Introduction
The maintenance of immune homeostasis requires a balance between stimulatory and inhibitory pathways. The herpesvirus entry mediator (HVEM; TNFRSF14) [1] serves as one of two key entry routes used by herpes simplex virus-1 and herpes simplex virus-2 to infect cells [2]. The selection of HVEM as a route of infection is intimately linked with the capacity of HSV to modulate immunity. As a cellular signaling receptor, HVEM functions as a molecular switch for pathways that can stimulate or inhibit hematopoietic cell activation. The HVEM-regulated pathways impact T and B cell activation [3, 4], dendritic cell proliferation [5], and protection of mucosal epithelia from damage during inflammation [6]. HVEM has five known functional ligands: two canonical TNF superfamily ligands (LIGHT: TNFSF14 and LTα: lymphotoxin-a) [7] and three unconventional ligands that belong to the Ig superfamily [B and T lymphocyte attenuator (BTLA), CD160, and the viral envelop protein of herpes simplex virus envelope glycoprotein D (HSV gD)] (Fig. 36.1a) [8]. The shared receptor usage of HVEM’s ligands, LIGHT and LTα, by the LTβ receptor and the two receptors for TNF suggests these molecules are part of a larger signaling network whose ramifications have not been fully elucidated [9]. Recent insights into the biophysics of the ligand–receptor interactions in the HVEM pathway suggest unanticipated functional consequences of this cosignaling network.
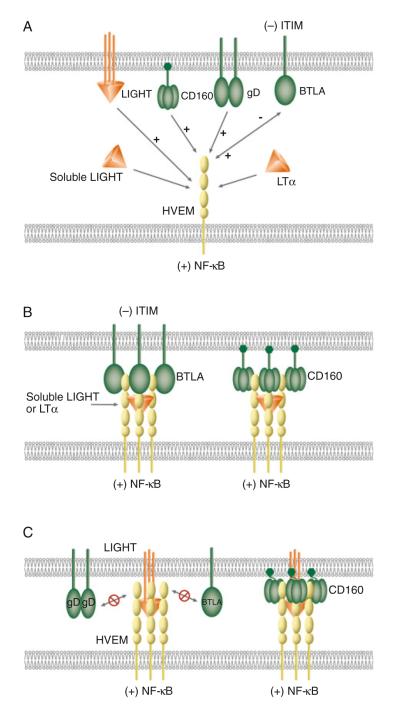
Fig. 36.1.
Schematic illustrations of molecular interactions between HVEM and its ligands. (a) Canonical and unconventional ligands of HVEM. LIGHT and LTα are the canonical ligands as well as positive activators of HVEM. Ligation of LIGHT or LTα activates HVEM-dependent NF-κB signaling. BTLA, CD160, and HSV gD serve as unconventional ligands for HVEM. Ligation of CD160 or gD to HVEM induces NF-κB activation. Bidirectional signaling occurs between HVEM and BTLA. HVEM-mediated BTLA signaling induces tyrosine phosphorylation of the ITIM in BTLA, providing an inhibitory signal to T cells. BTLA, CD160, and gD bind HVEM at the CRD1, but LIGHT and LTα interact with HVEM in the CRD2 and 3. The binding of gD to HVEM blocks HVEM binding to LIGHT or BTLA (not shown in diagram). (b) Cooperative binding between soluble LIGHT, LTα, HVEM, BTLA, and CD160. Left complex shows formation of a LIGHT–HVEM–BTLA trimolecular complex with three molecules of soluble LIGHT, HVEM, and BTLA. Right complex shows the assembling of LIGHT–HVEM–CD160 trimolecular complex with three molecules of soluble LIGHT and HVEM, and nine molecules of CD160. (c) Model of membrane LIGHT, HVEM, BTLA, and CD160 interaction. Membrane LIGHT competes with BTLA for the binding with HVEM. The binding of membrane LIGHT to HVEM prevents BTLA or gD from binding to HVEM due to membrane restriction (left complex). The flexibility of the GPI-link in CD160 may accommodate the membrane restraint for the formation of membrane LIGHT–HVEM–CD160 trimolecular complex (right complex)
Canonical Ligands: LIGHT and LTα
Lymphotoxin-α and LIGHT are members of the tumor necrosis factor superfamily (TNFSF) having a common structural motif that forms TNFR binding site. LIGHT was initially identified as cellular ligand for HVEM through the characterization of a distinct 30 kDa HVEM-binding protein on the surface of an activated human CD4+ T cell hybridoma (II-23) [7]. LTα is one of the original tumor necrosis factors [10]. LTα contains a classic signal cleavage site and is secreted as a homotrimer, while LIGHT is a type 2 transmembrane glycoprotein. The extracellular domain of LIGHT may be cleaved from the surface and released in a functional soluble form [11].
