Abstract
Growing evidence indicates that microRNAs play a significant role in tumor development and may constitute robust biomarkers for cancer diagnosis and prognosis. In the present study, we evaluated the clinical and functional relevance of miR-122 expression in human hepatocellular carcinoma. We report that miR-122 is specifically repressed in a subset of primary tumors which are characterized by poor prognosis. We further demonstrate that the loss of miR-122 expression in tumor cells segregates with specific gene expression profiles linked to cancer progression, namely the suppression of hepatic phenotype and the acquisition of invasive properties. We identify liver-enriched transcription factors as central regulatory molecules in the gene networks associated with loss of miR-122, and provide evidence suggesting that miR-122 is under the transcriptional control of HNF1A, HNF3A, and HNF3B. We further demonstrate that loss of miR-122 results in an increase of cell migration and invasion and that restoration of miR-122 reverses this phenotype. In conclusion, miR-122 is a marker of hepatocyte-specific differentiation and an important determinant in the control of cell migration and invasion. From a clinical point of view, our study emphasizes miR-122 as a diagnostic and prognostic marker for HCC progression.
Keywords: hepatocellular carcinoma, microRNA, differentiation, metastasis, microarray
Introduction
HCC is the third most common cause of death from cancer world-wide, accounting for at least 600,000 deaths annually (Parkin et al., 2005). The most prominent etiologies of HCC include chronic viral hepatitis B and C (HBV, HCV), alcoholic or nonalcoholic steatohepatitis, and aflatoxin intoxication (Thorgeirsson and Grisham, 2002; El-Serag, 2004). Although recent advances in functional genomics provide increasingly comprehensive portrayal of hepatocarcinogenesis (Thorgeirsson et al., 2006; Villanueva et al., 2007), the molecular pathogenesis of HCC remains poorly understood. Indeed, the clinical heterogeneity of HCC, the lack of good diagnostic markers and treatment strategies has rendered the disease a major challenge.
MicroRNAs (miRNAs) belong to an abundant family of endogenous, short, and non-coding RNAs which are believed to be key post-transcriptional regulators of gene activity. It has been widely demonstrated that miRNAs play important roles in the control of numerous biological processes, such as development, differentiation, proliferation, and apoptosis (Ambros, 2004). Therefore, it is not surprising that miRNAs have been implicated in cancer as well. Altered miRNA expression was observed in a large variety of neoplasms (Lu et al., 2005), including HCC (Murakami et al., 2006; Braconi and Patel, 2008; Budhu et al., 2008; Ladeiro et al., 2008; Li et al., 2008; Wong et al., 2008). Significance of miRNAs in cancer onset and progression has been supported by the experimental demonstrations of their oncogenic and tumor-suppressive properties (Esquela-Kerscher and Slack, 2006). In addition, miRNA genes have been found to be frequently located in cancer-associated genomic regions, such as fragile sites, minimal regions of loss of heterozygosity and minimal regions of amplification (Calin et al., 2004).
MicroRNA-122 (miR-122) is the most abundant miRNA in the liver accounting for approximately 70% of the total miRNA population (Lagos-Quintana et al., 2002; Chang et al., 2004). Although miR-122 has been described as a liver specific miRNA, slight expression has been also reported in heart (Tang et al., 2007). Several studies have emphasized the importance of miR-122 in liver homeostasis. In vivo, miR-122 has notably been implicated in fatty acid and cholesterol metabolism and HCV infection (Jopling et al., 2005; Krutzfeldt et al., 2005; Esau et al., 2006; Chang et al., 2008; Cheung et al., 2008; Elmen et al., 2008; Henke et al., 2008). Expression of miR-122 has been found to be repressed in HCC (Kutay et al., 2006; Murakami et al., 2006; Gramantieri et al., 2007; Braconi and Patel, 2008; Budhu et al., 2008; Girard et al., 2008; Ladeiro et al., 2008; Lin et al., 2008) as well as in nonalcoholic steatohepatitis (Cheung et al., 2008). Recent work suggested that miR-122 deregulation is associated either with specific clinical risk factors (HBV, HCV) or HCC metastasis (Braconi and Patel, 2008; Budhu et al., 2008; Girard et al., 2008; Ladeiro et al., 2008; Varnholt et al., 2008).
Given the important role of miR-122 in liver pathology, we aimed i) to evaluate the diagnostic and prognostic significance of miR-122 expression in human HCC, and ii) to determine the functional implication of miR-122 deregulation in liver cancer development. By investigating the expression of miR-122 in 64 human HCC and 28 matched non-neoplastic surrounding liver tissues, we show that miR-122 repression in HCC correlates with clinically relevant parameters, such as etiology, survival, tumor size and differentiation status. Furthermore, we demonstrate that the loss of miR-122 expression correlates with distinct gene expression profiles characteristic of tumor progression (i.e. suppression of hepatic differentiation phenotype and gain of metastatic properties). Finally, we identify liver-enriched transcription factors as central functional regulators in the gene networks associated with the loss of miR-122 in human liver cancer and show that miR-122 is an important factor in the control of cellular migration and invasion.
Results
Heterogeneous expression of miR-122 in human HCC-derived cell lines
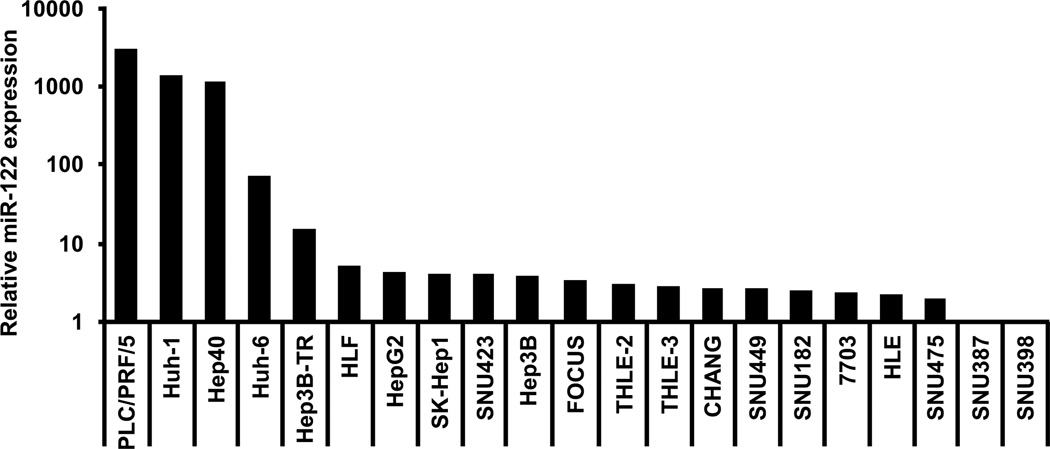
Natural properties of miRNAs, including their small size and the lack of polyadenylated tail, have made their quantification challenging. Here, the expression of miR-122 was first determined by real-time Q-RT-PCR in 21 HCC-derived cell lines (Lee and Thorgeirsson, 2002) using specific sets of primers (Figure 1 and SI Figure 1). Interestingly, the levels of miR-122 were extremely variable with more than 1000-fold expression differences among the HCC cell lines. Some cell lines including PLC/PRF/5, Huh-1, and Hep40 exhibited very high levels of miR-122, whereas others, such as Hep3B and HepG2, expressed little or did not express miR-122 at all (e.g. SNU387, and SNU398). Specificity and reproducibility of PCR assays were confirmed by modulating the expression of miR-122 in vitro (SI Figure 2). We conclude that Q-RT-PCR is a reliable assay for quantification of the miR-122 expression and that HCC cell lines represent a suitable model for functional studies of miR-122 in liver cancer.
