Abstract
To investigate the range of autoinhibitory mechanisms used by TKDs (tyrosine kinase domains) from the insulin receptor family of RTKs (receptor tyrosine kinases), we determined crystal structures of TKDs from TrkA (tropomyosin receptor kinase A, a nerve growth factor receptor) and Ror2 (receptor tyrosine kinase-like orphan receptor 2, an unconventional Wnt receptor). TrkA autoinhibition closely resembles that seen for the insulin receptor, relying on projection of an activation loop tyrosine residue into the substrate-binding site and occlusion of the ATP-binding site by the activation loop. Ror2 employs similar mechanisms, but the unusual replacement of the phenylalanine residue in its Asp-Phe-Gly motif with leucine necessitates occlusion of the ATP-binding site by other means. The unusual Asp-Leu-Gly motif in Ror2 is displaced compared with other inactive kinases, allowing the activation loop to interact directly with the TKD's αC helix, in another mode of autoinhibition that is characteristic of the other extreme of this receptor family: ALK (anaplastic lymphoma kinase) and Met. These findings provide insight into the expected range of activating mutations in these TKDs in cancer. We also describe symmetrical dimers of the inactive TrkA TKD resembling those found in other RTKs, possibly reflecting an arrangement of kinase domains in a pre-formed TrkA dimer.
Keywords: autoinhibition, insulin receptor kinase, receptor tyrosine kinase (RTK), receptor tyrosine kinase-like orphan receptor 2 (Ror2), tropomyosin receptor kinase A (TrkA), X-ray crystallography
Abbreviations: ALK, anaplastic lymphoma kinase; CDK, cyclin-dependent kinase; C-lobe, C-terminal lobe; COSMIC, Catalogue Of Somatic Mutations In Cancer; DTT, dithiothreitol; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; IGF1R, insulin-like growth factor 1 receptor; IRK, insulin receptor kinase; MuSK, muscle-specific kinase; Ni-NTA, Ni2+-nitrilotriacetate; N-lobe, N-terminal lobe; NSCLC, non-small-cell lung cancer; Ror, receptor tyrosine kinase-like orphan receptor; RTK, receptor tyrosine kinase; TCEP, tris-(2-carboxyethyl)phosphine; TKD, tyrosine kinase domain; Trk, tropomyosin receptor kinase
INTRODUCTION
Recent clinical success with targeted inhibitors of the kinases EGFR (epidermal growth factor receptor), B-Raf and ALK (anaplastic lymphoma kinase) underscore both the promise and the challenges of ‘personalized’ cancer medicine [1,2]. Well-defined activating mutations in each of these kinases have been identified as oncogenic ‘drivers’, promoting uncontrolled proliferative or cell survival signalling that can be curbed with specific kinase inhibitors. As genomic analysis of tumours burgeons, however, it is becoming increasingly clear that many mutations found in cancer are ‘passengers’ rather than drivers [1]. The ability to determine which mutations in a tumour are oncogenic drivers, and thus which signalling nodes should be targeted therapeutically, will be crucial for advancing personalized medicine in oncology.
For protein kinases, including RTKs (receptor tyrosine kinases), which are frequently found as oncogenic drivers in cancer [3], a key goal is to understand which mutations increase constitutive kinase activity and signalling. EGFR provides an excellent illustration of how structural understanding of kinase activation mechanisms [4] can explain driver mutations in NSCLC (non-small-cell lung cancer) [5]. The inactive EGFR TKD (tyrosine kinase domain) is autoinhibited by intramolecular interactions between a short α-helix in its activation loop and another important helix (αC) that is consequently displaced from the position it adopts in the active kinase. This mode of autoinhibition closely resembles that seen in the Src family and CDKs (cyclin-dependent kinases) [6]. Known driver mutations activate EGFR by destabilizing these intramolecular autoinhibitory interactions. Intriguingly, oncogenic driver mutations that destabilize very similar autoinhibitory interactions are also found in ALK [7–9], a member of the large IRK (insulin receptor kinase) subfamily of RTKs [10,11] that is important in neuroblastoma [12]. Despite the fact that the ALK TKD shows closest sequence similarity to IRK family members, the autoinhibitory interactions that define its inactive conformation most closely resemble those in EGFR, Src and CDKs [6,13]. As in EGFR, ALK TKD structures reveal hydrophobic interactions between a small α-helix in the activation loop and the crucial αC helix that stabilize the inactive kinase conformation [7,8]. The side chains of the ALK residues most frequently mutated in neuroblastoma [12] (Phe1174 and Arg1275) make important contributions to these autoinhibitory interactions, and their mutation causes constitutive ALK activation. Indeed, Phe1174 and Arg1275 in ALK are structurally equivalent to EGFR residues Val769 and Leu861 [9], at which activating mutations are also seen in NSCLC [5].