The LIGHT gene is located on human Chr 19p13 and a genetic paralog of LTβ, FasL, and TL1A [12]. LIGHT shares significant amino acid sequence homology with the C-terminal receptor-binding domains of LTβ (34% identity), and it shares binding to the LTβR, which engages the heterotrimer, LTα1β2. LIGHT, like all TNFSF members, forms a trimeric complex [13, 14] that allows multivalent binding with cell surface receptors. Receptor clustering is the key initiating step in the activation of TNF receptor signaling [15].
Both LIGHT and LTα bind to a similar region of HVEM. The binding site of LIGHT and LTα on HVEM were mapped on cysteine-rich domain-2 (CRD2) and CRD3 using HVEM mutants. Although the binding sites of LIGHT and LTα on HVEM are distinct, it is likely that their binding sites are topographically overlapping as the molecules are cross competitive [7]. It has been shown that HVEM has a stronger binding avidity to LIGHT than LTα [7], and LIGHT-induced HVEM signaling results in the recruitment of the TNF receptor-associated factor 2 (TRAF2) to the cytoplasmic tail of HVEM. The activation of a TRAF-dependent NF-κB pathway provides positive costimulation and prosurvival signal to T cells [13]. Although there have been many studies on the binding, structure and function of LTα in particular, in relation to TNFR1 and TNFR2, the distinct role of LTα on the HVEM signaling network remains unclear. However, LTα enhanced binding interactions between HVEM and BTLA [16, 17] presumably through oligomerization of HVEM. Additional studies are needed to further define the impact of LTα in the LIGHT–HVEM–BTLA/CD160 signaling system.
The expression of LIGHT is regulated at the transcriptional level [7, 14]. LIGHT is inducible but transiently expressed on the surface of activated T lymphocytes [7, 14]. Although both LIGHT and LTα are expressed in activated T cells, the transcriptional regulation of their genes appears to be mediated via different signaling pathways [7]. LIGHT expression was detected in MCF10A breast epithelial line [18] and melanoma cells [19]. Thus, LIGHT appears to have a broader range of expression compared to LTα, which is limited to activated T cells, B cells, NK cells, and LTi cells.
Unconventional Ligands: BTLA and CD160
BTLA and CD160 were originally identified as receptors for HVEM [20, 21] involved in activating inhibitory signaling. However, recent studies demonstrated that both BTLA and CD160 serve as activating ligands for HVEM [8]. BTLA or CD160 binding to HVEM induced HVEM-dependent NF-κB activation, demonstrating bidirectional signaling between HVEM and BTLA (Fig. 36.1a) in cells interacting in trans [8]. BTLA appears to form dimers as a membrane protein, providing a basis for oligomerizing HVEM that leads to TRAF2 recruitment and activation of NF-κB RelA. These results highlight the complexity of LIGHT– HVEM–BTLA/CD160 cosignaling networks.
In contrast to the TNFSF ligands, BTLA is a type 1 transmembrane protein with a single intermediate (I) type Ig domain. Three conserved tyrosine-based signaling motifs, two ITIM, and a Grb-2 recognition consensus are present in the cytoplasmic domain of BTLA in both mouse and human [22]. The utilization of the ITIM and recruitment of the protein tyrosine phosphatases, Src homology domain (SHP)-1 and SHP-2, into the cytoplasmic tail of BTLA appear to be HVEM dependent [23, 24]. However, the distinct role of the ITIM and the contributions of SHP-1 and SHP-2 in BTLA intracellular signaling have not been precisely defined. BTLA is expressed in a broad range of hematopoietic cells, including mature lymphocytes, splenic macrophages, dendritic cells, as well as T and B cells in the developing thymus [25, 26], and thus the effects of BTLA signaling may modulate many aspects of innate and adaptive immunity.
