to search a data set to find all examples of gene expression in a leaf. Any botanist doing this search would include experiments that described gene expression in petioles or midribs, but a computer would not know to include these results unless it was told by an ontology that every petiole and every midrib are parts of some leaf.
Relationships in OBO Foundry bio-ontologies generally are taken from either the BFO or the Relation Ontology (RO; Smith et al., 2005). The most basic relationship in anatomical ontologies such as the PO is the class–subclass relation, specified as is_a in OBOF or as SubClassOf in OWL (e.g., vascular leaf is_a leaf and epidermal cell is_a plant cell). The PO uses a number of other relations to describe both the spatial and temporal relationships among classes, including part_of (e.g., petiole part_of leaf), develops_from (e.g., root hair cell de-velops_from trichoblast), and participates_in (e.g., sporangium participates_in sporophyte development stage). A complete list of relations used by PO is available at the website http://wiki.plantontology.org/index.php/Relations_in_the_Plant_On-tology. All relationships in the PO are OWL “existential restrictions”, also known as all–some relationships (Smith et al., 2005). In the graphical representations in Fig. 1, each edge (that is, each relationship R, between any two terms A and B) should be read as “all instances of A stand in relation R to some instance of B”. For example, all instances of parenchyma cell are part of some instance of parenchyma tissue.
DISCUSSION
Challenges of building an ontology that spans an entire kingdom
While many reference bio-ontologies like the GO, PATO, and ChEBI (Table 1) are designed to be species neutral, most anatomical and developmental stage ontologies are species or clade specific. The PO is unique among anatomy ontologies in spanning such a broad taxonomic range while simultaneously providing highly detailed coverage of anatomy, morphology, and developmental stages. Inclusive ontologies such as the PO are crucial for comparative research, but they present a number of challenges connected with the need to incorporate the divergent vocabularies used for different taxa and to find commonalities within the phenotypic variation characteristic of large taxonomic groups. Land plants present a special challenge, because they are one of the few groups of organisms in which both life cycle phases (both the gametophyte stage and the sporophyte stage) are multicellular. Plants have structures (e.g., leaves) and development stages (e.g., dormant stages) that appear similar and even share similar functions across taxa, but, because they occur in different life cycle phases, may arise through very different developmental processes and may or may not be under control of similar genes. Fortunately, plants also have many commonalities, such as similarities in body plan, modular growth, and the organization of cells, tissues, and organs. Below, we discuss several of the challenges in creating an ontology for all plants and describe how they are addressed in PO.
There is no single common vocabulary for all of plant science
Botany, like any other traditional branch of biology, has a vocabulary that has grown up over many centuries. As the introduction of the International System of Units demonstrated, significant advantages flow from terminological uniformity. The ontology approach embraced by the PO does not, however, seek to impose a single, inflexible vocabulary across the whole of plant science. Rather, its strategy of using ontology terms to enhance existing data through annotations is compatible with an approach that involves the use of multiple terminologies by different communities of scientists. Moreover, although ontology definitions must be clear and precise to exploit ontological reasoning, a certain flexibility in the ontology itself is also necessary and is achieved in several ways. First, the graph-theoretic structure of the ontology allows for easy addition of new branches and for easy association of new community-specific systems of terms with existing branches. Second, the hierarchical nature of an ontology provides flexibility of a sort not found in a glossary, by allowing the use of a more general class whenever a more precise match cannot be made. For example, if a researcher is uncertain whether to classify a particular structure as a rhizome or a tuber, because it has some characteristics of each, the more general ontology class shoot axis can be used instead. Although the use of a maximally specific term is encouraged, it is never logically inconsistent to use a more general term to describe a particular structure.
It is common for the same biological entity to have different names in different taxa. For example, vascular leaf may be called “frond” in cycads, ferns, or palms and “needle” in some conifers. In another example, “BBCH principal growth stage 6” (BBCH Working Group, 2001) is used in a very specialized way by the Z. mays community for flowering stage. Synonymy (described under Term names and synonyms) is a straightforward way for ontologies to remain flexible to the needs of different communities, e.g., by providing synonyms such as BBCH principal growth stage 6, rice growth stage-6.1, or Sorghum growth stage 8 for flowering stage, or corymb, raceme, or Sorghum cob as synonyms of inflorescence.
Another challenge stems from the use of the same name for different entities. For example, the word “calyptra” is used for a covering of the sporangium derived from the archegonium in mosses, but it is also used for the fused perianth parts of Eucalypteae flowers. Although unique IDs are sufficient to disambiguate two different classes with the same name, a better practice is to give them different names. The use of taxonomic names in ontology term names is avoided, so rather than use names like “moss calyptra” and “eucalypt calyptra”, PO developers chose the names spore capsule calyptra for the moss term and calyptra corolla and calyptra calyx for the eucalypt terms. This leaves open the possibility that the same or similar structures could occur in other taxa of which the curators were not aware.
Terms that had taxon-specific names in previous versions of the PO and GO (based on the use of the sensu qualifier; Ilic et al., 2007) have been eliminated. In the PO, they were eliminated by merging them into their more general classes (e.g., Zea integument was merged into integument). Classes that occur in a limited set of species but have unique characteristics that merit their own subclass, such as the ear in Zea, were given new names that emphasize the structural aspects of the class, rather than the taxon name (e.g., ear inflorescence rather than Zea ear). The use of a certain class may be limited to specific taxa by specifying the restriction in a comment and marking that class as belonging to a taxon-specific subset, but this has the disadvantage that the taxonomic restriction is not computable. To make the restriction computable, taxonomic relations such as only_in_taxon or never_in_taxon (Deegan [nee Clark] et al., 2010) can be used to connect an anatomy ontology class to a class from an ontological representation of a resource such as the NCBI Taxonomy. These relations have been used in conjunction with ontology reasoners to detect mistakes in multispecies metazoan ontologies (Mungall et al., 2012). Selected taxonomic relations will be added to the PO in the future. For example, by adding the relation portion of vascular tissue only_in Tracheophyta, any attempt to associate to a nonvascular plant a term standing in a subclass of or part of relation to portion of vascular tissue, such as xylem or sieve element, will be detected as an error by the reasoner. Taxonomic restrictions are less useful for structures whose presence or absence is more variable within clades, such as compound leaf, so use of taxonomic restrictions for these types of structures are better applied in taxon-specific applications that use the PO, rather than being incorporated into the main ontology.