Figure 1.
Relative expression of miR-122 in human HCC-derived cell lines. Expression of miR-122 was evaluated in 21 human HCC-derived cell lines by Q-RT-PCR and normalized to the level of 5s RNA. Data are represented as fold induction relative to the lowest miR-122 expressing SNU-398 cell line using the 2−ΔΔCt method. The great heterogeneity of miR-122 expression indicated that HCC-derived cell lines represent excellent natural models for functional studies of miR-122 in liver cancer.
Loss of miR-122 expression in HCC is associated with poor prognosis
Next, the expression of miR-122 was analyzed in a panel of 64 primary liver tumors and 28 matched non-tumor surrounding liver tissues extensively characterized in our lab previously (Lee et al., 2004a; Lee et al., 2004b; Kaposi-Novak et al., 2006; Lee et al., 2006; Coulouarn et al., 2008). HCC samples were representative of patients with various clinical outcomes and pathological features (Table 1). Interestingly, miR-122 levels were highly variable among the clinical samples (CV>2), suggesting that miR-122 expression in primary tumors was as heterogeneous as found in cell lines.
Table 1.
Clinical and pathological features of HCC cases
| Gender | # cases |
|---|---|
| Female | 16 |
| Male | 48 |
| Age | |
| Median (range) | 62 (34–85) |
| < 60 | 27 |
| > 60 | 37 |
| Survival (months) | |
| Mean ± s.e.m. | 34.8 ± 4.9 |
| Cirrhosis | |
| No | 33 |
| Yes | 31 |
| Plasma α-fetoprotein | |
| < 300 ng/mL | 41 |
| > 300 ng/mL | 23 |
| Etiology | |
| HBV | 18 |
| HCV | 13 |
| HBV + HCV | 3 |
| ALD | 13 |
| ALD + Viral infection a | 4 |
| other | 2 |
| unknown | 11 |
| Tumor size | |
| < 5 cm | 34 |
| > 5 cm | 30 |
| Edmondson-Steiner grade | |
| Grade 1 | 1 |
| Grade 2 | 26 |
| Grade 3 | 33 |
| Grade 4 | 4 |
| Vascular invasion | |
| No | 14 |
| Yes | 29 |
| Gene signatures b | |
| Good prognosis | 40 |
| Bad prognosis | 24 |
| Hepatocyte | 49 |
| Hepatoblast | 15 |
| c-Met Negative | 45 |
| c-Met positive | 19 |
| TGF-β Early | 12 |
| TGF-β Late | 14 |
Either HVB or HCV infection
From our previously published HCC stratification (Lee et al., 2004a; Kaposi-Novak et al., 2006; Lee et al., 2006; Coulouarn et al., 2008)
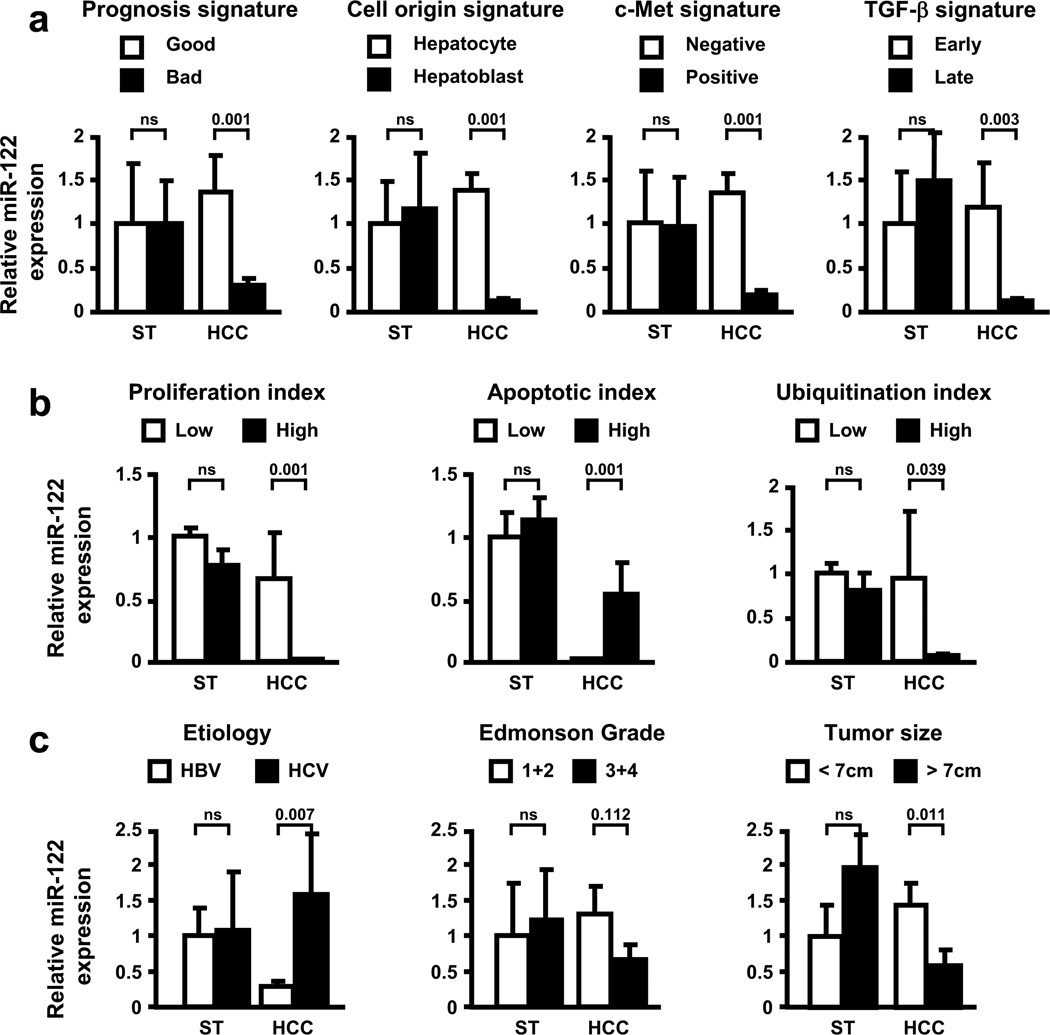
To further explore clinical relevance, the expression of miR-122 was analyzed as a function of various clinical parameters and tumor features (Figure 2). First, the expression levels of miR-122 were compared between HCC subtypes previously characterized by the experimentally well-defined gene expression signatures (Lee et al., 2004a; Lee et al., 2004b; Kaposi-Novak et al., 2006; Lee et al., 2006; Coulouarn et al., 2008). As shown in Figure 2a, miR-122 was specifically repressed in the tumors which exhibited a bad prognosis signature. Accordingly, the overall survival time of the patients with low and high miR-122 expression in HCC was 30.3±8.0 and 83.7±10.3 months, respectively (P<0.001). MiR-122 was also repressed in a subset of HCC which harbored a hepatoblast signature, as well as a c-Met and a late TGF-β signature (Figure 2a). The hepatoblast signature, derived from hepatic progenitor cells, is specific for tumors with a poor differentiation status (Lee et al., 2006) whereas c-Met and late TGF-β signatures are associated with subsets of tumors with poor prognosis and aggressive phenotype (Kaposi-Novak et al., 2006; Coulouarn et al., 2008). The loss of miR-122 expression was correlated with high proliferation and ubiquitination index along with low apoptotic index (Figure 2b). Supporting a recent observation (Varnholt et al., 2008), the expression of miR-122 was clearly etiology-dependent since miR-122 was repressed only in HCC arising in HBV-infected livers (Figure 2c). No significant correlations (P<0.05) were observed between the status of HBV infection and the presence of hepatoblast, TGF-β or c-Met signatures. Finally, miR-122 repression was found to be associated with poor differentiation status and large tumor size (Figure 2c). Taken together, these data demonstrate that miR-122 expression in human HCC correlates with clinically relevant parameters and that loss of miR-122 is a marker of poor prognosis.
Figure 2.
Repression of miR-122 correlates with clinically relevant parameters in human HCC. Expression of miR-122 was evaluated by Q-RT-PCR in 64 human HCC and 28 non-tumor surrounding liver tissues (ST). (a) Expression of miR-122 was evaluated in ST and HCC tissues as a function of experimentally well-defined gene expression signatures (described in Lee et al., 2004a; Kaposi-Novak et al., 2006; Lee et al., 2006; Coulouarn et al., 2008). Data indicated that miR-122 was specifically repressed in HCC with bad prognosis which harbor hepatoblast, c-Met and late TGF-β gene expression signatures. (b) The assessment of miR-122 expression as a function of biochemical parameters indicated that miR-122 was particularly repressed in HCC with high proliferation and ubiquitination index, and low apoptotic index (Lee et al., 2004b). (c) The evaluation of miR-122 expression as a function of etiology, Edmonson grade and tumor size indicated that miR-122 was repressed in HCC arising from HBV-infected livers and was associated with poorly differentiated and large size tumors.
miR-122 levels define specific gene expression profiles in HCC
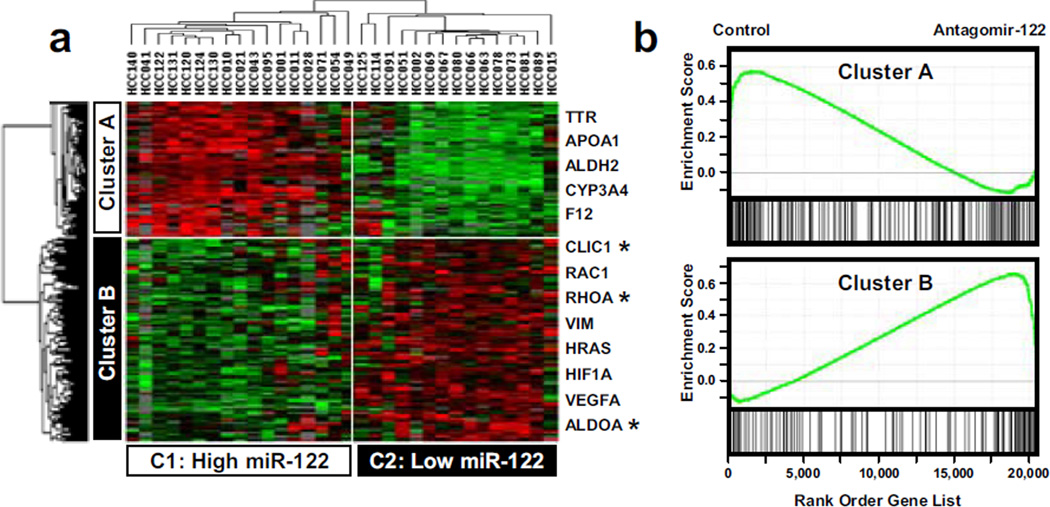
To get insights into the functional significance of miR-122 expression, tumors were divided into two groups. High and low miR-122 expressing tumors (15 and 17 cases, respectively) were defined by >2 fold changes in the median miR-122 expression across the samples. Gene expression profiling identified 509 non-redundant genes which were differentially expressed between the tumors with high vs. low miR-122 expression (SI Table 1). As expected, clustering analysis divided HCC into two distinct clusters (C1 and C2) defined by the level of miR-122 (Figure 3a). Differentially expressed genes were similarly organized in two clusters and included genes which displayed either positive (Cluster A) or negative (Cluster B) correlation with miR-122 expression (Figure 3a). Supporting our approach, cluster B included numerous predicted targets for miR-122 (e.g. ALDOA, CLIC1, and RHOA). The specific contribution of miR-122 to the HCC transcriptome was further validated by testing the enrichment of the genes embedded in clusters A and B in the gene expression profiles derived from the experimental modulation of miR-122 levels (Krutzfeldt et al., 2005). As shown in Figure 3b, genes highly expressed in the Clusters A and B (high and low miR-122 expressing HCC) were significantly enriched in the gene expression profiles corresponding to either control or mice treated with antagomir-122, respectively (Krutzfeldt et al., 2005). In addition, the restoration of miR-122 expression in vitro to a large extend was able to shift the expression patterns of Cluster A and B genes from C2 (Low miR-122) toward C1 (High miR-122) (SI Figure 3). These results demonstrate that the expression of genes embedded in the clusters A and B is highly dependent on miR-122 expression.
