Abstract
Histone H3-lysine79 (H3K79) methyl transferase DOT1L plays critical roles in normal cell differentiation as well as initiation of acute leukemia. We used structure and mechanism based design to discover several potent inhibitors of DOT1L with IC50 values as low as 38 nM. These inhibitors exhibit only weak or no activities against four other representative histone lysine and arginine methyltransferases, G9a, SUV39H1, PRMT1 and CARM1. The x-ray crystal structure of a DOT1L:inhibitor complex reveals that N6-methyl group of the inhibitor, located favorably in a predominantly hydrophobic cavity of DOT1L, provides the observed high selectivity. Structural analysis shows that it will disrupt at least one H-bond and/or have steric repulsion for other histone methyltransferases. These compounds represent novel chemical probes for biological function studies of DOT1L in health and disease.
Human genome is packed into chromatins, which are composed of millions of repetitive units known as nucleosomes. A single nucleosome includes a fragment of DNA (~147 bp) wound around a disc-like histone octamer consisting of two histone H2A, H2B, H3 and H4 proteins. Post-translational epigenetic modifications on several lysine and arginine residues of histones, such as methylation and acetylation, control the accessibility of the DNA, thereby regulating the expressing or silencing of a gene.1 It has been widely recognized that, in addition to gene mutations, aberrant epigenetic modifications play an important role in the initiation of many diseases, such as cancer.2–4 Great interest has therefore been generated to study histone modifying enzymes, such as histone methyltransferases, as well as their functions in pathogenesis. Histone methyltransferases include a large family of dozens of histone lysine methyltransferases (HKMT) and histone/protein arginine methyltransferases (PRMT),5,6 many of which have recently been found to play critical roles in cell differentiation, gene regulation, DNA recombination and damage repair.7 Therefore, small molecule inhibitors of histone methyltransferases represent useful chemical probes for these biological studies as well as potential therapeutics.8 However, very few inhibitors of histone methyltransferases (HKMT and PRMT) have been discovered and developed.8,9
We are particularly interested in human histone lysine methyltransferase DOT1L,10,11 which is highly conserved from yeasts to mammals. DOT1L is a unique HKMT in that, unlike all other HKMTs containing a SET domain (which are class V methyltransferases), it belongs to the class I methyltransferase family. In addition, DOT1L is the only known enzyme that specifically catalyzes methylation of the histone H3-lysine79 (H3K79) residue located in the nucleosome core structure, while other methylation sites are in the unordered N-terminal tail of histone. Moreover, clinical importance of DOT1L as well as the H3K79 methylation is that DOT1L has been found to be necessary and sufficient for the initiation and maintenance of leukemia with MLL (mixed lineage leukemia) gene translocations.12–14 This type of leukemia accounts for ~75% infant and ~10% adult acute leukemia with a particularly poor prognosis.15 DOT1L therefore represents a novel target for intervention. It is of interest that during the process of revising this manuscript for publication, a DOT1L inhibitor was disclosed, which possesses selective activity against MLL leukemia.16
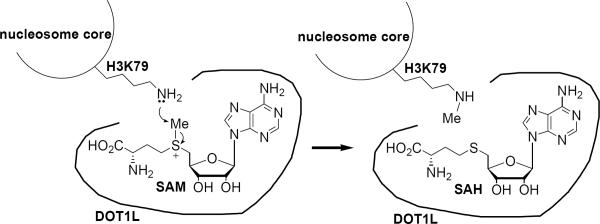
DOT1L catalyzes an SN2 reaction of the H3K79 ε-NH2 of the substrate nucleosome with the methyl group of S-(5'-adenosyl)-L-methionine (SAM), which is the cofactor of the enzyme, as schematically illustrated in Figure 1. One of the reaction products, S-(5'-adenosyl)-L-homocysteine (SAH) has been known to be a non-selective inhibitor of many methyltransferases, including DOT1L.17 We also found it inhibits recombinant human DOT1L (catalytic domain 1–472)10 with a Ki value of 160 nM (Table 1). However, SAH cannot be used as a probe in cell biology or in vivo, since it is quickly degraded to become adenosine and homocysteine by SAH hydrolase,18 keeping cellular SAM/SAH molar ratio of ~40:1.19 In addition, selectivity is of importance for a DOT1L inhibitor to be a useful probe, since other histone lysine and arginine methyltransferases also use SAM and histone/nucleosome as their cofactor and substrate.5,6
Figure 1.