Not all biologically significant entities fit neatly into categories
Traditional classifications of plant structures are organized around cell, tissue, and organ (e.g., Esau, 1977), but these categories are inadequate for describing many plant anatomical entities, such as flower, stele, or fruit septum. Structures such as these motivated the creation of three new classes in the PO: collective plant organ structure, cardinal organ part, and collective organ part structure, respectively (see earlier Overview of the Plant Ontology). Structures that span multiple ontology categories present another challenge. For example, in the PO, trichome and rhizoid can include both unicellular (plant cell) and multicellular (portion of plant tissue) subclasses, and ovary may be either the basal portion of a single carpel (a cardinal organ part) or the basal portion of group of fused carpels (a collective organ part structure). To address this variability while ensuring logical consistency, developers treat trichome, rhizoid, and ovary as direct subclasses of plant structure within the PO. Maintaining this level of logical consistency is necessary to support successful computational analysis.
Even with the creation of novel categories, ontology developers face the challenge of how to encompass the variation in anatomy and development found throughout the plant kingdom (Kirchoff et al., 2008). Take, for example flower, which the PO defines as “a determinate reproductive shoot system that has as part at least one carpel or at least one stamen and does not contain any other determinate shoot system as a part.” Because reproductive shoot system is a subclass of collective plant organ structure, a flower by definition must have the characteristics of a collective plant organ structure, that is, it must contain two or more plant organs. Although this is usually the case, PO curators are well aware that some species have flowers consisting of a single stamen (e.g., Ascarina) or a single carpel (e.g., Peperomia). However, the goal of a reference ontology like the PO is to describe the canonical form of any class, that is, the form that encompasses most of the variation across species and individuals under normal developmental conditions. Thus, there inevitably will be exceptions to some ontology classes. For example, a Peperomia flower may be scored in the PO as an instance of a carpel, rather than as a flower. Although this treatment may appear to contradict the homology of Peperomia’s single carpel being derived from more complex flowers, there is no logical inconsistency, since homology statements are outside the scope of anatomy ontologies (see below) and must be specified as an additional layer of information.
Homology of structures may or may not be known
There are many reasons for comparing two or more entities in biology, including analyzing their selective advantage under certain conditions, attempting to infer function or gene expression in unstudied species based on well-studied species, or identifying fossil taxa. The PO aims to serve the majority of these needs and therefore tries to group structures in a way that is based on established botanical knowledge and thereby maximize the possibilities for comparison. Anatomical entities in the PO are categorized on the basis of structural, positional, and developmental information. Because this information, taken together with phylogeny, is the same that is used to determine homology, some of the relationships in the PO may appear to represent homology. However, PO curators, like curators working on other anatomy ontologies (e.g., Mabee et al., 2007), made a conscious decision not to specify homology relations. In part, this is pragmatic, because homology is often not known, and because the understanding of homology still differs among scientists (Roux and Robinson-Rechavi, 2010; Nixon and Carpenter, 2012). Even when it is known, grouping by homology could limit the possibilities for comparative analysis; there are many times when one might want to compare structures that are known to be not homologous (e.g., vascular and nonvascular leaves) or to compare structures to test homology hypotheses. Homology statements require a phylogeny and should be considered as a separate layer of information that can be added on top of an ontology by mapping homology relations to ontology terms (Mabee et al., 2007). Similarly, users can create novel categories of ontology terms for other needs, based on criteria such as function or taxonomy.
Generality may cause loss of detail
It is challenging to create an ontology that is broad enough for all plants but also able to provide specialized terms for structures and development stages that occur in specific taxa. The strategy used in the PO is to create general classes that can be applied across all taxa in which the entity occurs, with more specific subclasses that may be instantiated in only a limited set of species. An example is the general class sporangium, which can be used for any land plant species, while the subclasses megasporangium and microsporangium are used only for heterosporous plants, and nucellus and pollen sac only for seed plants. In this example, there is no specific subclass for sporangia in bryophytes and other homosporous plants, because they have no features that distinguish them from sporangia in general. In contrast, the general term leaf has subclasses vascular leaf and nonvascular leaf. In this case, leaves of all plants can be classified into one of the two subclasses, and individual leaves should be described using one or the other of these subclasses or using a class at some lower level in the leaf branch of the ontology. It is important to note that, although classes are often congruent with particular clades, the definitions of classes are not based on affinity with a clade but rather with the structural, positional, or developmental similarities of entities that may happen to be conserved within a clade.
In an ontology that only covers one or a handful of species, it is possible to incorporate a large number of mereological statements—that is, statements concerning relations between parts and wholes—that could be seen by biologists to hold across all the species treated. As the coverage of plant species within the PO was expanded, many new terms describing a much larger variety of plant anatomical entities were incorporated, and in consequence, the PO lost some of its ability to describe plant structures specified by the part_of relation. For example, it is correct to say that flower is part_of some inflorescence for all flowers in Zea, Oryza, and Arabidopsis, but this is not true for all angiosperms, and this part_of assertion had to be removed when the PO was expanded. Similar problems arose for the assertion pollen sac part_of anther (due to the inclusion of gymnosperms) and for plant sperm cell part_of pollen (due to the inclusion of nonseed plants).