CD160 was originally identified as a binding partner of MHC class I molecules with weak binding affinity [27]. Recent studies by Cai et al. [21] demonstrated that CD160 exhibited specific binding to HVEM and that signal transduction mediated through HVEM and CD160 was shown to be inhibitory to T cells. CD160 is also a member of the Ig family with a single Ig V-like domain and a predominant glycosylphosphatidylinositol (GPI) motif, which enables anchorage to the cell surface. It has been shown that the GPI anchored CD160 on activated T cells was cleaved by a metalloprotease [28]. Furthermore, additional isoforms of CD160 have also been reported. Giustiniani et al. [29] have recently indentified three isoforms of CD160 (CD160ΔIg-GPI, CD160-TM, and CD160ΔIg-TM) that were generated by alternative splicing. Both CD160ΔIg-GPI and CD160ΔIg-TM have a deletion of the Ig domain, which is likely to abolish their binding capability to HVEM. Additionally, CD160-TM and CD160ΔIg-TM also contain transmembrane and intracellular domains. Although the distinct role of these new isoforms in LIGHT–HVEM–BTLA/CD160 cosignaling system remains to be determined, it is anticipated that the newly found isoforms, in particularly the CD160-TM, would allow intracellular signaling. Unlike BTLA, CD160 assemble as a trimer with interchain disulfide bridges [30]. The trimeric form of CD160 along with its high binding avidity to HVEM enables it to function as a highly effective ligand for HVEM. CD160-expressing EL4 cells specially activated HVEM-dependent NF-κB reporter in transfected 293T cells and induced nuclear translocation of RelA in HT29 colon cell line, which naturally express HVEM [8]. The expression of CD160 is restricted to the hematopoietic compartment. CD160 is expressed in NK and NKT cells, γδT cells, intestinal intraepithelial T cells, CD8+CD28− T cells, and a subset of CD4 T cells, but not in B cells [30]. CD160 mRNA was detected in spleen, peripheral blood, and lymphocytes in the small intestine [30].
Distinct Ligand Binding Sites on HVEM
HVEM is a typical TNFRSF member with four cysteine-rich domains (CRD). The 11 disulfide bonds in the ectodomain create an elongated structure. The presence of at least two distinct ligand-binding sites in topographically separate regions of HVEM allows this receptor to simultaneously interact with multiple ligands. LIGHT and BTLA bind on opposite sides of HVEM. BTLA and CD160 engage residues in the N-terminal CRD1 of HVEM, whereas the contacts of LIGHT and LTα are located in CRD2 and CRD3. The binding of soluble LIGHT to HVEM does not inhibit either BTLA or CD160 binding to HVEM [8, 16, 31], substantiating the conclusion that LIGHT and BTLA have distinct binding sites. Interestingly, soluble LIGHT and BTLA bind cooperatively to HVEM. Soluble LIGHT or LTα enhanced the binding of BTLA to HVEM [16, 17]. This result is consistent with a view that LIGHT clusters HVEM, which in turn, increases the avidity for BTLA (Fig. 36.1b). This interpretation suggests that soluble LIGHT, HVEM, and BTLA form a trimolecular complex.
We suggested a model in which the LIGHT–HVEM–BTLA trimolecular complex is likely to contain a trimer of soluble LIGHT with three molecules of HVEM and BTLA. However, the trimolecular complex of LIGHT–HVEM–CD160 would contain a trimer of LIGHT and three HVEM, and three trimers of CD160 (Fig. 36.1b). The formation of these higher ordered complexes not only enhances binding between HVEM, LIGHT, LTα, BTLA, and CD160, but also enhances HVEM clustering and, importantly, HVEM signaling.