Structural studies have revealed a spectrum of autoinhibitory mechanisms for IRK family RTKs. ALK lies at one extreme, with an EGFR/Src/CDK-like mode of autoinhibition. IRK lies at the other, with a very different autoinhibitory activation loop conformation that simultaneously occludes the ATP-binding site and projects a key tyrosine side chain (Tyr1162) into the substrate-binding site [13]. The autoinhibitory mechanisms of inactive IGF1R (insulin-like growth factor 1 receptor) [14] and MuSK (muscle-specific kinase) [15] kinases closely resemble that seen in IRK, with very similar activation loop configurations. However, structures of the inactive TKDs of the IRK family members Met [16] and Ron [17] have quite distinct activation loop conformations. Since mutations that disrupt autoinhibitory interactions are known to be important oncogenic drivers in ALK (and Met), it seems clear that understanding the mechanism of TKD autoinhibition will be important for predicting which mutations are likely to be significant for IRK family RTKs in cancer or other diseases in which RTK activation is important. Since the cases of Met, ALK and Ron demonstrate that not all IRK family RTKs are regulated in the same way as IRK, we were motivated to ask how other kinases in this family are regulated. Notably absent from the list of IRK family TKDs with known structure are members of the Trk (tropomyosin receptor kinase), Ror (receptor tyrosine kinase-like orphan receptor) and DDR (discoidin domain receptor) subfamilies, which have all been implicated in cancer [18–20] and other diseases. In the present paper, we describe crystal structures to 2.4 Å (1 Å=0.1 nm) resolution of the inactive TKDs from TrkA (a receptor for nerve growth factor [21]) and Ror2 (now known to be a Wnt receptor [22]). In the context of the other IRK family structures, these new results provide a useful framework for interpreting the consequences of TKD mutations in this family.
EXPERIMENTAL
Plasmid construction
DNA encoding the intracellular domain residues 498–796 of human TrkA (NCBI reference sequence NP_002520.2) and residues 452–753 of human Ror2 (NCBI reference sequence NP_004551.2) were amplified by PCR to include a coding sequence for an N-terminal hexahistidine tag and unique EcoRI and XhoI restriction sites for TrkA or SpeI and XhoI restriction sites for Ror2. Appropriately digested fragments were subcloned into the pFastBac1 vector (Invitrogen) using the indicated restriction sites. The Bac-to-Bac expression system (Invitrogen) was then used for generation of recombinant baculoviruses and for protein expression in Spodoptera frugiperda Sf9 insect cells.
Protein production and purification
Sf9 cells at (1.5–2)×106/ml were infected with recombinant baculovirus, and harvested by centrifugation after 3 days. Sf9 cells expressing histidine-tagged TrkA498–796 (~7 litres of medium) were lysed by sonication in 100 ml of 50 mM NaKPO4 (pH 8.0), containing 300 mM NaCl, 5% (w/v) glycerol, 10 mM imidazole, 10 mM 2-mercaptoethanol, 0.5 mM PMSF and protease inhibitor cocktail (Roche). The lysate was then mixed with Ni-NTA (Ni2+-nitrilotriacetate) beads (Qiagen) for 1 h at 4°C. Beads were washed in 50 column volumes of lysis buffer (described above), and bound TrkA498–796 was eluted with increasing concentrations of imidazole in 25 mM Mes (pH 6), containing 300 mM NaCl, 5% (w/v) glycerol, 10 mM 2-mercaptoethanol, 0.5 mM PMSF and protease inhibitor cocktail (Roche). Eluted protein was then purified further using a Fractogel SO3− cation exchange column (EMD) equilibrated with 25 mM Mes (pH 6), containing 5% (w/v) glycerol, 2 mM DTT (dithiothreitol) and eluting with a gradient from 10 mM to 1 M NaCl. TrkA498–796 was then applied to a HiTrap butyl-Sepharose HP column (GE Healthcare) in 25 mM Mes (pH 6), containing 150 mM NaCl and 2 mM DTT, eluting with a gradient from 0.8 M to 0 M (NH4)2SO4, and subjected to a final step of size-exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated in 25 mM Mes (pH 6), containing 250 mM NaCl and 2 mM DTT.
Sf9 cells expressing histidine-tagged Ror2452–753 (~8 litres of medium) were lysed by sonication in 150 ml of lysis buffer, composed of 20 mM NaKPO4 (pH 8.0), containing 200 mM NaCl, 10 mM 2-mercaptoethanol, 1 mM PMSF, 10 μM benzamidine, 2.3 μM leupeptin, 2 μM aprotinin and 3 μM pepstatin (Sigma). Cell lysates containing Ror2452–753 protein were mixed with Ni-NTA beads (Qiagen) for 30 min at 4°C, which were then washed with lysis buffer before elution of protein in lysis buffer containing 200 mM imidazole. Eluted protein was passed through a Fractogel TMAE (trimethylaminoethyl) column (EMD) equilibrated with 25 mM Tris/HCl (pH 8), containing 100 mM NaCl and 2 mM DTT to remove anionic contaminants, and was then passed through a CHT2.1 hydroxyapatite column (Bio-Rad Laboratories) equilibrated in 20 mM Hepes (pH 8), containing 2.5 mM NaKPO4, 200 mM NaCl, 2 mM DTT and 1 mM PMSF, before loading on to a HiTrap butyl-Sepharose HP column in 25 mM Tris/HCl (pH 8), containing 125 mM NaCl and 2 mM DTT. Ror2452–753 was eluted from butyl-Sepharose with a gradient from 0.5 M to 0 M (NH4)2SO4 in this same buffer, and then subjected to size-exclusion chromatography using a Superdex 200 column equilibrated in 20 mM Tris/HCl (pH 7.5), containing 120 mM NaCl and 1 μM TCEP [tris-(2-carboxyethyl)phosphine].