Fortunately, there are several ways to provide information about mereology while maintaining correct class–subclass relationships. The first is to create multiple subclasses that can then be used to make the correct part_of relationships. For example, two new subclasses of plant sperm cell were created: pollen sperm cell, which is part_of pollen, and antheridium sperm cell, which is part_of antheridium. The two unique subclasses signal that there is variation in the mereological relationships across taxa. The second approach is to use the has_part relation. For example, the PO states that every inflorescence has_part some flower and every anther has_part some pollen sac, both of which are true in every case.
Using ontologies to advance plant science
A set of well-developed ontologies that can be applied to plants is a valuable resource for many aspects of botanical research because it provides both a controlled vocabulary and a logical framework for semantic reasoning. Yoder et al. (2010) described five areas of inquiry for Hymenoptera anatomy that are currently inhibited by the lack of a consistent vocabulary and that arise equally in research in plant anatomy and development:
comparisons of gene expression patterns
description of phenotypic variability in the context of environment
computed reasoning based on a well-defined semantic framework
comparative morphology and phylogenetics
descriptive taxonomy and phenomics
The comparison of gene expression patterns under (1) is the most conspicuously successful use of bio-ontologies to date, having been applied primarily to the model species used in support of research on human health and disease. Researchers have begun using bio-ontologies for (2–5), but their application in these areas for plant science is still largely unexplored. In this section, we describe some of the ways that ontologies such as the PO could benefit plant science research, including illustrations from ontology-based research on other taxa that could be applied to plants. The utility of ontologies for ecological research, including plant ecology, has been reviewed by Madin et al. (2008), and so is not covered here.
Ontologies for comparative genetics, phenomics, and development
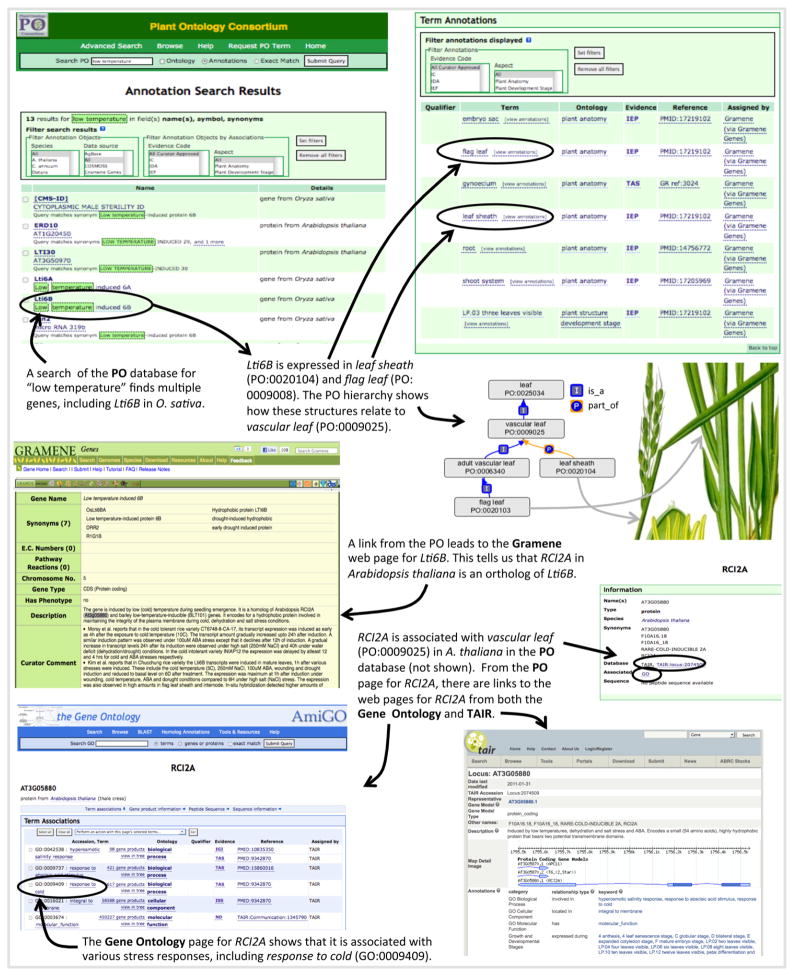
For almost 10 years, model species databases have been using ontologies to describe genetic and phenomic data in plant species such as A. thaliana (Berardini et al., 2004), O. sativa (Yamazaki and Jaiswal, 2005), and Z. mays (Vincent et al., 2003). By providing consistent vocabulary across multiple species, the PO enhances these annotations and increases the potential for comparative studies (Avraham et al., 2008). The use of ontologies such as the PO and GO ensures that different researchers working on different taxa are referring to comparable entities. The PO currently pools expression and phenotype data for 22 plant species, allowing scientists to take advantage of the efforts of multiple databases in one location. In the latest PO release (release 17), there are 2 175 694 annotations for 110 950 unique objects (e.g., genes, germplasm) associated with PO terms—an almost 10-fold increase since 2008. Users can browse or search the PO annotation database for information on genes, proteins, RNA, QTLs, and germplasm associated with a particular structure or development stage (Avraham et al., 2008). This data then can be used to generate or answer biological hypotheses on the role of different genes in development and morphology or the response of plants to environmental conditions (Fig. 3).
Fig. 3.