Viral Ligands of HVEM and BTLA: HSV gD and UL144
HSV-1 gD is a type 1 transmembrane glycoprotein. The N-terminal ectodomain contains three N-glycosylation sites and six cysteine residues for the formation of three disulfide bridges. Sequence structure analysis revealed an IgV-like domain at the N- and C-terminal extensions [32, 33]. Although there is no significant sequence homology between gD and other cellular ligands of HVEM, gD shows direct binding with HVEM. The binding site of HSV gD was mapped in the CRD1 of HVEM [34]. The gD site is topographically close to the BTLA site on HVEM, but their binding sites are not identical [16]. The unique location of the gD site on HVEM enables gD to perform two distinctive functions. Firstly, the binding of gD-Fc on HVEM prevents HVEM from binding to both LIGHT and BTLA. The ectodomain of gD serves as a multi-function inhibitor, which not only blocks binding of HVEM to LIGHT, but also to BTLA. Secondly, gD forms a stable dimer with a disulfide link between two subunits [35, 36]. The dimeric nature of gD allows it to oligomerize HVEM, serving as a functional ligand. The gD–HVEM complex is likely to contain two molecules of gD and two HVEM. Direct evidence for supporting this hypothesis came from the observation that the binding of gD-Fc to HVEM activated NF-κB [8].
UL144 is a herpes virus ortholog of HVEM [37] that binds BTLA [16], but not to the canonical TNF ligands, LIGHT and LTα. UL144 is encoded within the ULb’ region of human cytomegalovirus, a β-herpesvirus. The protein contains two CRD homologous to CRD1 and 2 of HVEM explaining the lack of binding of the TNF-related ligands. The protein is highly variable in the ectodomain [38] but contains a conserved short intracellular domain. The role of UL144 in the pathogenesis of CMV is unknown, but the engagement of BTLA suggests a possible role in immune evasion.
The Trans and Cis of HVEM Signaling
In general, HVEM is viewed as a positive immune regulator since it activates NF-κB transcriptional programs that are involved in cell survival and proliferative responses. In addition, ligation of BTLA or CD160 to HVEM also activated NF-κB in a TRAF2-dependent pathway, providing a prosurvival signal for T cells [8]. In this setting, the ligands and receptors function in trans between interacting cells. However, our recent studies indicate that HVEM and BTLA can interact in cis, laterally within the same membrane [8]. Flow cytometric analysis demonstrated that cis-interaction between HVEM and BTLA is the predominant complex expressed on the surface of naïve human and mouse T cells, and the formation of HVEM–BTLA cis-complex inhibited HVEM-dependent NF-κB activation. The heterodimeric complex of HVEM and BTLA uses the same site in CRD1 as for trans-interaction, with only the ectodomain of BTLA required to form the cis-complex. The HVEM–BTLA cis-complex competitively inhibits trans-signaling by all its cellular ligands, providing a mechanism for maintaining T cells in a resting state. As the binding sites of BTLA and CD160 on the CRD1 of HVEM are topographically close to each other, the receptor binding domains of BTLA and CD160 also act as a competitive inhibitors blocking BTLA or CD160 from interacting with HVEM in trans and inducing NF-κB activation.
Interestingly, herpes simplex virus envelope glycoprotein gD also forms a cis-complex with HVEM. Cis-association between gD and HVEM blocks trans-interaction of LIGHT, BTLA, or gD to HVEM in the cis-complex. This is consistent with the observation that gD expressing cells were resistant to HSV infection, indicating that gD might interfere with its endogenous receptors [39]. We propose that the formation of cis-complexes between HVEM and BTLA, CD160 or gD competitively (against BTLA, CD160 and gD) or non-competitively (against LIGHT and LTα) inhibits HVEM activation by ligands expressed in the surrounding microenvironment, allowing T cell to remain in the naïve state.
Although LIGHT interacts with HVEM in a topographically distinct site, which differs from the BTLA binding site, membrane LIGHT inhibits HVEM–BTLA trans-interaction [16]. These results suggested that the proximity of the membrane may sterically exclude HVEM from binding BTLA when membrane LIGHT occupies its binding site in the CRD2 and 3 regions. Promoted by high affinity binding, the LIGHT–HVEM trans-complex, may in turn, sterically compete with membrane BTLA from binding HVEM, thus acting in a noncompetitive fashion to disrupt inhibitory signaling by BTLA (Fig. 36.1c, left complex). We propose that the proximity of TNF homology domain of LIGHT to the membrane surface could prevent BTLA from engaging HVEM when HVEM engages LIGHT in trans. Similarly, the close proximity of the base of the LIGHT trimer to the membrane surface would also prevent access of gD to CRD1 of HVEM (Fig. 36.1c, left complex). However, the inherent flexibility of the GPI linkage in CD160 might allow CD160 to accommodate the steric requirement for the formation of the membrane LIGHT–HVEM–CD160 trimolecular complex (Fig. 36.1c, right complex).