Figure 3.
miR-122 expression defines specific gene expression profiles in human HCC. (a) Hierarchical cluster analysis of 509 genes differentially expressed in liver primary tumors as a function of miR-122 expression. Data are presented in a matrix format in which rows and columns represent genes and HCC samples, respectively. Differentially expressed genes are grouped in two clusters, A and B. HCC samples cluster in two groups, C1 and C2, which greatly differ in term of miR-122 level (C1: 2.7±0.4; C2: 0.6±0.4; mean±s.e.m; P<0.001). Genes included in cluster A and cluster B are up- and down-regulated in cluster C1 HCC, respectively. As expected cluster B includes several predicted miR-122 target genes (*). (b) Gene Set Enrichment Analysis using the genes embedded in the clusters A and B as signatures and the gene expression profiles derived from the experimental inhibition of miR-122 in mice (Krutzfeldt et al., 2005). Cluster A and B genes are significantly enriched in the profiles corresponding to control mice (Enrichment Score ES=0.578, P<0.001), and mice treated with antagomir-122 (ES=0.658, P<0.001), respectively.
Loss of miR-122 in HCC coincides with suppression of the hepatic phenotype
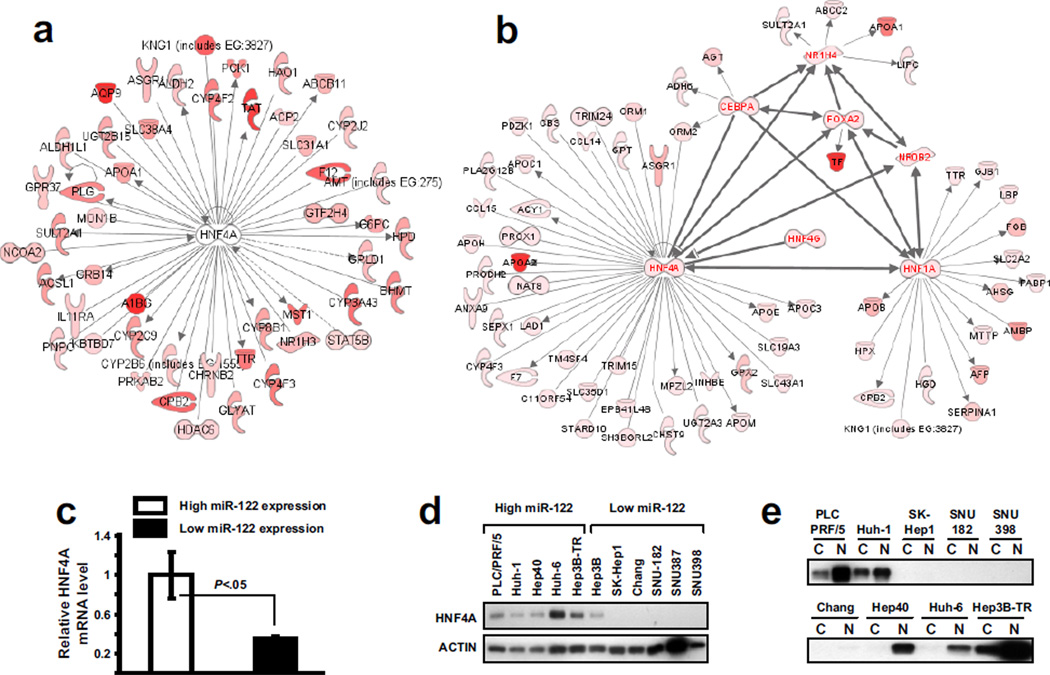
As shown in Figure 3a, the gene expression profiles were remarkably homogeneous within each subgroup of HCC, suggesting that miR-122 deregulation represents an important event during the course of HCC progression. To reveal the dominant signaling networks associated with the loss of miR-122 expression in HCC, systematic pathway and gene ontology analyses were performed using genes which either positively (Cluster A) or negatively (Cluster B) correlated with miR-122 expression. Ingenuity Pathway Analysis revealed a clear relationship between the miR-122 associated gene expression signature and the functional categories linked to liver cancer and liver metabolism (Table 2). Genes embedded in Cluster A were implicated in molecular and cellular functions characteristic of well differentiated hepatocytes (Table 2 and SI Table 1). These genes were involved in lipid metabolism (APO-A1, -C4, -F), acute phase response (ITIH3, SERPIN-C1, -F2), detoxification (CYP-2C9, -3A4, 4A11), and coagulation (F12, F13B). Moreover, high expression of miR-122 significantly correlated with the global activation of an extensive regulatory network centered on HNF4A (Figure 4a). Enrichment of HNF4A targets among miR-122-positive HCC suggests the involvement miR-122 in maintaining hepatocyte differentiation thereby implying that the loss of miR-122 contributes to suppression of the hepatic phenotype in HCC with poor prognosis (Figure 2).
Table 2.
Ingenuity Pathway Analysis of genes correlated with miR-122 expression in HCC
| Positive correlation | Negative correlation |
|---|---|
| Disease and disorders | |
| Hepatic system disease (P<10−6) | Cancer (P<10−9) |
| Molecular and cellular functions | |
| Lipid metabolism (P<10−9) | Cell cycle (P<10−5) |
| Amino acid metabolism (P<10−3) | Cell. assembly & org. (P<10−5) |
| Physiological system development and functions | |
| Tumor morphology (P<10−3) | Immune response (P<10−3) |
| Embryonic development (P<10−3) | Connective tissue dev. (P<10−3) |
| Tumor morphology (P<10−3) | |
| Top canonical pathways | |
| Fatty acid metabolism (P<10−10) | Actin cystosk. Signaling (P<10−4) |
| Xenobiotic metabolism (P<10−5) | Hypoxia signaling (P<10−3) |
| Coagulation system (P<10−5) | |
| FXR/ RXR Activation (P<10−4) | |
| Top Tox functions: Hepatotoxicity | |
| Liver cholestasis (P<10−6) | Hepatocellular carcinoma (P<10−3) |
Figure 4.
