Abstract
The study of intracellular bacteria and nanometer-size membrane vesicles within infected host cells poses an important challenge as it is difficult to identify each distinct population in the context of the complex populations generated from active host-pathogen interactions. Here, suspension cultures of L929 cells infected with the prevalent obligate intracellular bacterium Chlamydia trachomatis strain F/Cal-IC-13 are utilized for the large scale preparation and isolation of natural membrane vesicles and bacterial forms. Cell lysis with nitrogen cavitation in combination with differential centrifugation, OptiPrep™ density gradient separation, and immunoenrichment using anti-chlamydial lipopolysaccharide antibodies and MagnaBind beads allows for the isolation of both productive and persistent bacterial forms, as well as membrane vesicles derived from the host and pathogen. We have evaluated these populations by electron microscopy and Western blot analysis for identification of biomarkers. In addition, purified persistent forms of C. trachomatis induced by ampicillin display adenosine-5'-triphosphate (ATP) transport activity, suggesting that ampicillin-induced persistent C. trachomatis organisms, at least in part, rely upon host ATP as an energy source. Importantly, several chlamydial cytotoxic and/or secreted proteins are demonstrated to be associated with these vesicles, supporting the idea that membrane vesicles are generated by Chlamydia as a means of carrying and delivering virulence factors necessary for pathogenesis. The ability to produce large-scale infections and generate distinct bacteria and host-derived populations for biochemical analysis, while reducing the burdens of time and cost have implications in all areas of chlamydiology. These protocols can be applied to other strains of C. trachomatis or other intracellular bacteria.
Keywords: Chlamydia trachomatis, membrane vesicles, intracellular bacterial infection
1.0 Introduction
We present here an optimized method for the isolation of intracellular chlamydial forms, as well as the isolation of vesicles from an intracellular infection with Chlamydia trachomatis. Vesicle isolation is coupled to the enrichment of a specific population of membrane vesicles that can easily be applied to a subset of either host or bacterial vesicles. C. trachomatis, an obligate intracellular bacterium, accounts for the majority of bacterial sexually transmitted diseases worldwide and represents a significant public health burden (Brunham and Rey-Ladino, 2005). C. trachomatis growth takes place exclusively within a specialized membrane-bound parasitophorous vacuole, termed an inclusion and is typified by a unique biphasic lifestyle consisting of non-replicative, but infectious elementary bodies (EBs) and replicative, noninfectious reticulate bodies (RBs) (Moulder, 1991; Hatch, 1999). The inclusion is comprised of both bacterial and host components that are delivered by host exocytic vesicles, although the exact mechanisms employed by chlamydiae to highjack host vesicles are not fully understood (Hackstadt et al., 1996; Van Ooij et al., 1997; Van Ooij et al., 2000; Fields and Hackstadt, 2002). Chlamydia relies on host vesicles not only to deliver components of the inclusion, but also nutrients that are required for normal infection. It has been shown that chlamydiae coordinate the trafficking of specific subsets of the host vesicle population to and from the inclusion. For example, protein complexes from the Golgi and endoplasmic reticulum (ER) are involved in inclusion membrane biogenesis (Hackstadt et al., 1995; Van Ooij et al., 2000; Elwell et al., 2011; Subtil, 2011; Pokrovskaya et al., 2012). Additionally, it has been observed by electron microscopy that there are abundant vesicles both at and within the chlamydial inclusion during infection; however, the precise derivation of these vesicles remains unknown (Giles et al., 2006; Wang et al., 2011). Evidence has been found that a sub-population of these vesicles contain chlamydial antigens such as lipopolysaccharide (LPS) and major outer membrane protein (MOMP). Furthermore, trafficking of these chlamydial antigens to the ER of infected epithelial cells has also been observed (Giles and Wyrick, 2008). Taken together, one can conclude that the total vesicle population within a cell changes during infection and that a portion of the vesicles observed in infected cells are produced by C. trachomatis.
Bacterial membrane vesicles play a role in the pathogenesis of a number of free-living bacteria via delivery of virulence factors to the environment or host cells. Bacterial membrane vesicles likely serve as part of a critical survival system as their production is generally increased upon exposure to chemical stressors, biofilm formation, bacteriophage infection and intracellular replication (Kulp and Kuehn, 2010; Deatherage and Cookson, 2012). Additionally, bacterial vesicles have proven to be an invaluable tool in the production of safe vaccines against a variety of bacteria (Sanchez et al., 2001; Muralinath et al., 2011; Nieves et al., 2011; Su and Snape, 2011). Characterizing bacterial vesicles produced during intracellular infections will further advance our understanding of host-pathogen interaction. However, the study of intracellularly produced bacterial vesicles has been hampered, not only because these small membrane vesicles are difficult to identify directly, but also because the complex populations derived from both host and pathogen within a single cell are not easily separated. Therefore, there is an urgent need to better define sub-populations of vesicles such that detailed biochemical analyses and functional characterization can be applied.
In C. trachomatis it has been observed that stressors such as ampicillin or interferon gamma (IFN-γ) not only increase vesicle production, but can also induce an aberrant bacterial form known as the persistent form in vitro (Giles et al., 2006; Wyrick, 2010; Wang et al., 2011). This persistent form is characterized by an enlarged, less-dense RB appearance by electron microscopy that undergoes DNA replication, but not binary fission (Beatty et al., 1994; Hogan et al., 2004; Lambden et al., 2006; Wyrick, 2010). Based on transcriptional analysis, it has been suggested that the metabolic and transport profiles of the persistent form more closely resemble that of the RB form rather than the EB form (Gerard et al., 2002). It has also been shown that persistent forms possess an altered secretion profile (Wang et al., 2011). Similar aberrant forms are observed in vivo suggesting that the formation of these altered C. trachomatis forms may be a component of infection ((Hogan et al., 2004) and our unpublished observations). Surprisingly, the exact role and nature of the persistent form is largely unknown, partially due to the difficulty of harvesting a relatively pure population.