The LIGHT–HVEM–BTLA/CD160 cosignaling system has the potential to simultaneously deliver stimulatory and inhibitory signals between interacting cells. Clear evidence indicates that HVEM–BTLA play a counter-regulatory role with the LTβR system in controlling dendritic cell proliferation within lymphoid tissues [5]. Cis-association between HVEM and its unconventional ligands, as well as the formation of the higher ordered trimolecular complexes with soluble LIGHT and LTα add additional levels of regulatory complexity in the LIGHT–HVEM– BTLA/CD160 system. The outcome of the signal transduction process mediated via HVEM not only depends on the timing of expression of individual signaling molecules, but also the distinctive combinations of molecules within the cis- and trans-complexes.
Acknowledgments
Support for this work was provided by grants from the National Institutes of Health (R37AI33068, AI067890, AI048073, and CA069381).
Contributor Information
Timothy C. Cheung, Department of Neurosciences, University of California - San Diego, La Jolla, CA 92093, USA; Mycobacterial Research Division, Centenary Institute, Missenden Road, Camperdown, NSW 2050, Australia.
Carl F. Ware, Division of Molecular Immunology, La Jolla Institute for Allergy and Immunology, 9420 Athena Circle, La Jolla 92037, CA, USA; Laboratory of Molecular Immunology, Sanford Burnham Medical Research Institute, 10901 N. Torrey Pines Road, La Jolla, CA 92037, USA.
References
- 1.Montgomery RI, Warner MS, Lum B, Spear PG. Herpes simplex virus 1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JM, Lin E, Susmarski N, Yoon M, Zago A, Ware CF, Pfeffer K, Miyoshi J, Takai Y, Spear PG. Alternative entry receptors for herpes simplex virus and their roles in disease. Cell Host Microbe. 2007;2:19–28. doi: 10.1016/j.chom.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaye J. CD160 and BTLA: LIGHTs out for CD4+ T cells. Nat Immunol. 2008;9:122–124. doi: 10.1038/ni0208-122. [DOI] [PubMed] [Google Scholar]
- 4.Murphy KM, Nelson AC, Sedy JR. Balancing co-stimulation and inhibition with BTLA and HVENat M. Rev Immunol. 2006;6:671–681. doi: 10.1038/nri1917. [DOI] [PubMed] [Google Scholar]
- 5.De Trez C, Schneider K, Potter K, Droin N, Fulton J, Norris PS, Ha SW, Fu YX, Murphy T, Murphy KM, Pfeffer K, Benedict CA, Ware CF. The inhibitory HVEM-BTLA pathway counter regulates lymphotoxin receptor signaling to achieve homeostasis of dendritic cells. J Immunol. 2008;180:238–248. doi: 10.4049/jimmunol.180.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinberg MW, Turovskaya O, Shaikh RB, Kim G, McCole DF, Pfeffer K, Murphy KM, Ware CF, Kronenberg M. A crucial role for HVEM and BTLA in preventing intestinal inflammation. J Exp Med. 2008;205:1463–1476. doi: 10.1084/jem.20071160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauri DN, Ebner R, Montgomery RI, Kochel KD, Cheung TC, Yu GL, Ruben S, Murphy M, Eisenberg RJ, Cohen GH, Spear PG, Ware CF. LIGHT, a new member of the TNF superfamily, and lymphotoxin alpha are ligands for herpesvirus entry mediator. Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 8.Cheung TC, Steinberg MW, Oborne LM, Macauley MG, Fukuyama S, Sanjo H, D’Souza C, Norris PS, Pfeffer K, Murphy KM, Kronenberg M, Spear PG, Ware CF. Unconventional ligand activation of herpesvirus entry mediator signals cell survival. Proc Natl Acad Sci U S A. 2009;106:6244–6249. doi: 10.1073/pnas.0902115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ware CF. Targeting lymphocyte activation through the lymphotoxin and LIGHT pathways. Immunol Rev. 2008;223:186–201. doi: 10.1111/j.1600-065X.2008.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pennica D, Nedwin GE, Hayflick JS, Seeburg PH, Derynck R, Palladino MA, Kohr WJ, Aggarwal BB, Goeddel DV. Human tumour necrosis factor: precursor structure, expression and homology to lymphotoxin. Nature. 1984;312:724–729. doi: 10.1038/312724a0. [DOI] [PubMed] [Google Scholar]