High level of miR-122 in HCC defines a specific gene expression profile linked to HNF4A. (a–b) Ingenuity Pathway Analysis applied to the genes which expression was positively correlated with the expression of miR-122 in human primary HCC (a) and HCC-derived cell lines (b). Liver-enriched transcription factors, HNF4A in particular, were identified as the central regulators in the gene signature associated with a high miR-122 expression. (c–e) In HCC-derived cell lines the loss of miR-122 is associated with a loss of HNF4A. (c) The evaluation of HNF4A mRNA level in HCC-derived cell lines indicated a positive correlation between HNF4A and miR-122. (d) The assessment of HNF4A expression in protein extracts isolated from the cell lines which exhibit high vs. low miR-122 further indicates a positive correlation between miR-122 and HNF4A expression. High miR-122 expressing cell lines, such as PLC/PRF/5, Huh-1, Hep40 or Huh-6, exhibit a considerably higher HNF4A protein levels as compared to SNU-182, SNU-387, and SNU-398 lines which exhibit low miR-122levels. (e) Analysis of HNF4A expression in cytoplasmic (C) and nuclear (N) compartments indicates a nuclear enrichment of HNF4A in the high miR-122 expressing cell lines.
Relationship between miR-122 and liver-enriched transcription factors
As a paradigm, HCC-derived cell lines were divided into two groups based on the miR-122 expression levels. Gene expression profiling identified 300 genes which were differentially expressed as a function of miR-122 expression (SI Figure 4). Consistent with our findings in primary tumors, high levels of miR-122 in HCC-derived cell lines were correlated with high expression of genes involved in the pathways characteristic of fully differentiated hepatocytes (SI Table 2). These included genes encoding apolipoproteins (APO-A1), coagulation factors (F2), and acute phase proteins (AHSG). More importantly, miR-122 exhibited a strong positive correlation with the expression level of numerous liver-enriched transcription factors essential for hepatocytic differentiation, such as HNF1A, HNF3A, HNF3B, HNF4A, HNF4G, and HNF6 (SI Figure 5). Gene network analysis not only confirmed activation of the HNF4A signature in HCC-derived cell lines expressing miR-122 but also revealed strong interactions with other liver-enriched transcription factors (Figure 4b).
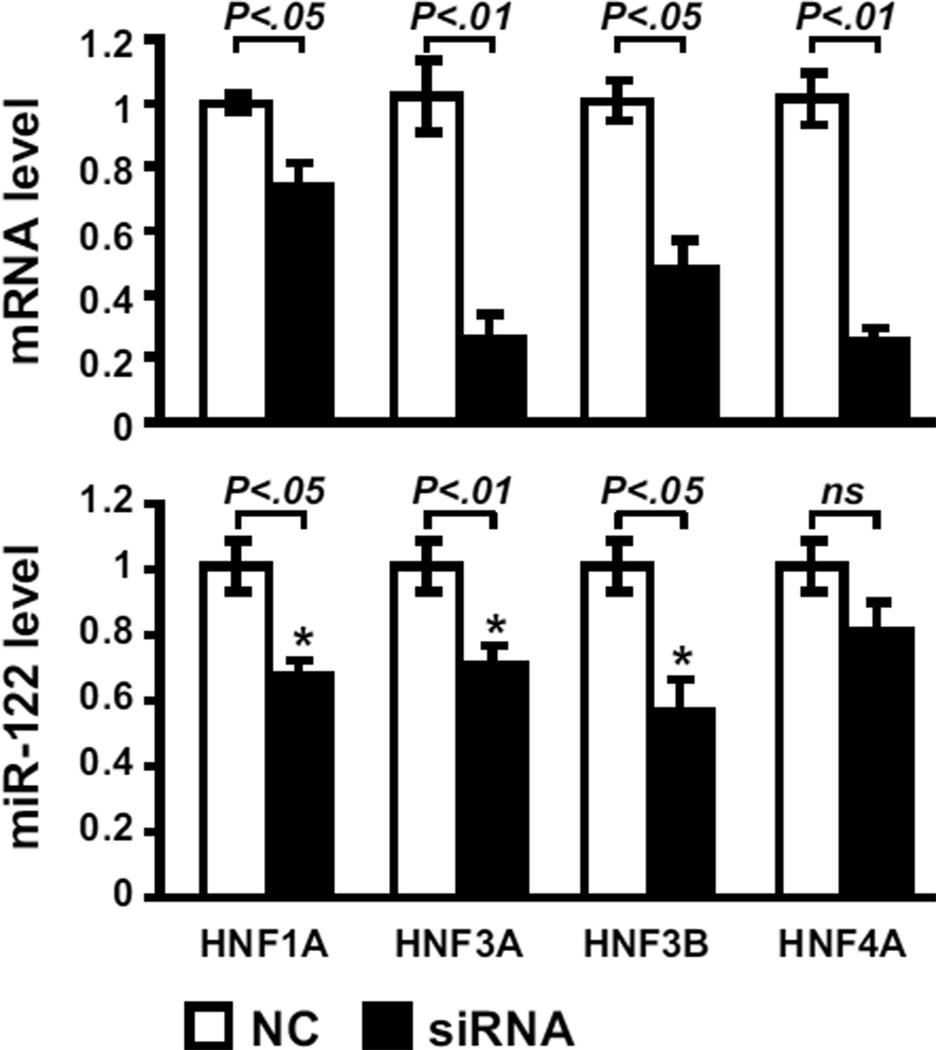
In this context, HNF4A was directly evaluated as a function of miR-122 expression. As shown in Figure 4c–e, HNF4A expression was directly correlated with the expression of miR-122, both on mRNA and protein levels. Furthermore, in the high miR-122 expressing cell lines, HNF4A was particularly enriched in the nuclear fraction as compared to the cytoplasmic fraction reflecting its greater transcriptional activity (Figure 4e). The latter is consistent with a central role of HNF4 revealed by the network analysis in the miR-122 overexpressing HCC cells lines (Figure 4b). Despite a strong positive correlation, the exact relationship between miR-122 and HNF4A appears to be highly complex. The lack of miR-122 seed region in the 3’UTR of HNF4A transcript makes HNF4A an unlikely direct target of miR-122. Accordingly, the experimental modulation of miR-122 expression did not affect HNF4A mRNA and protein levels (data not show). It is also dubious that HNF4A directly drives the expression of miR-122 since the silencing of HNF4A by siRNA did not significantly change the expression of miR-122 (Figure 5). Conversely, the knock-down of HNF1A, HNF3A or HNF3B was able to reduce miR-122 expression implying that mir-122 may be under transcriptional control of these transcription factors (Figure 5). These results provide new insight into the transcriptional regulation of miR-122 gene and implicate miR-122 as a marker of hepatocyte differentiation.