Data exploration with the Plant Ontology (PO). Suppose a researcher wanted to identify genes that are involved in a response to low temperature in the leaves of a nonmodel species. A search of PO annotations for “low temperature” returns a list of genes with those words in their descriptions (upper left). Selecting Lti6B from the list of results takes the user to the PO page for that gene (upper right), where it is shown that Lti6B is expressed in both flag leaf and leaf sheath in Oryza sativa. Because flag leaf and leaf sheath are respectively a subclass of and a part of a vascular leaf (see ontology diagram, center right) we can infer that Lti6B is expressed in vascular leaf. From the PO gene page, there is a direct link to the database that supplied the annotation (Gramene in this case, center left), which provides more detail on Lti6B, including the fact that RCI2A is an ortholog in Arabidopsis. A new search of PO annotations leads to the PO page for RCI2A, which is expressed in vascular leaf. From the RCI2A page, users can link to the TAIR locus page (lower right) or the Gene Ontology page (lower left) for RCI2A. This evidence, which spans the monocot–dicot divide, suggests that Lti6B and its orthologs are important for a response to low temperature in leaves across angiosperms, and provides a candidate for genetic analysis in the nonmodel species. By linking resources from multiple databases, the PO makes this type of information much more accessible. Orzya sativa image modified from http://en.wikipedia.org/wiki/File:Oryza_sativa_-_K%C3%B6hler%E2%80%93s_Medizinal-Pflanzen-232.jpg (image in the public domain).
Although not all individual researchers will face the need to annotate their data with ontology terms to do their research, a collective annotation effort by many researchers can greatly enhance plant science. The scientific community recognizes that major progress in scientific discovery often demands sophisticated data sharing and has supported initiatives like the requirement to share sequence data through public repositories such as GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Similarly, the mapping of botanical data sets to ontology terms will advance the discovery of information for the scientific community, but comes with the costs to scientists of additional time spent describing the content of their data (Madin et al., 2008). So far, this burden has fallen largely on professionally curated databases such as TAIR, MaizeGDB, and Gramene, but the potential for greater community input exists. For example, the Sol Genomics Network (SGN) uses a community curation model in which over 100 researchers add and edit information on Solanaceae using simple web-based tools (http://solgenomics.net/; Bombarely et al., 2010). As genomic and phenomic data become available for more species, we anticipate that the work of semantically tagging data with ontology terms will become more dispersed, while at the same time, the need for cross-species queries will become more common, leading more researchers in plant science to turn to ontology resources.
Ontologies for taxonomy and systematics
The cheap, fast generation of molecular sequence data has revolutionized the field of systematics as well as genomics, yet a similar revolution has not occurred with morphological data. Despite advances in imaging technology and other methods of collecting morphological data, the challenge of storing and accessing data, and synthesizing it across studies, remains daunting (Ramírez et al., 2007). Furthermore, vast stores of legacy data for plant characters exist in journal articles and monographs, in the form of free text descriptions and character matrices that are more than 10–15 yr old and may only be available in a noncomputable form, such as print or portable document format (PDF). Making morphological characters and character states computable offers a way to connect them to similar characters in other matrices as well as to genomic information (Mabee et al., 2007; Deans et al., 2012). Interoperability of morphological data is essential for advancing our understanding of the tree of life, especially for analyzing character evolution, incorporating fossil taxa into phylogenies, and reusing existing data.
Ontologies offer an effective way to build interoperability into character descriptions, without restricting systematists’ ability to describe characters as they see fit. The entity-quality (EQ) formalism has been used for describing mutant phenotypes of organisms such as mouse (Gkoutos et al., 2004) and teleost fish (Dahdul et al., 2010), and its use has been recommended for systematic descriptions of phenotypes (Mabee et al., 2007; Balhoff et al., 2010). An EQ statement (Table 2) describes a phenotype as a combination of an entity (E) and a quality (Q), such as leaf lamina ovate (E=leaf lamina + Q=ovate, where ovate is known to be a subclass of shape). More complex phenotypes can be described using multiple entities and qualities. EQ statements can be used to describe both characters and character states, provided the quality ontology is structured so that characters are more general classes of character states. For example, if ovate has an is_a relation to shape, and character states take the form “leaf lamina ovate”, then a reasoner can automatically infer that the character is “leaf lamina shape”. Likewise, if the character is first specified as “leaf lamina shape” an ontology can be used to restrict the possible character states to terms such as ovate and obovate (subclasses of shape).
Table 2.
Examples of entity-quality (EQ) statements for plant systematic characters, using ontology terms. The entity term is always the same for a character and it corresponding states, but the quality terms for states are subclasses of the term for the character in the Phenotypic Quality Ontology (PATO), e.g., palmate and pinnate are subclasses of arrangement.
| Character or State | Entity | Qualities |
|---|---|---|
| Character: primary venation arrangement | PO:primary vein | PATO:arrangement |
| States: palmate, pinnate | PO:primary vein | PATO:palmate PATO:pinnate |
| Character: stigma symmetry | PO:stigma | PATO:symmetry |
| States: symmetric, asymmetric | PO:stigma | PATO:symmetrical PATO:asymmetrical |
| Character: style type | PO:style | PATO:structure |
| States: solid, hollow | PO:style | PATO:solid (unlumenized) PATO:hollow |
| Character: cyanogenic compounds | ChEBI:cyanogenic compound | PATO:count |
| States: present, absent | ChEBI:cyanogenic compounds | PATO:present PATO:absent |
Balhoff et al. (2010) developed the application Phenex for annotating character matrix files with ontology terms. Phenex loads the relevant ontologies, pertaining, for example, to anatomical entities, qualities, and taxonomic names, and allows users to create EQ statements for their characters and states. Both a traditional character matrix and ontological annotations are output in Nexus-Extensible Markup Language (NeXML) format (http://www.nexml.org/). Dahdul et al. (2010) have used Phenex to curate legacy data from phylogenetic studies on ostariophysan fishes, and similar work is possible for plants. Ontological descriptions of characters and character states also can be combined with natural language processing to extract characters from historical literature and detect logically inconsistent descriptions (Cui, 2010a, b).