Crystallization and structure determination
Crystals were obtained using the hanging-drop vapour-diffusion method, by mixing equal volumes of protein and reservoir solutions and equilibrating over the reservoir solution at 21°C. For TrkA498–796, protein was concentrated to ~6 mg/ml in 25 mM Mes (pH 6), containing 250 mM NaCl and 2 mM DTT and then diluted with water to 3.25 mg/ml. Crystals were obtained with a reservoir solution of 1.5 M NaCl, 0.1 M Mes (pH 6.5) and 0.2 M NaKPO4. For Ror2452–753, protein was concentrated to 7.2 mg/ml in 20 mM Tris/HCl (pH 7.5), containing 125 mM NaCl and 1 μM TCEP, and crystals were obtained over a reservoir containing 20% PEG [poly(ethylene glycol)] 3350 and 0.2 M Mg(NO3)2 (Hampton Research PEG Ion Screen 16). Before flash-freezing in liquid nitrogen, TrkA498–796 crystals were cryoprotected in reservoir solution containing 40% (w/v) dextrose, and Ror2452–753 crystals were cryoprotected in reservoir solution containing 20% (w/v) glycerol. Diffraction data were collected at beamline 23ID-B of GM/CA@APS (Advanced Photon Source) and were processed using HKL2000 [23] (Table 1). TrkA498–796 crystallized in space group H32 with one molecule in the asymmetric unit, and Ror2452–753 crystallized in space group C2221 with two molecules in the asymmetric unit.
Table 1. Data collection and refinement statistics (molecular replacement).
Each dataset was collected from a single crystal. Values for the highest-resolution shell are shown in parentheses.
| TrkA | Ror2 | |
|---|---|---|
| Data collection | ||
| Space group | H32 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 105.0, 105.0, 203.3 | 102.8, 112.9, 114.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.4 | 46.9–2.4 |
| Rsym | 0.065 (0.553) | 0.112 (0.565) |
| I/σ | 52.9 (5.3) | 21.9 (4.1) |
| Completeness (%) | 99.9 (100) | 100 (99.9) |
| Redundancy | 11.1 (11.3) | 7.5 (7.3) |
| Refinement | ||
| Resolution (Å) | 2.4 | 2.4 |
| Number of reflections | 17195 | 26251 |
| Rwork/Rfree | 0.20/0.25 | 0.17/0.20 |
| Number of atoms | ||
| Protein | 2261 | 4274 |
| Ion | 0 | 16 (4×NO3−) |
| Water | 77 | 228 |
| B-factors | ||
| Protein | 76.3 | 39.1 |
| Ion | 49.6 | |
| Water | 62.6 | 38.3 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.004 | 0.003 |
| Bond angles (°) | 0.758 | 0.643 |
Structures were solved by molecular replacement with Phaser [24], using co-ordinates for the MuSK TKD (PDB code 1LUF) [15] as a search model. Cycles of manual building/rebuilding using Coot [25] were alternated with rounds of refinement employing REFMAC [24], plus composite omit maps calculated with CNS [26]. Later stages employed PHENIX [27], with TLS (Translation–Libration–Screw-rotation) refinement [28]. PROCHECK [29] identified no residues in the disallowed region of the Ramachandran plot. Structure figures were generated using PyMOL (version 1.5.0.2; http://www.pymol.org). Data collection and refinement statistics are shown in Table 1. The final refined TrkA model includes amino acids 498–534, 537–548, 550–610 and 614–793. There is additional density close to the N-terminus of the TrkA structure that we could not model confidently, projecting towards the active site. The final Ror2 structure is lacking density for residues 452–463 at the N-terminus. In addition, residues 512, 516, 576–580 and 752–753 in chain A could not be confidently modelled, and neither could residues 515, 573–580 and 753 in chain B. Cys694 in both chain A and chain B of the Ror2 TKD appears to form a disulfide bond with an equivalent residue in a symmetry mate.
RESULTS AND DISCUSSION
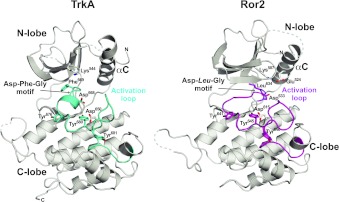
Both the TrkA and Ror2 TKD structures were solved in their unphosphorylated inactive forms (Figure 1), with no bound nucleotide. The TrkA and Ror2 structures exhibit the common kinase topology consisting of an N-terminal lobe and a larger C-terminal lobe. The N-terminal lobe consists of a twisted five-stranded β-sheet plus one conserved α-helix, called αC, that plays a key role in the transition from an inactive to an active conformation [30]. The larger C-terminal lobe is predominantly helical, and also contains the activation loop that undergoes key conformational changes upon activation, contributing to the catalytic and substrate-binding sites in the active conformation of the TKD, but instead playing an autoinhibitory role in the inactive conformation.
Figure 1. Structures of TrkA and Ror2 TKDs.
Structures of inactive TKDs from TrkA (left) and Ror2 (right) in cartoon representation. The N-lobe and C-lobe are labelled, as is the catalytically important αC helix. The activation loops are labelled and highlighted in cyan (TrkA) and magenta (Ror2). Side chains in the DFG motif of TrkA (Asp668-Phe669-Gly670) and the equivalently located DLG motif in Ror2 (Asp633-Leu634-Gly635) are shown, and tyrosine residues from the YXXXYY motif are shown in stick representation. Also shown is the invariant lysine residue in strand β3 (Lys544 in TrkA, Lys507 in Ror2), which forms a salt bridge with αC Glu524 in Ror2 (but not TrkA). The catalytic base aspartate (Asp650 in TrkA, Asp615 in Ror2) is also shown and labelled. Molecule A was used for Ror2 TKD. The N- and C-terminal residues of the modelled structure are marked where visible in the orientation shown.