Figure 5.
The silencing of HNF1A, HNF3A, and HNF3B results in a decrease of miR-122 level. The high miR-122 expressing PLC/PRF/5 cell line was transfected with 40 nM of specific siRNA directed against HNF-1A, -3A, -3B, and -4A (siRNA, black column) vs. negative control (NC, white column). Forty eight hours after the transfection, the efficiency of the silencing was confirmed by the decreased level of respective mRNA (upper panel). As shown in the lower panel, the silencing of HNF-1A, -3A, and -3B significantly reduces the level of miR-122.
Loss of miR-122 coincides with acquisition of invasive phenotype
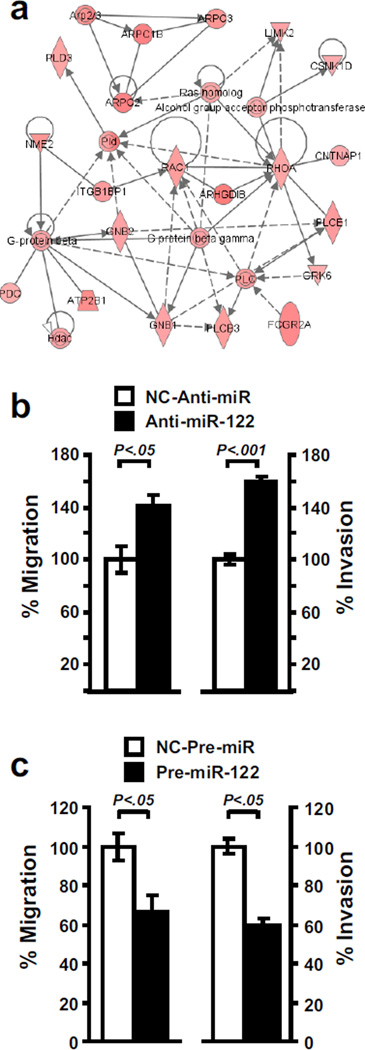
Lastly, we analyzed the genes which were negatively correlated with miR-122 (Cluster B in Figure 3a). Expression profiling indicated that loss of miR-122 expression was associated with a higher expression of numerous genes related to metastasis, including genes involved in cell motility (RAC1, RHOA), angiogenesis (VEGF), hypoxia (HIF1A) and epithelial-mesenchymal transition (vimentin) (SI Table 1, SI Figure 6). Notably, RHOA and RAC1 were two central molecules in a network associated with cell motility (Figure 6a). Therefore, we investigated the impact of miR-122 loss on cell motility. As shown in Figure 6b, the experimental inhibition of miR-122 in Huh-1 cells resulted in an increase of migration and invasion. Further validating the importance of miR-122 in the control of cell motility, the ectopic expression of miR-122 was able to reduce the migration and invasion of SK-Hep1 cells (Figure 6c). These data emphasize the critical role of the loss of miR-122 expression during tumor progression and the potential relevance of miR-122 restoration for HCC therapy.
Figure 6.
miR-122 impacts the propensity of cancer cells to migrate and invade. (a) Ingenuity Pathway Analysis was performed on the genes up-regulated in HCC samples which exhibit a low miR-122 expression (Cluster B in Figure 3). This procedure revealed a network associated with cell motility and identified RAC1 and RHOA as two central regulators. (b) Inhibition of miR-122 results in the acquisition of migrating and invasive properties. The effect of miR-122 inhibition on cellular migration and invasion was assessed in Huh-1 by transfecting 20 nM of either anti-miR-122 (black column) or negative control (NC-Anti-miR, white column). Invasive and migrating activity was measured by using the BD BioCoat Matrigel Invasion Chambers. Data indicate that the loss of miR-122 increases the migrating and invasive properties of Huh-1 cells. (c) Conversely, in the miR-122 negative SK-Hep1 cell line the restoration of miR-122 by transfecting miR-122 precursor molecules results in a decreased migration and invasion (Pre-miR-122, black column) as compared to cells transfected with negative control (NC-Pre-miR, white column).
Discussion
Accumulating evidence suggests a causal role for miRNAs in human cancer (Esquela-Kerscher and Slack, 2006). In this study, we have evaluated the clinical and functional relevance of liver specific miR-122 for human HCC. We show that the repression of miR-122 correlates with clinically relevant parameters, such as etiology, tumor size and differentiation grade. More importantly, miR-122 repression is specific for liver tumors which are characterized by poor prognosis and coincides with suppression of hepatic phenotype as well as gain of metastatic properties.
Our findings demonstrate a link between the expression levels of miR-122 and HBV or HCV infection. By analyzing the expression of miR-122 in a cohort of HCC with different etiology, we have established that down-regulation of miR-122 occurs mainly in the HCC arising in the HBV-infected livers. In the HCC developed with HCV infection, miR-122 expression was either maintained or increased, in agreement with recent observations (Varnholt et al., 2008). Interestingly, the efficiency of HCV replication has been found to be closely correlated with the miR-122 expression levels (Jopling et al., 2005; Chang et al., 2008), and further studies have implicated miR-122 in HCV translation, suggesting involvement of miR-122 in HCV tropism to the liver (Henke et al., 2008). Therefore, we speculate that HCV not only requires the host-expressed miR-122 for its replication, but it may also either directly induce miR-122 expression or indirectly inhibit miR-122 down-regulation.