- 11.Granger SW, Butrovich KD, Houshmand P, Edwards WR, Ware CF. Genomic Characterization of LIGHT reveals linkage to an immune response locus on chromosome 19p13.3 and distinct isoforms generated by alternate splicing or proteolysis. J Immunol. 2001;167:5122–5128. doi: 10.4049/jimmunol.167.9.5122. [DOI] [PubMed] [Google Scholar]
- 12.Granger SW, Ware CF. Commentary: turning on LIGHT. J Clin Invest. 2001;108:1741–1742. doi: 10.1172/JCI14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooney IA, Butrovich KD, Glass AA, Borboroglu S, Benedict CA, Whitbeck JC, Cohen GH, Eisenberg RJ, Ware CF. The lymphotoxin-beta receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. Biol J (Chem) 2000;275:14307–14315. doi: 10.1074/jbc.275.19.14307. [DOI] [PubMed] [Google Scholar]
- 14.Harrop JA, McDonnell PC, Brigham-Burke M, Lyn SD, Minton J, Tan KB, Dede K, Spampanato J, Silverman C, Hensley P, DiPrinzio R, Emery JG, Deen K, Eichman C, Chabot-Fletcher M, Truneh A, Young PR. Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. J Biol Chem. 1998;273:27548–27556. doi: 10.1074/jbc.273.42.27548. [DOI] [PubMed] [Google Scholar]
- 15.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 16.Cheung TC, Humphreys IR, Potter KG, Norris PS, Shumway HM, Tran BR, Patterson G, Jean-Jacques R, Yoon M, Spear PG, Murphy KM, Lurain NS, Benedict CA, Ware CF. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A. 2005;102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez LC, Loyet KM, Calemine-Fenaux J, Chauhan V, Wranik B, Ouyang W, Eaton DL. A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc Natl Acad Sci U S A. 2005;102:1116–1121. doi: 10.1073/pnas.0409071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai Y, Guo R, Hsu LT, Yu LG, Ni J, Kwon BS, Jiang G-w, Lu J, Tan J, Ugustus M, Carter K, Rojas L, Zhu F, Lincoln C, Endress G, Xing L, Wang S, Oh OK, Gentz R, Ruben S, Lippman ME, Hsieh LS, Yang D. LIGHT, a novel ligand for lymphotoxin b receptor and TR2/HVEM induces apoptosis and suppresses in vivo tumor formation via gene transfer. J Clin Invest. 1998;102:1142–1151. doi: 10.1172/JCI3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortarini R, Scarito A, Nonaka D, Zanon M, Bersani I, Montaldi E, Pennacchioli E, Patuzzo R, Santinami M, Anichini A. Constitutive expression and costimulatory function of LIGHT/TNFSF14 on human melanoma cells and melanoma-derived microvesicles. Cancer Res. 2005;65:3428–3436. doi: 10.1158/0008-5472.CAN-04-3239. [DOI] [PubMed] [Google Scholar]
- 20.Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, Hildner K, Scheu S, Pfeffer K, Ware CF, Murphy TL, Murphy KM. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 21.Cai G, Anumanthan A, Brown JA, Greenfield EA, Zhu B, Freeman GJ. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol. 2008;9:176–185. doi: 10.1038/ni1554. [DOI] [PubMed] [Google Scholar]
- 22.Gavrieli M, Watanabe N, Loftin SK, Murphy TL, Murphy KM. Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of B and T lymphocyte attenuator required for association with protein tyrosine phosphatases SHP-1 and SHP-2. Biochem Biophys Res Commun. 2003;312:1236–1243. doi: 10.1016/j.bbrc.2003.11.070. [DOI] [PubMed] [Google Scholar]
- 23.Chemnitz JM, Lanfranco AR, Braunstein I, Riley JL. B and T lymphocyte attenuator-mediated signal transduction provides a potent inhibitory signal to primary human CD4 T cells that can be initiated by multiple phosphotyrosine motifs. J Immunol. 2006;176:6603–6614. doi: 10.4049/jimmunol.176.11.6603. [DOI] [PubMed] [Google Scholar]