Autoinhibition of TrkA and Ror2
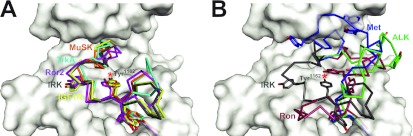
Both the TrkA and Ror2 TKDs share one of the key autoinhibitory features of IRK [13], projecting the second tyrosine of the YXXXYY (Tyr-Xaa-Xaa-Xaa-Tyr-Tyr) motif found in the activation loop of IRK family kinases into the (tyrosine) substrate-binding site. As shown in Figure 2(A), Tyr680 of TrkA and Tyr645 of Ror2 occupy the same position as Tyr1162 in IRK [13] (marked with an asterisk in Figure 2A), and make similar interactions with both the catalytic base aspartate residue (Asp650 and Asp615 in TrkA and Ror2 respectively; Asp1132 in IRK) and an arginine residue conserved in IRK family kinases (Arg654 and Arg619 in TrkA and Ror2 respectively; Arg1136 in IRK). The activation loop conformations of the TrkA and Ror2 TKDs are very similar to those seen in IRK, MuSK and IGF1R [13–15] (Figure 2A), whereas the ALK, Met and Ron activation loops are quite different (Figure 2B), as discussed below.
Figure 2. Comparison of activation loop configurations in IRK family kinases.
Known inactive conformation structures of IRK family kinases (in addition to TrkA and Ror2) were overlaid on that of the insulin receptor (PDB code 1IRK [13]; grey, with the TKD shown in surface representation), and their activation loops coloured. (A) The activation loops of MuSK (PDB code 1LUF [15]; orange), TrkA (cyan), Ror2 molecule A (magenta) and IGF1R (PDB code 1M7N [14]; yellow) align well with that of IRK. (B) The activation loops of Ron (PDB code 3PLS [17]; red), Met (PDB code 2G15 [16]; blue) and ALK (PDB code 3L9P [8]; green) are more widely variable and are compared with that of IRK (grey). Tyrosine side chains in the YXXXYY motif are shown, and the substrate-mimicking tyrosine residue (Tyr1162 in IRK) is marked with a red asterisk.
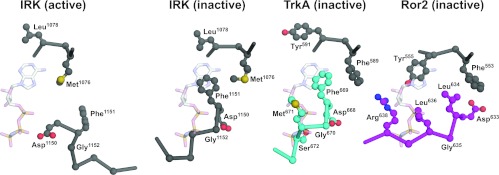
The TrkA and Ror2 TKDs in their inactive conformations also mirror other key aspects of the autoinhibitory mechanisms originally described for IRK [13], although Ror2 has several unique features that suggest a slightly different mode of autoinhibition. The activation loop conformation in both the TrkA and Ror2 TKDs is such that it directly occludes the space in which the phosphate moieties of ATP would bind to an active kinase, as described previously for inactive IRK, MuSK and IGF1R [13–15]. As shown in Figure 3, backbone and/or side-chain atoms in the region following the DFG (Asp-Phe-Gly) motif occlude the phosphate-binding site, as seen in particular for Met671 and Ser672 of TrkA and Leu636 of Ror2. A switch from this so-called ‘DFG-out’ (inactive) conformation seen in TrkA, Ror2 and inactive IRK to a ‘DFG-in’ (active) configuration reverses this inhibition in the active IRK structure, as seen on the left in Figure 3. Like IRK, inactive TrkA also places the side-chain of its DFG motif phenylalanine residue (Phe669) into the binding site for the adenine ring to further block ATP binding. This feature is common to IRK (Phe1151), MuSK and IGF1R. However, Ror2 does not mimic this aspect of the autoinhibitory interactions, and has the phenylalanine residue of its DFG motif replaced with leucine. The unusual DLG (Asp-Leu-Gly) motif in Ror2 adopts an ‘DFG-out’-like conformation (Figures 1 and 3), but it is displaced approximately 4–4.5 Å away from the ATP-binding site compared with that in IRK and TrkA, and therefore does not affect the location adopted by the adenine ring in active kinases. Instead, the side chain of Tyr555, two positions from the gatekeeper residue (Phe553) at the end of strand β5 (and quite distant from the DFG motif), occupies an unusual position for inactive kinases, and occludes the predicted binding site for the ATP adenine ring (Figure 3). The corresponding residue is tyrosine in numerous other TKDs, although it is replaced by phenylalanine or leucine in others. Regardless of the residue identity, however, the side-chain orientation in all other inactive insulin receptor family TKDs, as well as EGFR, FGFRs (fibroblast growth factor receptors) and Src family TKDs is orthogonal to that of Tyr555 in Ror2, suggesting that the ability of this tyrosine residue to occlude the ATP-binding site may be a feature unique to Ror2. It will be interesting to determine whether Tyr555 is a phosphorylation site in Ror2, and whether phosphorylation (or mutation) of this residue enhances kinase activity.
Figure 3. ATP-binding-site occlusion by the activation loop in inactive IRK, TrkA and Ror2.
Structures of the inactive TKDs from IRK [13], TrkA and Ror2 were overlaid on the structure of active IRK (PDB code 1IR3) determined with bound peptide substrate and a non-hydrolysable ATP analogue AMP-PNP (adenosine 5′-[β,γ-imido]triphosphate) [41]. Close-up views of residues surrounding the binding site for AMP-PNP (shown from the 1IR3 structure) are shown for inactive IRK (activation loop coloured grey), TrkA (activation loop coloured cyan) and Ror2 (activation loop coloured magenta). Whereas the 1IR3 (active) structure readily accommodates AMP-PNP, the nucleotide-binding site is occluded directly by the activation loop in inactive IRK, TrkA and Ror2, with backbone and side-chain clashes with the phosphate groups. In addition, the DFG motif phenylalanine residue (Phe1151 in IRK and Phe669 in TrkA) blocks the binding site for the adenine ring of AMP-PNP. This inhibitory interaction is not maintained in Ror2, which has a phenylalanine-to-leucine substitution in the DFG motif (yielding DLG), and Tyr555 from elsewhere in the TKD takes an a similar role.