Mechanism of catalysis of DOT1L.
Table 1.
| DOT1L | CARM1 | PRMT1 | G9a | SUV39H1 | |
|---|---|---|---|---|---|
| SAHa | 0.16 | 0.40 | 0.86 | 0.57 | 4.9 |
| 1 a | 0.29 | >100 | 22.7 | >100 | >100 |
| 2 a | 1.1 | 18 | 21.2 | >100 | >100 |
| 3 b | 15.7 | 46.4 | 22.0 | >100 | >100 |
| 4 b | 0.038 | 1.1 | 2.7 | 1.8 | >100 |
| 5 b | 0.12 | >100 | >100 | >100 | >100 |
| 6 b | 0.11 | >100 | >100 | >100 | >100 |
Ki values for competitive inhibitors SAH, 1 and 2;
IC50 values for inhibitors 3 – 6.
We analyzed the crystal structure of the DOT1L:SAM complex11 as well as those of all other histone methyltransferases available in Protein Data Bank and found one structural feature that is unique to the binding of SAM to DOT1L, which can be exploited to design selective DOT1L inhibitors. As shown in Supporting Information Figure S1a, the 6-NH2 group of SAM forms only one H-bond with DOT1L with a large hydrophobic cavity nearby. However, the 6-NH2 group of bound SAM or SAH has two H-bonds with PRMTs (which also belong to class I methyltransferases), such as CARM1 (also known as PRMT4) as shown in Figure S1b. All other HKMTs, such as G9a, are SET-domain methyltransferases having a completely different structure. The binding conformation of SAM/SAH to these enzymes is distinct from that of DOT1L, with the 6-NH2 group facing towards the protein and forming two H-bonds (Figure S1c). We thus hypothesized that N6-substituted SAH analogs, such as 1 and 2 (Chart 1), are potent and selective DOT1L inhibitors. This turned out to be the case. Compounds 1 and 2 were synthesized from N6-substituted adenosine (Supporting Information Experimental Section). Compound 1, having only one extra -CH3 group compared to SAH, was found to be still a potent DOT1L inhibitor with a Ki value of 290 nM (Table 1 and Figure S2), but it possesses only weak or no inhibitory activities against two PRMTs (CARM1 and PRMT1) and two HKMTs (G9a and SUV39H1) with Ki values of 22.7 – >100 μM (Table 1). In contrast, SAH remains an inhibitor of all these enzymes with Ki values of 0.4 – 4.9 μM. Similarly, compound 2, N6-benzyl-SAH, has good activity on DOT1L (Ki: 1.1 μM), but is very weak against CARM1 and PRMT1 (Ki = 18 and 21.2 μM) and inactive on G9a and SUV39H1 (Table 1).
Chart 1.
Structures of DOT1L inhibitors.
Next, x-ray crystallography was used to investigate how compound 1 binds to DOT1L, with a particular interest in the binding site of the N6-methyl group that provides excellent selectivity. We determined the crystal structure of the DOT1L:1 complex at 2.5 Å. Details of data processing and refinement are shown in Table S1 and the overall structure and protein-ligand interactions of the DOT1L:1 complex illustrated in Figure S3. As shown in Figure 2a, the protein as well as the SAH moiety of the inhibitor superimpose with those of the previously reported DOT1L:SAM structure11 with a rms (root mean square) deviation of 0.2 Å. As a result, all of the 10 H-bonds as well as other interactions between the ligand and the protein remain essentially intact (Figure 2b), which is in agreement with the potent inhibitory activity of 1. The N6-methyl group is nicely inserted into a hydrophobic cavity, surrounded by Phe223, Leu224, Val249, Lys187 and Pro133 (Figures 2b,c). In addition, its orientation allows the 6-NH group to form a H-bond with Asp222 that is important to the binding of the adenine ring.
Figure 2.
X-ray crystal structure of the human DOT1L:1 complex. (A) Superposition of the structures of DOT1L:1 (with C atoms in green) and DOT1L:SAM (in purple) with a rms deviation of 0.2 Å. For clarity, only protein backbones are shown; (B) Close-up view of the active site of DOT1L:1 structure, with 10 H-bonds shown in dotted lines; (C) Electrostatic potential surface (with 25% transparency) of the DOT1L:1 complex, showing the N6-methyl group of 1 is located in a hydrophobic cavity. 1 is shown as a space-filling model.