Similar to the transcription factors, tissue specificity of miRNA expression may imply a role in differentiation. Due to the high abundance in the liver, it has been suggested that miR-122 may maintain the hepatic differentiation status by repressing the genes which are normally not expressed by hepatocytes (Girard et al., 2008). The temporal expression pattern of miR-122 during liver development is consistent with its involvement in liver differentiation (Chang et al., 2004). In support of this notion, recent work showed that promotion of endodermal differentiation of human embryonic stem cells (hESC) by NaButyrate was associated with an increase of miR-122 expression and a parallel activation of several hepatocyte-specific genes (Tzur et al., 2008). According to our findings in human HCC and HCC-derived cell lines, the expression of miR-122 was tightly correlated with the expression of numerous genes characteristic of the differentiated hepatocytes. In agreement with the current view of miR-122 as a regulator of lipid and cholesterol metabolism, the expression of abundant apolipoprotein genes was found to be correlated with miR-122 levels (Krutzfeldt et al., 2005; Esau et al., 2006; Elmen et al., 2008). The specific down-regulation of miR-122 in poorly differentiated tumors as well as in tumors characterized by the hepatoblast gene signature supported miR-122 as an important determinant of hepatic differentiation. Moreover, in two independent datasets, we identified HNF4A as the central regulatory transcription factor in the network of genes which were differentially expressed as a function of miR-122 expression. HNF4A is a liver-enriched transcription factor controlling the differentiation-dependent gene expression in developing and adult livers (Odom et al., 2004). Although we initially speculated that HNF4A was the missing link between the differentiation status and the transcription of miR-122 gene, the additional gene silencing experiments suggested a more important role for other liver-enriched transcription factors, such as HNF1A, HNF3A, and HNF3B. The promoter region of miR-122 gene has not been yet identified but a binding site for HNF3B has been located 5kb upstream of miR-122 (SI Figure 7). Although further experiments are needed to address the functionality of this binding site, our data provide the first evidence for the tissue specificity of miR-122 expression.
Using a comparative genomic approach based on the experimentally well defined gene expression signatures, we have previously identified several clinically relevant subtypes of HCC with significant differences in the biological properties and clinical outcome (Lee et al., 2004a; Lee et al., 2004b; Kaposi-Novak et al., 2006; Lee et al., 2006; Coulouarn et al., 2008). Importantly, we showed here that the repression of miR-122 was characteristic of HCC which displayed a hepatoblast, c-Met or late TGF-β signature. These HCC subtypes were characterized by increased invasiveness, tumor recurrence and decreased patient survival whereby raising a possibility than the loss of miR-122 may predispose to the acquisition of invasive phenotype. Consistent with this, miR-122 was recently described as a metastasis-related miRNA in HCC (Budhu et al., 2008), and repression of miR-122 was reported in highly metastatic human breast cancer cell lines (Tavazoie et al., 2008). Supporting these data, our results demonstrate that HCC cell lines known to exhibit a more invasive phenotype also displayed a decreased miR-122 expression (Coulouarn et al., 2008). Additionally, a significant enrichment of genes related to metastasis (RHOA, VEGF, HIF1A, Vimentin) was observed both in HCC and cell lines lacking miR-122 expression (SI Table 1). Accordingly, we provide direct evidence that the modulation of miR-122 greatly influenced the propensity of cancer cells to migrate and invade. Indeed, the loss of miR-122 resulted in the acquisition of migrating and invasive properties and the restoration of miR-122 was able to reverse this phenotype. In an attempt to identify the underlying molecular mechanisms responsible for the impact of miR-122 on cell motility, we used microRNA target prediction algorithms on genes negatively correlated with miR-122. Interestingly, an evolutionary conserved binding site for miR-122 was detected in the 3’UTR of RHOA mRNA (SI Figure 8a), and the analysis of the secondary structure of RHOA 3’UTR indicated that the predicted binding site coincided with a hairpin-loop structure, making this sequence likely accessible for an interaction with miR-122 (SI Figure 8b). RHOA is frequently overexpressed in HCC and several groups correlated the increase of RHOA expression with tumor progression, cell differentiation, and metastasis (reviewed in Grise et al., 2009). Moreover, RHOA overexpression in liver has been directly correlated with the acquisition of an aggressive phenotype, both in vitro and in vivo (Yoshioka et al., 1995; Xue et al., 2008). In addition, gene reporter experiments suggested a direct functional interaction between miR-122 and RHOA 3’UTR (SI Figure 8c). Nevertheless, the experimental inhibition or induction of miR-122 failed to demonstrate a consistent modulation of RHOA on a protein level. Also, the acquisition of invasive properties observed upon miR-122 inhibition (Figure 6b) appeared to be independent of RHOA since the silencing of RHOA did not significantly abrogated this response. The absence of RHOA modulation on a protein level could be due to an enhanced stability of the protein. Alternatively, RHOA transcript may be tightly regulated by positive and negative signals. For example, the increase in the oncogenic miR-155, which was recently reported to target and reduce the level of RHOA (Kong et al., 2008), may counteract the miR-122 effect. These observations raise important questions regarding the regulation of transcript stability by microRNAs and suggest that the integration of activating and inhibitory signals driven by different microRNAs may define the level of a particular transcript. Interestingly, while writing this manuscript, Tsai et al., published that miR-122 regulates intrahepatic metastasis by controlling the disintegrin and metalloprotease ADAM17 (Tsai et al., 2009). Together, these data not only emphasize the critical role of miR-122 in migration and invasion but suggest that miR-122 contribution to tissue remodeling is broader than a direct effect on actin cytoskeleton organization through a RHOA-dependant mechanism.
In conclusion, our work sheds new light on the clinical and functional implications of miR-122 expression in liver cancer. We identified miR-122 as a new diagnostic and prognostic microRNA marker for HCC progression. Given that the modulation of miR-122 expression became a reality in animal models (Czech, 2006; Elmen et al., 2008) our study provides a rationale for classification and development of novel therapy for human HCC.
Methods
Human tissue samples
A total of 64 HCC tissues and 28 matched non-tumor surrounding liver tissues were obtained from patients undergoing partial hepatectomy as treatment for HCC. Tissue banking was approved by the Institutional Review Board of US National Cancer Institute. Determination of proliferation, apoptosis and ubiquitination index was described previously (Lee et al., 2004b). Total RNAs were isolated using the CsCl density gradient centrifugation method (Coulouarn et al., 2006).
Cell lines
HCC-derived cell lines were obtained from American Type Culture Collection (ATCC, Rockville, MD) and RINKEN Cell Bank (Japan) and maintained in Dulbecco’s modified Eagle medium/F12 medium supplemented with 2mM glutamine and 10% fetal bovine serum, as described (Lee and Thorgeirsson, 2002). Total RNAs were extracted from cells at 80% confluence using RNeasy mini kit (Qiagen, Valencia, CA).