Comparison with ALK, Met, Ron and IRK autoinhibition
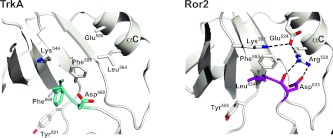
In contrast with the TKDs depicted in Figure 2(A), the activation loops of inactive ALK, Met and Ron do not interfere directly with ATP binding, and adopt a diverse array of conformations (Figure 2B). Tyrosine residues in the YXXXYY motif of ALK and Met do not project into the substrate-binding site in the manner described above for TrkA, Ror2 and IRK. In Ron, Tyr1238 does occupy the substrate tyrosine site, but approaches from quite a different orientation than is seen in IRK and the other TKDs shown in Figure 2. Autoinhibition of ALK, Met and Ron instead relies substantially on direct interactions between the activation loop and the αC helix, both displacing the αC helix from the position it adopts in active kinases and distorting the activation loop. This mode of autoinhibition is not employed by IRK [13] and is also not seen for MuSK or TrkA. Ror2 lies between these two scenarios. In addition to occluding the ATP-binding site like IRK, the Ror2 TKD activation loop also contacts the αC helix directly (Figure 4). It does so through predicted hydrogen bonds between Asp633 from the DLG motif and an αC arginine side chain (Arg528), made possible because of the 4–4.5 Å displaced position of this motif away from the active site in Ror2. It is interesting that the αC helix is displaced further from the position adopted in active kinases in the case of inactive Ror2 than in the TrkA, IRK, MuSK or IGF1R counterparts, and is closer to the position for the αC helix seen in Met and Ron. Thus Ror2 appears to employ a ‘hybrid’ mode of autoinhibition that involves both direct occlusion of the ATP-binding site and activation loop–αC interactions. By contrast, TrkA very closely resembles IRK.
Figure 4. DFG motif conformation and activation loop–αC interactions in TrkA and Ror2.
Left: close-up of the DFG motif region in TrkA TKD, with the DFG motif itself coloured cyan. Note the absence of salt bridge between the β3 invariant lysine residue (Lys544) and αC glutamate residue (Glu560), for which complete side-chain density was not seen. Van der Waal's contacts between the Phe589 and Leu564 side chains are likely to contribute to stabilization of the αC position. Right: close-up of the DFG motif equivalent in Ror2 TKD (coloured magenta), which has the sequence DLG, with Leu634 replacing the normal phenylalanine residue at this position in other kinases. Note that the β3 invariant lysine residue (Lys507) forms a salt bridge with the αC helix glutamate residue (Glu524) even in the inactive Ror2 kinase (as also seen in ALK and Met), although the displaced αC helix position prevents Lys507 from contributing to an active-like ATP-binding site. The DLG motif of Ror2 TKD in the activation loop also interacts directly with the αC helix, thus stabilizing the inactive state through a mode that more closely resembles autoinhibition in Met, ALK or EGFR. Arg528 in the αC helix interacts directly with both backbone and side chain of Asp633 in the DLG motif to stabilize this conformation. Mutations at Arg528 might be predicted to activate Ror2.
The different modes of IRK family kinase autoinhibition are also reflected in their normal (ligand-dependent) activation mechanisms. The TKDs in Figure 2(A) resemble IRK, with the initial activating event likely to be autophosphorylation of the tyrosine side chain projecting into the substrate-binding site (Tyr680 in TrkA, Tyr645 in Ror2, Tyr1162 in IRK, Tyr1135 in IGF1R and Tyr754 in MuSK), thus reversing autoinhibition [13,15]. Phosphorylation of other tyrosine residues in the characteristic activation loop YXXXYY motif in these kinases disrupts additional autoinhibitory interactions [mostly with C-lobe (C-terminal lobe) residues] and/or stabilizes the active conformation. The TKDs depicted in Figure 2(B) vary. Since Ron places the side chain of Tyr1238 from its activation loop in the substrate-binding site to inhibit the kinase, autophosphorylation of this tyrosine residue is likely to play a key role in receptor activation [17]. In both ALK and Met, YXXXYY motif tyrosine residues instead stabilize autoinhibitory activation loop–αC interactions in the inactive state, without directly affecting the substrate-binding site. In Met, the Tyr1234 side chain (equivalent to IRK Tyr1162) contacts αC directly, and its phosphorylation is required for Met activation [16]. Similarly, Tyr1278 in ALK (the first tyrosine residue in the YXXXYY motif) contributes directly to autoinhibitory activation loop–αC interactions, and its phosphorylation has been reported to be the initial event in ALK activation [31].
Possible relevance of TrkA and Ror2 TKD dimers
RTKs are well known to signal as dimers, with extracellular ligand binding inducing either dimerization or the transition from an inactive dimeric form to an active dimer [3]. Accordingly, functional significance has been attached to dimers of both active and inactive TKDs. Since the Ror2 TKD crystallized with two molecules in the asymmetric unit, we were therefore interested to ask whether any Ror2 or TrkA TKD dimer, either non-crystallographic or crystallographic, might be functionally relevant, perhaps as an ‘inactive dimer’ of the sort suggested for a symmetrical EGFR TKD structure [32]. During the course of the present study, other structures of Trk receptor kinases [33–35] and Ror2 were published or released in the PDB. It was therefore important to determine whether these other structures share any crystal packing modes with the TrkA and Ror2 structures described in the present paper.