It is therefore clear that introducing a N6-substituent does not significantly affect the binding of SAH to DOT1L. However, our experiments show the N6-substituted SAH analogs 1 and 2 cannot bind to other HKMTs and PRMTs strongly (Table 1), suggesting any substitution on this position will disrupt at least one H-bond and/or change the binding conformation of the adenine ring, thereby causing a considerable affinity loss. In addition, for SET-domain HKMTs, any N6-substituent will lead to intolerable steric repulsion with the protein, preventing these compounds from binding. These results show N6-substituted SAH analogs are selective inhibitors of DOT1L and provide a structural basis for further inhibitor design and development.
A mechanism based inhibitor design was exploited to find selective DOT1L inhibitors with improved potency. Compound 3 (Chart 1) was initially synthesized. The rationale is that it can undergo intramolecular cyclization at neutral pH to form a reactive aziridinium intermediate,20, 21 which may be covalently bound to the ε-NH2 group of H3K79 (Figure S4). Compound 3 was found to exhibit only weak enzyme inhibition against DOT1L with an IC50 value of 15.7 μM. We reasoned that compound 4 with one more -CH2- could be a better inhibitor, since the two C–N bonds (~1.47 Å each) in 3 are considerably shorter than the C–S bonds (~1.82 Å) in SAM/SAH. The crystal structures of DOT1L show that SAM as well as 1 bind to the protein in a fully extended conformation, suggesting the amino acid moiety of 3 might not be able to reach its optimal binding site in DOT1L. Compound 4 has not been made before and our synthetic route is shown in Scheme 1. The 2',3'-dihydroxyls of adenosine were selectively protected with an acetonide and the 5'-hydroxyl was converted to a -NH2, via a Mitsunobu reaction followed by treatment with hydrazine. The product was alkylated with ethyl bromoacetate and reduced with LiAlH4 to afford compound 7. tert-Butyl ester of L-glutamic acid was first protected with one tert-butoxycarbonyl (BOC) group and its δ-carboxyl converted to a methyl ester. It is necessary to protect the amino group with a second BOC before reduction to give aldehyde 8. Compounds 7 and 8 subjected to a reductive amination to produce compound 9, whose free hydroxyl group was converted to an iodide with PPh3/I2, affording, after acidic deprotection, compound 4.
Scheme 1.
General synthesis for compounds 4 – 6.a
aReagents and conditions: (i) acetone, SOCl2; (ii) phthalimide, PPh3, diisopropyl azodicarboxylate; (iii) NH2NH2, 80 °C; (iv) ethyl bromoacetate, NEt3; (v) LiAlH4; (vi) BOC2O; (vii) ClCOOMe, DMAP, NEt3; (viii) BOC2O, DMAP; (ix) DIBAL, −78 °C; (x) NaCNBH3, HCl, MeOH; (xi) PPh3, I2, imidazole, 0 °C; (xii) HCl-dioxane.
Compound 4 was found to be an extremely potent inhibitor of DOT1L with an IC50 value of 38 nM (Table 1), almost quantitatively inactivating DOT1L. Interestingly, it possesses relatively weak or no inhibitory activity on other methyltransferases with IC50 values of 1.1 – >100 μM, respectively, showing a high selectivity (>29-fold). It is remarkable that, due to complicated enzyme kinetics of histone methyltransferases involving covalent binding of inhibitor 4 (or 3) to the substrate, we measured IC50 values for each enzyme using a minimal enzyme concentration (50 – 100 nM), Km of SAM, as well as saturated concentration of the substrate. Under these assay conditions, the IC50 values may be used to compare the relative inhibitory ability of each compound across these enzymes. Although 4 does not have an N6-substituent, the locally more hydrophobic environment at the binding site of the putative aziridinium intermediate of 4 in DOT1L might account for the selectivity, since it could protect the highly reactive aziridinium cation from non-specific hydrolysis. The corresponding sites in other histone methyltransferases are either exposed to the solvent (for SET domain HKMTs) or polar (for PRMTs). We synthesized compounds 5 and 6, which are N6-substituted analogs of 4, using the general approach in Scheme 1. These two compounds also exhibit potent activity against DOT1L with IC50 values of 120 and 110 nM, respectively (Table 1). As expected, their N6-methyl and benzyl group provide excellent selectivity: 5 and 6 are essentially inactive against other methyltransferases, showing these compounds could have wide applications in probing the biological functions of DOT1L.