Cell transfection
Transfections were carried out using siPORT™ NeoFX™ according to the manufacturer's instruction (Ambion, Austin, TX). RNA and protein analysis were conducted after 48 hours incubation in normal culture condition. miRIDIAN microRNA Hairpin Inhibitors and Mimics from Dharmacon (Waltham, MA) were used for the specific inhibition and overexpression of miR-122. Amount of small RNA was adjusted to a final concentration of 20 nM. Efficiency and specificity of the transfections were evaluated by confocal microscopy and Q-RT-PCR (SI Figure 9). Silencer select siRNAs along with the Negative Control #1 (Ambion, Austin, TX) were used for the specific knock-down of HNF1A (s13870), HNF3A (s6689), HNF3B (s6692), HNF4A (s6698), FOXO1 (s5259), and RHOA (s760).
Real time RT-PCR of miR-122
Expression of miR-122 was measured using mirVana Q-RT-PCR miRNA Detection Kit and SYBR® Green I (Ambion, Austin, TX). Total RNA were reverse transcribed at 37°C for 30 min with an ArrayScript Enzyme mix including miR-122 specific RT primers according to the manufacturer’s instructions. Quantitative PCR analysis was performed with an IQ-5 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) in a 96-well reaction plate. Melting analysis of the PCR products was conducted to validate the amplification of the specific product. Quantitative values were determined using the 2−ΔΔCt method, where the ΔCt value of a given RNA sample was calculated by subtracting the Ct value of miR-122 from the Ct value of 5S small RNA used for normalization.
DNA microarray analysis
Gene expression data used in this study (GEO accession numbers: Human microarray platform, GPL1528; human HCC microarray data, GSE1898) were generated using a genome-wide set of long-mer human oligonucleotides obtained from Illumina and printed at the Advanced Technology Center (National Cancer Institute). Target preparation and hybridization on microarrays were previously described (Lee and Thorgeirsson, 2002). Differentially expressed genes were identified by a univariate two-sample t-test with a random variance model. Permutation P values for significant genes (P < 0.01) were computed based on 10.000 random permutations as previously described (Coulouarn et al., 2006). Expression profiles were clustered by the Cluster 3.0 program using an unsupervised hierarchical procedure and visualized with the TreeView program (Coulouarn et al., 2006).
Gene Set Enrichment Analysis (GSEA) and pathway analysis
GSEA was performed using the web-based tool developed by Broad Institute (www.broad.mit.edu/gsea/). Enrichment scores were determined after 1,000 permutations. Ingenuity Pathway Analysis tool was used to examine the functional associations between differentially expressed genes and to generated the highest significant gene networks (www.ingenuity.com). Biologically relevant networks were identified using the scoring system provided by Ingenuity.
miRNA target prediction
Potential miR-122 target among genes negatively correlated with miR-122 expression were determined using the publicly available miRanda (www.microrna.org) and TargetScan (www.targetscan.org) algorithms. Secondary structure of potential target transcripts was explored using the web-based RNA analyzer tool (www.wb2x01.biozentrum.uni-wuerzburg.de).
RHOA 3’ UTR reporter analysis
The human sequence of the RHOA 3’ UTR was amplified by PCR using Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) and cloned into a SacI- and HindIII-cut pMIR-Report luciferase vector (Ambion, Austin, TX). A mutant construct was generated by introducing mutations in the putative miR-122 binding site. Mutations were introduced by stand overlap PCR using mutagenic primers. All constructs were sequenced to confirm that the desired mutations had been obtained. Firefly luciferase constructs containing the wild-type or the mutant variant of RHOA 3’ UTR were co-transfected with a β-galactosidase control vector in the presence of 20 nM miR-122 inhibitor or negative control. Transfections were performed with 0.1 μg firefly luciferase constructs and 0.01 μg β-galactosidase normalization vector using Lipofectamine 2000. Luciferase and β-galactosidase activities were assessed 48 hours after transfection using a dual-light chemiluminescent reporter gene assay system (Applied biosystems, Foster City, CA).
Invasion and migration assays
The effect of miR-122 on invasion and migration was evaluated by using the BD BioCoat Matrigel Invasion Chambers as described (Coulouarn et al., 2008).
Immunoblotting
Protein expression analysis was as previously described (Coulouarn et al., 2006). Protein extraction was performed using M-PER and NE-PER reagents (Pierce, Rockford, IL). Twenty micrograms of protein extract were subjected to SDS/PAGE and transferred to PVDF membranes. Membranes were probed with a goat polyclonal HNF4A antibody and a rabbit polyclonal RHOA antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Detection was performed by ECL (Amersham Biosciences, Piscataway, NJ) after incubation of a HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Supplementary Material
Acknowledgments
Financial Support: This project was supported in part by the Intramural Research Program of the Center for Cancer Research.
Abbreviations
- HCC
hepatocellular carcinoma
- miRNA
microRNA
Footnotes
Supporting Information is linked to the online version of the paper.
Contributor Information
Cédric Coulouarn, Email: cedric_coulouarn@nih.gov.
Valentina M. Factor, Email: valentina_factor@nih.gov.
Jesper B. Andersen, Email: jesper_boje_andersen@nih.gov.
Marian E. Durkin, Email: marian_durkin@nih.gov.
Snorri S. Thorgeirsson, Email: snorri_s_thorgeirsson@nih.gov.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–1809. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215–8223. doi: 10.1128/JVI.02575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR- 122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulouarn C, Gomez-Quiroz LE, Lee JS, Kaposi-Novak P, Conner EA, Goldina TA, et al. Oncogene-specific gene expression signatures at preneoplastic stage in mice define distinct mechanisms of hepatocarcinogenesis. Hepatology. 2006;44:1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. 2006;354:1194–1195. doi: 10.1056/NEJMcibr060065. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127:S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR- 122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Grise F, Bidaud A, Moreau V. Rho GTPases in hepatocellular carcinoma. Biochim Biophys Acta. 2009;1795:137–151. doi: 10.1016/j.bbcan.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA- 155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004a;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004b;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- Lee JS, Thorgeirsson SS. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134–1143. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
- Li W, Xie L, He X, Li J, Tu K, Wei L, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006;43:S145–S150. doi: 10.1002/hep.21063. [DOI] [PubMed] [Google Scholar]
- Tzur G, Levy A, Meiri E, Barad O, Spector Y, Bentwich Z, et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS ONE. 2008;3:e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, et al. MicroRNA- 223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Imamura F, Shinkai K, Miyoshi J, Ogawa H, Mukai M, et al. Participation of rhop21 in serum-dependent invasion by rat ascites hepatoma cells. FEBS Lett. 1995;372:25–28. doi: 10.1016/0014-5793(95)00937-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.