The Ror2 structure (PDB code 3ZZW) contains two molecules in the asymmetric unit, in common with our structure. In both cases, the two Ror2 TKD molecules form an antiparallel dimer, burying a total surface area of 1725 Å2 (our structure) and 2640 Å2 (3ZZW) on the two TKDs. However, the surfaces involved in each interface are quite different. Within the Ror2 monomer, all of the structural features described in the present paper are maintained in 3ZZW, with the exception that molecule B in 3ZZW has a different DLG motif configuration and position, slightly closer to the ‘DFG-out’ conformation seen in inactive IRK, so that the characteristic Asp633/Arg528 hydrogen bond shown in Figure 4 (between the activation loop and αC helix) cannot form.
Of the two TrkA TKD structures in the PDB, one (code 4AOJ) has an inhibitor bound (AZ-23), which forces the DFG motif into an ‘in’ (rather than ‘out’) conformation, and much of the activation loop is lacking. Otherwise, this structure closely resembles the TrkA structure described in the present paper. There are three molecules in the 4AOJ asymmetric unit, but no crystallographic or non-crystallographic packing mode is shared with our TrkA structure. Recently published inhibitor-bound TrkB [34] and TrkC [33] structures also closely resemble our TrkA structure, although all but one (PDB code 4ASZ [34]) has an inhibitor bound, either displacing the DFG motif or flipping it into an ‘in’ configuration. A TrkA structure with no inhibitor bound [34] was very recently released (PDB code 4F0I). This closely resembles our structure and, interestingly, does appear to share one crystallographic dimer interface as described below.
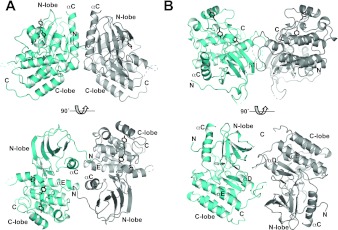
We used the PISA (Protein Interfaces, Surfaces and Assemblies) server [36] to search the PDB for crystal packing modes between TKDs similar to those seen for our Ror2 and TrkA structures. No examples were seen of TKDs that pack like Ror2, but close relatives of one of the TrkA crystallographic dimers were seen both in inhibitor-bound TrkC (PDB code 3V5Q [33]) and eight PDB entries describing structures of the inactive TKD from FGFR1. This crystallographic TrkA dimer is shown in Figure 5(A). A total surface area of 1314 Å2 is buried in the interface of this dimer, in which the two copies of the αC helix are parallel and contact one another directly. Further contacts are made between the C-terminus of the αC helix in one molecule and the C-terminus of the αE helix in its neighbour. In addition, the N-terminus of each molecule projects towards the active site of its neighbour, terminating with uninterpretable electron density within the active site, possibly reflecting a mode of intermolecular autoinhibition in an inactive dimer. The N-terminus shows similar behaviour in the crystallographic TrkC dimer seen in PDB code 3V5Q, which buries a surface area of 1136 Å2. Moreover, Mohammadi et al. [37] highlighted a remarkably similar dimer when they first reported the structure of the inactive FGFR1 TKD (PDB code 1FGK), burying a surface area of 1657 Å2. The fact that this dimer is seen for three different inactive TKDs, including one (FGFR1) that is only distantly related, suggests that it may be functionally relevant for inactive RTK TKDs. A second non-crystallographic TrkA TKD dimer, shown in Figure 5(B), is shared by our TrkA structure and that of Bertrand et al. [34] (PDB code 4F0I), although was not seen for other TKDs in the PDB. This dimer buries ~1500 Å2, primarily contributed by side-chains from the ‘hinge’ region that links β5 in the N-lobe (N-terminal lobe) and αD in the C-lobe, the kinase insert domain (between helices αD and αE), and strand β1 in the N-lobe. These interactions could also contribute directly to TKD autoinhibition in an inactive dimer.
Figure 5. Possible inactive TrkA TKD dimers.
Two orthogonal views of crystallographic dimers of TrkA TKDs. One TrkA TKD is coloured cyan, and the other grey. (A) A dimer that is also found in an inhibitor-bound TrkC TKD crystal structure (PDB code 3V5Q [33]) and in inactive FGFR1 TKD structures. The αC helix, which is responsible for the majority of the interactions is labelled, as is the αE helix, the C-terminus of which also makes contributions to the dimer interface. The N- and C-termini of the TKD are marked. Note the projection of the N-terminus towards the TKD active site, a feature that is also seen in the TrkC crystallographic dimer from 3V5Q. (B) a dimer that is also found in a recently released TrkA TKD structure (PDB code 4F0I [34]), which involves the ‘hinge’ region, kinase insert domain and strand β1. This dimer could help to maintain the inactive configuration through restraints on the hinge region, for example. No evidence for TrkA TKD dimerization in solution was obtained in analytical ultracentrifugation studies (not shown). Tyrosine residues in the YXXXYY motif within the activation loop are coloured black, to highlight the fact that they are quite distant in these dimers from the active site of the neighbouring TKD that may transphosphorylate them. The tyrosine trans-autophosphorylation events that lead to TrkA activation could not occur in the context of these inactive dimers, but would require their reorganization upon extracellular ligand binding or association with additional receptor molecules.
As for many other RTKs, pre-formed inactive dimers of TrkA [38] and TrkB [39] have been reported on the basis of cross-linking and complementation experiments, formation of which appears to require the intracellular domain. It is possible that one or both of the symmetrical dimers of TrkA TKDs shown in Figure 5 plays a role in stabilizing such dimers, along similar lines to a proposed symmetric inactive dimer of EGFR that was observed crystallographically [32]. Since the tyrosine residues of the activation loop YXXXYY motif are distant from the dimer interface in both dimers shown in Figure 5, their trans-autophosphorylation following extracellular nerve growth factor binding would require a substantial rearrangement of such an inactive dimer if it exists in cells.