In summary, this work is of interest for a number of reasons. First, DOT1L, a specific histone H3K79 methyltransferase, plays a critical role in normal cell differentiation as well as the initiation and maintenance of acute leukemia with MLL gene translocations. DOT1L inhibitors therefore represent novel chemical probes for its functional studies as well as potential therapeutics for leukemia. Second, we used structure and mechanism based design to synthesize and identify several potent DOT1L inhibitors with IC50 values as low as 38 nM. These compounds exhibit only weak or no inhibitory activities on four other representative histone lysine and arginine methyltransferases. Third, we determined the crystal structure of the DOT1L:1 complex, revealing the structural basis for the excellent selectivity. The methyl group of the inhibitor is located favorably in a hydrophobic cavity of DOT1L, while it will disrupt at least one H-bond and/or have steric repulsions for all other histone methyltransferases. This finding should provide implications for future DOT1L inhibitor design and development.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant (RP110050) from Cancer Prevention and Research Institute of Texas (CPRIT) to Y.S. and a grant (Q1279) from Robert Welch Foundation to B.V.V.P. The recombinant human DOT1L (1–472) expression plasmid was kindly provided by Dr. Yi Zhang (U. of North Carolina). We thank the staff of the X-ray Crystallography Facility at Baylor College of Medicine for assistance in data collection.
ABBREVIATIONS
- HKMT
histone lysine methyltransferases
- PRMT
histone/protein arginine methyltransferases
- H3K79
histone H3 lysine 79
- MLL
mixed lineage leukemia
- SAM
S-(5'-adenosyl)-L-methionine
- SAH
S-(5'-adenosyl)-L-homocysteine
- BOC
tert-butoxycarbonyl
Footnotes
Supporting Information Available. Supplementary figures 1 – 4, Supplementary table, Experimental Section and the full author list of the Reference 16. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Kouzarides T. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- (2).Jones PA, Baylin SB. Cell. 2007;128:683. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wilson CB, Rowell E, Sekimata M. Nat. Rev. Immunol. 2009;9:91. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- (4).Tsankova N, Renthal W, Kumar A, Nestler EJ. Nat. Rev. Neurosci. 2007;8:355. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- (5).Cheng X, Collins RE, Zhang X. Biophys. Biomol. Struct. 2005;34:267. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Schubert HL, Blumenthal RM, Cheng X. Trends Biochem. Sci. 2003;28:329. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bhaumik SR, Smith E, Shilatifard A. Nat. Struct. Mol. Biol. 2007;14:1008. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- (8).Copeland RA, Solomon ME, Richon VM. Nat. Rev. Drug Discov. 2009;8:724. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- (9).Cole PA. Nat. Chem. Biol. 2008;4:590. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Curr. Biol. 2002;12:1052. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- (11).Min J, Feng Q, Li Z, Zhang Y, Xu RM. Cell. 2003;112:711. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- (12).Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. Cell. 2005;121:167. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- (13).Krivtsov AV, Armstrong SA. Nat. Rev. Cancer. 2007;7:823. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- (14).Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, Kutok JL, Kung AL, Armstrong SA. Cancer Cell. 2008;14:355. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, McGlennen R, Smith FO, Woods WG, Salzer WL, Johnstone HS, Dreyer Z, Reaman GH. Blood. 2006;108:441. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Daigle SR, et al. Cancer Cell. 2011;20:53. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA. Chem. Biol. Drug Des. 2011;78:199. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- (18).Borchardt RT, Keller BT, Patel-Thombre U. J. Biol. Chem. 1984;259:4353. [PubMed] [Google Scholar]
- (19).Chiba P, Wallner C, Kaiser E. Biochem. Biophys. Acta. 1988;971:38. doi: 10.1016/0167-4889(88)90159-0. [DOI] [PubMed] [Google Scholar]
- (20).Osborne T, Weller Roska RL, Rajski SR, Thompson PR. J. Am. Chem. Soc. 2008;130:4574. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]
- (21).Weller RL, Rajski SR. ChemBioChem. 2006;7:243. doi: 10.1002/cbic.200500362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.