The use of ontologies to precisely define characters and character states allows morphologists and systematists to increase the interoperability of their data, but words alone may not provide sufficient guidance for future scientists. Images are also needed to document characters, states, and matrix cell scores. Organizing images based on ontology annotations makes it easier to access them for phylogenetic or other types of studies (Ramírez et al., 2007; Yoder et al., 2010). Several existing repositories are oriented toward storing images for systematics, such as MorphoBank (O’Leary and Kaufmann, 2012), Plant-Systematics.org (http://www.plantsystematics.org/), and Morphbank (http://www.morphbank.net/), but descriptions in these databases are still in a free-text form. PO curators are working with the curators of Morphobank to incorporate tools that will allow users to construct ontology-based characters and states and automatically associate ontology terms with the images documenting homology statements. They are also working with the curators of PlantSystematics.org to provide reciprocal links between PO terms and corresponding images on PlantSystematics. org.
The ability to construct EQ statements depends on the existence of well-structured and comprehensive ontologies for both entities and qualities. Most entities for plants are covered through the Plant Ontology (PO), although some may be in the domain of other ontologies (for example, cellular components like plastid or root hair are covered by the Gene Ontology). A recent analysis, in which we compared terms in the Flora of North America (FNA) glossary (FNA Editorial Committee, 1993) to PO terms, suggests that the PO is fairly complete in its coverage of the morphological entities needed to describe vascular plants. The FNA glossary contains 839 unique concepts under the classifications “structure” and “feature”, which roughly correspond to PO’s classification of plant anatomical entity. Of these, 126 match exactly to PO term names or close variants (e.g., FNA cell matches exactly to PO plant cell) and 193 match to existing PO synonyms. 333 FNA terms had matching classes in the PO and have been added as synonyms to existing PO terms (e.g., acorn was added as a narrow synonym for fruit and chalazae was added as an exact synonym for chalaza). There were 92 FNA terms representing 50 unique classes (e.g., punctum) that are too general for the PO, because they can be applied ambiguously to many plant structures that are distinguished in PO. These classes are better modeled in terms of the corresponding quality (e.g., punctate) in the Phenotypic Quality Ontology (PATO). Based on the FNA term list, only 143 distinct plant structures are missing from the PO, many of which are specialized structures that only occur in a few taxa (e.g., pollinium or phylloclade). Corresponding terms are being added to the PO.
Compared to plant anatomical entities, ontologies for plant qualities may be still significantly under-developed for systematic descriptions of plant characters, as is shown by a review of existing glossaries and ontologies (Cui, 2010a). PO curators are working with members of the scientific community, including FNA and PATO curators, members of the International Association of Wood Anatomists (IAWA; Lens et al., 2012), and the databases that contribute to the PO, to enrich the list of quality descriptors for plants. A number of terms for plant qualities already exist in PATO, along with many terms relating to generally applicable qualities such as shape, size, or texture, and the ability to construct meaningful EQ statements for systematic work using PATO and other existing ontologies has already been clearly demonstrated (Dahdul et al., 2010).
The Plant Ontology as an educational resource
Ontologies provide novel tools for plant science education at the middle school to college level. The hierarchical nature of the PO, along with the graphical view available in the browser, allows students to visualize the relationships among plant parts or plant development stages in a way that is not possible with a traditional glossary. Ongoing work on image annotation for PO terms will provide visual references for students, and associating PO plant terms with image or video collections would be a good educational exercise. Because an ontology is a way of modeling knowledge in a domain, it is in some ways comparable to a concept map. The PO could be used to develop a lesson plan that allows students to create their own plant ontology with a limited set of anatomical terms, then compare it to the existing ontology structure in the PO. Comparing how the students’ ontologies differ from the PO can generate discussions on how botanists understand plant structures and how people organize knowledge. For more advanced students, ontology annotations provide a source of genetic and genomic data that can be used to generate or test hypotheses, as in Fig. 3.
Ontologies and semantic applications
The power of ontologies lies in their utility for reasoning by means of software applications. The PO is currently integrated into a number of web-based applications such as Virtual Plant (Katari et al., 2010), Bio-Array Resource for Plant Biology (Brady and Provart, 2009), and VPhenoDBS (Green et al., 2011), as well as knowledge bases such as Plant Expression Database (Wise et al., 2008) and Phenopsis DB (Fabre et al., 2011). Although these applications represent relatively simple uses of ontologies, they give some indication of the potential for data reuse. More sophisticated applications such as Semantic J-SON (Kobayashi et al., 2011) can be used for biological applications including genome design, sequence processing, inference over phenotype databases, and full-text search indexing. The PO project has also developed an application to make PO data available over the web, in real time, making it more accessible to developers wanting to incorporate it into their own applications (http://plantontology.org/docs/otherdocs/web_services_guide.html).