Activating mutations in IRK family TKDs
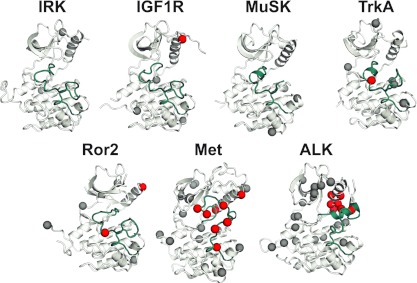
Figure 6 summarizes mutations found in the TKDs of IRK family RTKs in cancer patients, from the COSMIC (Catalogue Of Somatic Mutations In Cancer) database (http://www.sanger.ac.uk/genetics/CGP/cosmic/) [40] and a survey of the literature. Patient mutations are found in all of the TKDs discussed in the present paper, except for Ron (not pictured in Figure 6). Two main points emerge from this inspection. First, ALK and Met, which are IRK family TKDs with autoinhibitory interactions that do not involve direct occlusion of substrate- or ATP-binding sites, have been found mutated most frequently in cancer patients. With the caveat that this is a limited dataset, this observation may suggest that these TKDs are the most ‘vulnerable’ to mutational activation in cancer. Secondly, many of the cancer-associated mutations in ALK and Met are found in the activation loop and αC helix (red in Figure 6), presumably reflecting an activating disruption of activation loop–αC helix interactions. Only two such mutations have been reported in these regions of Ror2, whereas just a single example is seen in TrkA and IGF1R, and no examples have been reported in MuSK. Since stabilization of the autoinhibitory activation loop–αC interactions seen in ALK and Met (and in EGFR) is distributed over a large number of residues, we suggest that there are many positions at which activating oncogenic driver mutations can occur, which may increase their frequency. By contrast, where autoinhibitory interactions involve projection of an activation loop tyrosine residue into the substrate-binding site, they are more ‘focused’ on that particular tyrosine residue, which will reduce the probability of oncogenic mutation. Accordingly, just as activating mutations in the TKD itself are rare in IGFR1R and MuSK, we expect them also to be quite infrequent in TrkA and Ror2 (and close relatives) on the basis of the structures of the present study.
Figure 6. Somatic mutations in IRK family kinases in cancer.
IRK family TKDs in their inactive conformation (PDB codes listed in Figure 2) are shown in the same orientation as in Figure 1, with the activation loop coloured cyan. Sites at which mutations have been reported in cancer patients, in the literature or COSMIC (http://www.sanger.ac.uk/genetics/CGP/cosmic/) database [40], are represented as spheres, coloured red if the mutations lie in the activation loop or αC helix. Note that the region of contact between the activation loop and αC helix is the site of many mutations in Met and ALK. TKDs without this autoinhibitory feature have fewer known mutations to date, and fewer in the activation loop or αC. These analyses identify a vulnerability in ALK and Met for oncogenic mutations as described in the text. Ron is not included because no mutations in this TKD have yet been described. IRK is included for comparison.
Conclusions
In conclusion, we describe crystal structures of the TrkA and Ror2 TKDs in their inactive states. The structures reveal that TrkA closely resembles IRK in its mode of autoinhibition (and presumably activation), relying only on occlusion of the substrate- and ATP-binding sites by the activation loop. On the other hand, Ror2 autoinhibition combines several elements; its activation loop occludes the substrate- and ATP-binding sites as seen in IRK, but with additional unique contributions from a tyrosine side chain close to the gatekeeper residue. Moreover, interactions between the activation loop and αC contribute to Ror2 autoinhibition, suggesting that additional sites of vulnerability for activating mutations in Ror2 may exist that are not present in IRK, IGF1R and TrkA. We also describe a model for inactive TrkA TKD dimers that closely resemble inactive dimers seen for the TKDs of TrkC and FGFR1.
AUTHOR CONTRIBUTION
Stephen Artim and Jeannine Mendrola performed all experimental and crystallographic work (on TrkA and Ror2 respectively). Stephen Artim, Jeannine Mendrola and Mark Lemmon interpreted results. Stephen Artim and Mark Lemmon wrote the paper.
ACKNOWLEDGEMENTS
We thank Kate Ferguson and members of the Lemmon and Ferguson laboratories for advice and a reading of the paper before submission, and Dr Garrett Brodeur for providing TrkA cDNA. Crystallographic data were collected at beamline 23ID-B of GM/CA@APS.
FUNDING
This work was funded in part by the National Institutes of Health [grant number R01-GM099891 (to M.A.L.)] and training grant funds to S.C.A. [grant number T32-GM008275]. Beamline 23ID-B of GM/CA@APS used to collect crystallographic data is funded by the National Cancer Institute [grant number Y1-CO-1020], National Institute of General Medical Sciences [grant number Y1-GM-1104], and the U.S. Department of Energy [contract number DE-AC02-06CH11357].