- 24.Gavrieli M, Murphy KM. Association of Grb-2 and PI3K p85 with phosphotyrosile peptides derived from BTLBiochem A. Biophys Res Commun. 2006;345:1440–1445. doi: 10.1016/j.bbrc.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 25.Han P, Goularte OD, Rufner K, Wilkinson B, Kaye J. An inhibitory Ig superfamily protein expressed by lymphocytes and APCs is also an early marker of thymocyte positive selection. J Immunol. 2004;172:5931–5939. doi: 10.4049/jimmunol.172.10.5931. [DOI] [PubMed] [Google Scholar]
- 26.Otsuki N, Kamimura Y, Hashiguchi M, Azuma M. Expression and function of the B and T lymphocyte attenuator (BTLA/CD272) on human T cells. Biochem Biophys Res Commun. 2006;344:1121–1127. doi: 10.1016/j.bbrc.2006.03.242. [DOI] [PubMed] [Google Scholar]
- 27.Le Bouteiller P, Barakonyi A, Giustiniani J, Lenfant F, Marie-Cardine A, Aguerre-Girr M, Rabot M, Hilgert I, Mami-Chouaib F, Tabiasco J, Boumsell L, Bensussan A. Engagement of CD160 receptor by HLA-C is a triggering mechanism used by circulating natural killer (NK) cells to mediate cytotoxicity. Proc Natl Acad Sci U S A. 2002;99:16963–16968. doi: 10.1073/pnas.012681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giustiniani J, Marie-Cardine A, Bensussan A. A soluble form of the MHC class I-specific CD160 receptor is released from human activated NK lymphocytes and inhibits cell-mediated cytotoxicity. J Immunol. 2007;178:1293–1300. doi: 10.4049/jimmunol.178.3.1293. [DOI] [PubMed] [Google Scholar]
- 29.Giustiniani J, Bensussan A, Marie-Cardine A. Identification and characterization of a transmembrane isoform of CD160 (CD160-TM), a unique activating receptor selectively expressed upon human NK cell activation. J Immunol. 2009;182:63–71. doi: 10.4049/jimmunol.182.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anumanthan A, Bensussan A, Boumsell L, Christ AD, Blumberg RS, Voss SD, Patel AT, Robertson MJ, Nadler LM, Freeman GJ. Cloning of BY55, a novel Ig superfamily member expressed on NK cells, CTL, and intestinal intraepithelial lymphocytes. J Immunol. 1998;161:2780–2790. [PubMed] [Google Scholar]
- 31.Compaan DM, Gonzalez LC, Tom I, Loyet KM, Eaton D, Hymowitz SG. Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J Biol Chem. 2005;280:39553–39561. doi: 10.1074/jbc.M507629200. [DOI] [PubMed] [Google Scholar]
- 32.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Molecular Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 33.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Eisenberg RJ, Cohen GH. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM) J Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarrias MR, Whitbeck JC, Rooney I, Ware CF, Eisenberg RJ, Cohen GH, Lambris JD. The three HveA receptor ligands, gD, LT-alpha and LIGHT bind to distinct sites on HveMol A. Immunology. 2000;37:665–673. doi: 10.1016/s0161-5890(00)00089-4. [DOI] [PubMed] [Google Scholar]
- 35.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handler CG, Cohen GH, Eisenberg RJ. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J Virol. 1996;70:6076–6082. doi: 10.1128/jvi.70.9.6076-6082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benedict C, Butrovich K, Lurain N, Corbeil J, Rooney I, Schenider P, Tschopp J, Ware C. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. Immunol J. 1999;162:6967–6970. [PubMed] [Google Scholar]
- 38.Lurain NS, Kapell KS, Huang DD, Short AJ, Paintsil J, Winkfield E, Benedict WC, CA, Bremer WJ. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J Virol. 1999;73:10040–10050. doi: 10.1128/jvi.73.12.10040-10050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campadelli Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]