Applications like these are part of the semantic web, an extension of the current World Wide Web that enables automatic navigation and use of digital resources (Berners-Lee et al., 2001; Ruttenberg et al., 2007). The semantic web depends on very specific means of data storage. All data items, including semantic relations, must be specified with a Uniform Resource Identifier (URI). A relationship between two data items is written as “triple” of three URIs: subject, predicate, and object (Kobayashi et al., 2011). An example of a triple from the PO is: ABCG11 expressed_in epidermal cell, where the URI for the subject ABCG11 is its TAIR (The Arabidopsis Information Resource) locus ID, the URI for the predicate expressed_in is its Relation Ontology ID, and the URI for the object epidermal cell is its PO ID. The Resource Description Framework (RDF) and RDF/XML syntax can be used to create triples for any document or database that is available over the web, which in turn allows semantic applications to query those documents or data (http://www.w3.org/TR/rdf-primer/; Carroll et al., 2005). All data in the PO, including terms, attributes of terms, and annotation data, can be accessed in this way. By semantically tagging data (e.g., character matrices, experimental results, images) to ontology terms using RDF/XML, researchers expose that data to automatic web searches, greatly enhancing its utility to the scientific and broader community.
Semantic web technologies have the potential to greatly enhance both basic science and translational research—the movement of discoveries from basic science to applications such as medicine or agriculture—but the lack of uniformly structured data are still a major obstacle. Ontologies provide a key element in removing this obstacle (Ruttenberg et al., 2007). The use of ontologies and semantic web applications is being widely explored for medical research (e.g., Ciccarese et al., 2008; Das et al., 2009), and similar approaches could be used for many aspects of applied plant sciences, such as plant disease control and crop breeding.
Ontologies are also an important component of natural language processing tools. These tools can be used for text mining of journal articles, historical and current taxonomic treatments, and museum labels (Spasic et al., 2005; Krallinger et al., 2008; Cui, 2010b). Semantic tagging of botanical data using ontology terms has the potential to revolutionize plant biodiversity research, when coupled with the rapid development of digital resources for botany, such as the Biodiversity Heritage Library (http://www.biodiversitylibrary.org), online herbarium specimen records, and online floras, monographs, and anatomical atlases (Brach and Boufford, 2011).
Conclusions
Bio-ontologies like the PO offer a flexible framework for comparative biology, based on common biological understanding. The PO is not intended to replace the wealth of existing botanical glossaries, whose scope goes far beyond the domain of the PO and provides the legacy on which the PO is built. However, ontologies offer something that glossaries do not: a controlled vocabulary for annotation of data and an associated logical framework for semantic reasoning. The PO, like any resource that attempts to summarize the knowledge in a given domain, will always be a work in progress. It must be able to describe, query, and visualize botanical knowledge that is constantly changing and at different stages of completeness (Ashburner et al., 2000), and it must be open to the needs of the scientific community and respond to scientific progress. Ontologies like the PO are community efforts, and collaborators—both individuals and databases—are instrumental in supplying data and suggesting new terms. Feedback and contributions from members of the botany and plant sciences community can ensure that ontologies for plants meet the needs of all potential users.
Acknowledgments
The authors thank Chris Sullivan (Center for Genome Research and Biocomputing at the Oregon State University) for hosting and maintenance of the Plant Ontology project web servers; Kevin C. Nixon (Cornell University) for access to and development of the PlantSystematics.org and Cornell University Plant Anatomy Collection (CU-PAC) databases and image servers; Hong Cui and James Macklin (Flora of North America), Naama Menda (Sol Genomics Network), Mary Schaeffer (MaizeGDB), Rosemary Shrestha (Consultative Group on International Agricultural Research, CGIAR), Rex Nelson (SoyBase), and the curators of The Arabidopsis Information Network (TAIR) for their work on term enrichment of the Plant Ontology; and numerous volunteers and reviewers of the Plant Ontology. Funding for this project came from the U. S. National Science Foundation, award IOS: 0822201.
LITERATURE CITED
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Lewis S. On ontologies for biologists: The Gene Ontology—Untangling the web. Novartis Foundation Symposium. 2002;247:66–88. [PubMed] [Google Scholar]
- Avraham S, Tung CW, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, et al. The Plant Ontology Database: A community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Research. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baader F. The description logic handbook: Theory, implementation, and applications. Cambridge University Press; New York, New York, USA: 2003. [Google Scholar]
- Balhoff JP, Dahdul WM, Kothari CR, Lapp H, Lundberg JG, Mabee P, Midford PE, et al. Phenex. Ontological Annotation of Phenotypic Diversity. 2010;5:e10500. doi: 10.1371/journal.pone.0010500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U, editor. BBCH Working Group. Growth stages of mono-and dicotyledonous plants. Federal Biological Research Centre for Agriculture and Forestry; Berlin, Germany: 2001. [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berners-Lee T, Hendler J, Lassila O. The Semantic Web—A new form of Web content that is meaningful to computers will unleash a revolution of new possibilities. Scientific American. 2001;284:34. [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, et al. The Sol Genomics Network (solgenomics.net): Growing tomatoes using Perl. Nucleic Acids Research. 2010;39:D1149–D1155. doi: 10.1093/nar/gkq866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brach AR, Boufford DE. Why are we still producing paper floras? Annals of the Missouri Botanical Garden. 2011;98:297–300. [Google Scholar]