References
- 1.Dancey J. E., Bedard P. L., Onetto N., Hudson T. J. The genetic basis for cancer treatment decisions. Cell. 2012;148:409–420. doi: 10.1016/j.cell.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Yauch R. L., Settleman J. Recent advances in pathway-targeted cancer drug therapies emerging from cancer genome analysis. Curr. Opin. Genet. Dev. 2012;22:45–49. doi: 10.1016/j.gde.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M. A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S. V., Bell D. W., Settleman J., Haber D. A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Jura N., Zhang X., Endres N. F., Seeliger M. A., Schindler T., Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossi R. T., Saccardo M. B., Ardini E., Menichincheri M., Rusconi L., Magnaghi P., Orsini P., Avanzi N., Borgia A. L., Nesi M., et al. Crystal structures of anaplastic lymphoma kinase in complex with ATP competitive inhibitors. Biochemistry. 2010;49:6813–6825. doi: 10.1021/bi1005514. [DOI] [PubMed] [Google Scholar]
- 8.Lee C. C., Jia Y., Li N., Sun X., Ng K., Ambing E., Gao M. Y., Hua S., Chen C., Kim S., et al. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem. J. 2010;430:425–437. doi: 10.1042/BJ20100609. [DOI] [PubMed] [Google Scholar]
- 9.Bresler S. C., Wood A. C., Haglund E. A., Courtright J., Belcastro L. T., Plegaria J. S., Cole K., Toporovskaya Y., Zhao H., Carpenter E. L., et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci. Transl. Med. 2011;3:108ra114. doi: 10.1126/scitranslmed.3002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 11.Morris S. W., Kirstein M. N., Valentine M. B., Dittmer K. G., Shapiro D. N., Saltman D. L., Look A. T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 12.Carpenter E. L., Mossé Y. P. Targeting ALK in neuroblastoma-preclinical and clinical advancements. Nat. Rev. Clin. Oncol. 2012;9:391–399. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard S. R., Wei L., Ellis L., Hendrickson W. A. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 14.Munshi S., Kornienko M., Hall D. L., Reid J. C., Waxman L., Stirdivant S. M., Darke P. L., Kuo L. C. Crystal structure of the apo, unactivated insulin-like growth factor-1 receptor kinase: implication for inhibitor specificity. J. Biol. Chem. 2002;277:38797–38802. doi: 10.1074/jbc.M205580200. [DOI] [PubMed] [Google Scholar]
- 15.Till J. H., Becerra M., Watty A., Lu Y., Ma Y., Neubert T. A., Burden S. J., Hubbard S. R. Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation. Structure. 2002;10:1187–1196. doi: 10.1016/s0969-2126(02)00814-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Marimuthu A., Tsai J., Kumar A., Krupka H. I., Zhang C., Powell B., Suzuki Y., Nguyen H., Tabrizizad M., et al. Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3563–3568. doi: 10.1073/pnas.0600048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Steinbacher S., Augustin M., Schreiner P., Epstein D., Mulvihill M. J., Crew A. P. The crystal structure of a constitutively active mutant RON kinase suggests an intramolecular autophosphorylation hypothesis. Biochemistry. 2010;49:7972–7974. doi: 10.1021/bi100409w. [DOI] [PubMed] [Google Scholar]
- 18.Brodeur G. M., Minturn J. E., Ho R., Simpson A. M., Iyer R., Varela C. R., Light J. E., Kolla V., Evans A. E. Trk receptor expression and inhibition in neuroblastomas. Clin. Cancer Res. 2009;15:3244–3250. doi: 10.1158/1078-0432.CCR-08-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebagay G., Yan S., Liu C., Cheung N. K. ROR1 and ROR2 in human malignancies: potentials for targeted therapy. Front. Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerman P. S., Sos M. L., Ramos A. H., Xu C., Dutt A., Zhou W., Brace L. E., Woods B. A., Lin W., Zhang J., et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang E. J., Reichardt L. F. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 22.Green J. L., Kuntz S. G., Sternberg P. W. Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 2008;18:536–544. doi: 10.1016/j.tcb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winn M. D., Isupov M. N., Murshudov G. N. Use of TLS anisotropic displacements in macromolecular refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 30.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 31.Tartari C. J., Gunby R. H., Coluccia A. M., Sottocornola R., Cimbro B., Scapozza L., Donella-Deana A., Pinna L. A., Gambacorti-Passerini C. Characterization of some molecular mechanisms governing autoactivation of the catalytic domain of the anaplastic lymphoma kinase. J. Biol. Chem. 2008;283:3743–3750. doi: 10.1074/jbc.M706067200. [DOI] [PubMed] [Google Scholar]
- 32.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albaugh P., Fan Y., Mi Y., Sun F., Adrian F., Li N., Jia Y., Sarkisova Y., Kreusch A., Hood T., et al. Discovery of GNF-5837, a selective TRK inhibitor with efficacy in rodent cancer tumor models. ACS Med. Chem. Lett. 2012;3:140–145. doi: 10.1021/ml200261d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertrand T., Kothe M., Liu J., Dupuy A., Rak A., Berne P. F., Davis S., Gladysheva T., Valtre C., Crenne J. Y., Mathieu M. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J. Mol. Biol. 2012;423:439–453. doi: 10.1016/j.jmb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Wang T., Lamb M. L., Block M. H., Davies A. M., Han Y., Hoffmann E., Ioannidis S., Josey J. A., Liu Z.-Y., Lyne P. D., et al. Discovery of disubstituted imidazo[4,5-b]pyridines and purines as potent TrkA inhibitors. ACS Med. Chem. Lett. 2012;3:705–709. doi: 10.1021/ml300074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Mohammadi M., Schlessinger J., Hubbard S. R. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 38.Shen J., Maruyama I. N. Nerve growth factor receptor TrkA exists as a preformed, yet inactive, dimer in living cells. FEBS Lett. 2011;585:295–299. doi: 10.1016/j.febslet.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 39.Shen J., Maruyama I. N. Brain-derived neurotrophic factor receptor TrkB exists as a preformed dimer in living cells. J. Mol. Signaling. 2012;7:2. doi: 10.1186/1750-2187-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forbes S. A., Tang G., Bindal N., Bamford S., Dawson E., Cole C., Kok C. Y., Jia M., Ewing R., Menzies A., et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbard S. R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]