- Brady SM, Provart NJ. Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell. 2009;21:1034–1051. doi: 10.1105/tpc.109.066050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman R, Courtot M, Derom D, Fostel J, He Y, Lord P, Malone J, et al. Modeling biomedical experimental processes with OBI. Journal of Biomedical Semantics. 2010;1(supplement 1):S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult C, Drabkin H, Evsikov A, Natale D, Arighi C, Roberts N, Ruttenberg A, et al. The representation of protein complexes in the Protein Ontology (PRO) BMC Bioinformatics. 2011;12:371. doi: 10.1186/1471-2105-12-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JJ, Bizer C, Hayes P, Stickler P. Named graphs. Web Semantics: Science, Services and Agents on the World Wide Web. 2005;3:247–267. [Google Scholar]
- Ciccarese P, Wu E, Wong G, Ocana M, Kinoshita J, Ruttenberg A, Clark T. The SWAN biomedical discourse ontology. Journal of Biomedical Informatics. 2008;41:739–751. doi: 10.1016/j.jbi.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté RG, Jones P, Apweiler R, Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M, Gibson F, Lister AL, Malone J, Schober D, Brinkman R, Ruttenberg A. MIREOT: The minimum information to reference an external ontology term. Applied Ontology. 2011;6:23–33. [Google Scholar]
- Cowell L, Smith B. Infectious Disease Ontology. In: Sintchenko V, editor. Infectious disease informatics. Springer; New York, New York, USA: 2010. pp. 373–395. [Google Scholar]
- Cui H. Competency evaluation of plant character ontologies against domain literature. Journal of the American Society for Information Science and Technology. 2010a;61:1144–1165. [Google Scholar]
- Cui H. Semantic annotation of morphological descriptions: An overall strategy. BMC Bioinformatics. 2010b;11:278. doi: 10.1186/1471-2105-11-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul WM, Balhoff JP, Engeman J, Grande T, Hilton EJ, Kothari C, Lapp H, et al. Evolutionary characters, phenotypes and ontologies: Curating data from the systematic biology literature. PLoS ONE. 2010;5:e10708. doi: 10.1371/journal.pone.0010708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Girard L, Green T, Weitzman L, Lewis-Bowen A, Clark T. Building biomedical web communities using a semantically aware content management system. Briefings in Bioinformatics. 2009;10:129–138. doi: 10.1093/bib/bbn052. [DOI] [PubMed] [Google Scholar]
- de Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, et al. Chemical entities of biological interest: An update. Nucleic Acids Research. 2009;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AR, Yoder MJ, Balhoff JP. Time to change how we describe biodiversity. Trends in Ecology & Evolution. 2012;27:78–84. doi: 10.1016/j.tree.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Deegan (nee Clark) J, Dimmer E, Mungall C. Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinformatics. 2010;11:530. doi: 10.1186/1471-2105-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, et al. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Research. 2007;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of seed plants. 2. Wiley, Hoboken; New Jersey, USA: 1977. [Google Scholar]
- Fabre J, Dauzat M, Nègre V, Wuyts N, Tireau A, Gennari E, Neveu P, et al. PHENOPSIS DB: An information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biology. 2011;11:77. doi: 10.1186/1471-2229-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FNA Editorial Committee. Flora of North America North of Mexico. Oxford University Press; New York, New York, USA: 1993. [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology in 2010: Extensions and refinements. Nucleic Acids Research. 2009;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos GV. Towards a phenotypic Semantic Web. Current Bioinformatics. 2006;1:235–246. [Google Scholar]
- Gkoutos GV, Green ECJ, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biology. 2004;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JM, Harnsomburana J, Schaeffer ML, Lawrence CJ, Shyu C-R. Multi-source and ontology-based retrieval engine for maize mutant phenotypes. Database. 2011:bar012. doi: 10.1093/database/bar012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon P, Smith B. SNAP and SPAN: Towards dynamic spatial ontology. Spatial Cognition and Computation. 2004;4:69–103. [Google Scholar]
- Haendel M, Neuhaus F, Osumi-Sutherland D, Mabee PM, Mejino JLVJ, Mungall CJ, Smith B. CARO—The Common Anatomy Reference Ontology. In: Burger A, Davidson D, Baldoc R, editors. Anatomy ontologies for bioinformatics: Principles and practice, computational biology series. Springer; New York, New York, USA: 2007. pp. 327–349. [Google Scholar]
- Hill DP, Smith B, McAndrews-Hill MS, Blake JA. Gene Ontology annotations: What they mean and where they come from. BMC Bioinformatics. 2008;9:S2. doi: 10.1186/1471-2105-9-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horridge M, Drummond N, Goodwin J, Rector A, Stevens R, Wang H. The Manchester OWL Syntax. Proceedings of the 2006 OWL Experiences and Directions Workshop (OWL-ED2006); University of Georgia; Athens, Georgia, USA. November 2006; 2006. [accessed 13 July 2012]. Available online at http://www.webont.org/owled/2006/accepted06. html. [Google Scholar]
- Ilic K, Kellogg EA, Jaiswal P, Zapata F, Stevens PF, Vincent L, Avraham S, et al. The Plant Structure Ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiology. 2007;143:587–599. doi: 10.1104/pp.106.092825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P. Gramene Database: A hub for comparative plant genomics. In: Pereira A, editor. Plant reverse genetics. Humana Press; Totowa, New Jersey, USA: 2011. pp. 247–275. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Bork P. Ontologies in quantitative biology: A basis for comparison, integration, and discovery. PLoS Biology. 2010;8:e1000374. doi: 10.1371/journal.pbio.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint Genome Institute. [accessed 30 April 2012];JGI Plant Genomics Program—Home [online]. Website. 2012 http://genome.jgi.doe.gov/genome-projects/
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, et al. VirtualPlant: A software platform to support systems biology research. Plant Physiology. 2010;152:500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchoff BK, Pfeifer E, Rutishauser R. Plant structure ontology: How should we label plant structures with doubtful or mixed identities? Zootaxa. 2008;1950:103–122. [Google Scholar]
- Kobayashi N, Ishii M, Takahashi S, Mochizuki Y, Matsushima A, Toyoda T. Semantic-JSON: A lightweight web service interface for Semantic Web contents integrating multiple life science databases. Nucleic Acids Research. 2011;39:W533–W540. doi: 10.1093/nar/gkr353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, Valencia A, Hirschman L. Linking genes to literature: Text mining, information extraction, and retrieval applications for biology. Genome Biology. 2008;9:S8. doi: 10.1186/gb-2008-9-s2-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens F, Cooper L, Gandolfo MA, Groover A, Jaiswal P, Lachenbruch B, Spicer R, et al. An extension of the Plant Ontology project supporting wood anatomy and development research. International Association of Wood Anatomists Journal. 2012;33:113–117. [Google Scholar]
- Mabee PM, Ashburner M, Cronk Q, Gkoutos GV, Haendel M, Segerdell E, Mungall CJ, Westerfield M. Phenotype ontologies: The bridge between genomics and evolution. Trends in Ecology & Evolution. 2007;22:345–350. doi: 10.1016/j.tree.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Madin JS, Bowers S, Schildhauer MP, Jones MB. Advancing ecological research with ontologies. Trends in Ecology & Evolution. 2008;23:159–168. doi: 10.1016/j.tree.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Madin JS, Bowers S, Schildhauer M, Krivov S, Pennington D, Villa F. An ontology for describing and synthesizing ecological observation data. Ecological Informatics. 2007;2:279–296. [Google Scholar]
- Mungall CJ, Gkoutos GV, Smith C, Haendel M, Lewis S, Ashburner M. Integrating phenotype ontologies across multiple species. Genome Biology. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biology. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D, Arighi C, Barker W, Blake J, Chang TC, Hu Z, Liu H, et al. Framework for a Protein Ontology. BMC Bioinformatics. 2007;8:S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon KC, Carpenter JM. More on homology. Cladistics. 2012;28:161–169. doi: 10.1111/j.1096-0031.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- O’Leary MA, Kaufmann SG. MorphoBank 3.0: Web application for morphological phylogenetics and taxonomy. 2012 Available at website http://www.morphobank.org.
- Pujar A, Jaiswal P, Kellogg EA, Ilic K, Vincent L, Avraham S, Stevens P, et al. Whole-plant growth stage ontology for angiosperms and its application in plant biology. Plant Physiology. 2006;142:414–428. doi: 10.1104/pp.106.085720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez MJ, Coddington JA, Maddison WP, Midford PE, Prendini L, Miller J, Griswold CE, et al. Linking of digital images to phylogenetic data matrices using a morphological ontology. Systematic Biology. 2007;56:283–294. doi: 10.1080/10635150701313848. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Roux J, Robinson-Rechavi M. An ontology to clarify homology-related concepts. Trends in Genetics. 2010;26:99–102. doi: 10.1016/j.tig.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen H, Doherty D, et al. Advancing translational research with the Semantic Web. BMC Bioinformatics. 2007;8(supplement 3):S2. doi: 10.1186/1471-2105-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober D, Smith B, Lewis S, Kusnierczyk W, Lomax J, Mungall CJ, Taylor C, et al. Survey-based naming conventions for use in OBO Foundry ontology development. BMC Bioinformatics. 2009;10:125. doi: 10.1186/1471-2105-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, et al. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ceusters W. Ontological realism: A methodology for coordinated evolution of scientific ontologies. Applied Ontology. 2010;5:139–188. doi: 10.3233/AO-2010-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall CJ, et al. Relations in biomedical ontologies. Genome Biology. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic I, Ananiadou S, McNaught J, Kumar A. Text mining and ontologies in biomedicine: Making sense of raw text. Briefings in Bioinformatics. 2005;6:239–251. doi: 10.1093/bib/6.3.239. [DOI] [PubMed] [Google Scholar]
- Tirmizi S, Aitken S, Moreira DA, Mungall CJ, Sequeda J, Shah NH, Miranker DP. Mapping between the OBO and OWL ontology languages. Journal of Biomedical Semantics. 2011;2:S3. doi: 10.1186/2041-1480-2-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent L, Coe EH, Polacco ML. Zea mays ontology—A database of international terms. Trends in Plant Science. 2003;8:517–520. doi: 10.1016/j.tplants.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Walls RL, Smith B, Elser J, Goldfain A, Stevenson DW, Jaiswal P. A plant disease extension of the Infectious Disease Ontology. Proceedings 3rd International Conference on Biomedical Ontology; Medical University of Graz, Graz, Austria. July 2012; [accessed 13 July 2012]. In press. Available online at http://kr-med.org/icbofois2012/icbopapers.htm. [Google Scholar]
- Washington NL, Lewis SE. Ontologies: Scientific data sharing made easy. Nature Education. 2008;1 [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2007;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon E, Dickerson JA. BarleyBase/PLEXdb. In: Edwards D, editor. Plant bioinformatics. Humana Press; Totowa, New Jersey, USA: 2008. pp. 347–363. [Google Scholar]
- Xiang Z, Courtot M, Brinkman RR, Ruttenberg A, He Y. OntoFox: Web-based support for ontology reuse. BMC Research Notes. 2010;3:175. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Mungall CJ, Ruttenberg A, He Y. Ontobee: A linked data server and browser for ontology terms. Proceedings of 2nd International Conference on Biomedical Ontology. 2011;1:279–281. [Google Scholar]
- Yamazaki Y, Jaiswal P. Biological ontologies in rice databases: An introduction to the activities in Gramene and Oryzabase. Plant & Cell Physiology. 2005;46:63–68. doi: 10.1093/pcp/pci505. [DOI] [PubMed] [Google Scholar]
- Yoder MJ, Mikó I, Seltmann KC, Bertone MA, Deans AR. A gross anatomy ontology for Hymenoptera. PLoS ONE. 2010;5:e15991. doi: 10.1371/journal.pone.0015991. [DOI] [PMC free article] [PubMed] [Google Scholar]