As part of our efforts to characterize the roles of diverse chlamydial forms and infectionassociated vesicles in regard to the pathogenesis of C. trachomatis, we optimized and developed purification procedures for each of these distinct populations. These protocols are based on the large-scale culture of suspension cells infected with the clinically prevalent C. trachomatis serovar F. Purified persistent forms were assayed for their ability to transport radiolabeled adenosine-5'-triphosphate (ATP), an unusual and important transport component of this obligate intracellular parasite. We also identify specific bacterial- or host-derived membrane vesicles by transmission electron microscopy (TEM) and Western blot analysis to facilitate the identification of biomarkers related to productive or persistent infection. Importantly, several chlamydial cytotoxic and/or secreted proteins are demonstrated to be associated with the vesicles, supporting the idea that membrane vesicles are generated by C. trachomatis a as a means of carrying and delivering virulence factors necessary for pathogenesis. This optimized procedure provides a maximum amount of product for the biochemical analysis of chlamydial forms and vesicles throughout the infection cycle.
2.0 Material and methods
2.1 Cell suspension culture
Mouse fibroblast L929 cells (L cell, CCL-1, ATCC) that are widely used for cultivation of Chlamydia, such as the invasive strains C. trachomatis lymphogranuloma venereum (LGV) L2 (Schachter and Wyrick, 1994), C. psittaci (Matsumoto and Manire, 1970; Tamura et al., 1971) and C. muridarum (Ramsey et al., 2009), were used in this study. To obtain single cell suspensions, freshly trypsinized L929 cells harvested from 162-cm2 flasks (Costa) were diluted to 1 ×105 cells/ml in a glass spinner flask (Corning) with culture medium composed of RPMI-1640 medium (Sigma) supplemented with 10% (v/v) deactivated fetal bovine serum (Sigma), 20 mM glutamine and 10 µg/ml gentamicin (Gibco). Suspension cells were cultured at 37 °C in a humid 5% CO2 atmosphere with gentle rotation by a magnetic stirrer (Thermolyne Model 45600) at a frequency of 60 rpm. One liter suspension cultures were begun with a minimal volume of 200 ml in a 1 liter flask. Morphology and cell concentration were monitored at 2 to 3-day intervals and fresh culture medium was supplemented as needed to maintain a cell density between 1 ×105 and 1×106 cells/ml. High cell densities will result in cell clumping.
2.2 C. trachomatis infection of L929 cell suspensions and exposure to ampicillin.
For infection of suspension cultures, monolayers of L929 cells in 6-well plates were inoculated with C. trachomatis serovar F/Cal-I-13 purified EBs stored at −80 °C (Carlson et al., 2005) with a dose resulting in 80–100% infection. This mixture was centrifuged at 1,825 xg for 40 minutes (min) at 37 °C and the supernatant was replaced with infection medium (RPMI-1640 culture medium supplemented with 10% (v/v) deactivated fetal bovine serum, 10 µg/ml gentamicin, 0.4% glucose, 1X nonessential amino acids (Gibco, 100X solution)) containing 1 µg/ml cycloheximide. At 40 hours post-infection (h pi) the infected cells were harvested by scraping each well into 0.5 ml sucrose phosphate glutamic acid buffer (0.2 M sucrose, 3.8 mM KH2PO4, 6.7 mM Na2HPO4, 5 mM L-glutamic acid, pH 7.4) (SPG) and gently sonicated (Braun-Sonic U, Braun, Germany) at an intensity of 40W at 0.9 cycle for 5 pulses on ice to release infectious C. trachomatis particles. The cell debris was pelleted by centrifugation at 233 xg for 5 min at 4 °C. The C. trachomatis-containing supernatant was placed on ice and used immediately. L929 suspension cells at a cell density of ~1×106 cells/ml were collected by centrifugation at 3,830 xg for 5 min and resuspended in a total volume of 10 ml SPG per 1 L suspension culture. Concentrated L929 cells were infected with C. trachomatis product in 10 ml total volume per liter suspension culture and incubated at 37 °C, rocking for 90 min to allow bacterial adsorption and entry. One 6-well plate of C. trachomatis product was used to infect 1 L of L929 cells. After the infection incubation, the host-bacteria mixture was resuspended to half the original volume in infection medium and returned to 37 °C in 5% CO2 for 36 h pi. In some experiments, infected L929 cells were exposed to 10 µg/ml ampicillin (Sigma) at 16 h pi (Wang et al., 2011). Generally, 4 L of infected suspension culture were used per purification.
2.3 Instant cytotoxicity assay and endpoint infection assays
The instant cytotoxicity assay was carried out by co-incubation of suspension L929 cells and purified EBs stored at −80 °C (MOI 1:10) at 37 °C for 1 hr. The mixture was then placed into a 96-well microtitre plate (Corning) and incubated at 37 °C to allow cell adherence to the wells. The L929 cells were examined for adherence at multiple time points (1, 2 and 24 hr). Morphology such as rounding and granulation observed by phase microscopy was also used as an indication of cytotoxicity (Moulder et al., 1976; Kellogg et al., 1977). Endpoint infection assays determining the inclusion forming units (IFU) were performed by infecting 96-well plates of L929 cells by serial dilution of each sample using the adherent cell infection protocol detailed in section 2.2.
2.4 Purification of vesicles and C. trachomatis particles from infected L929 cells
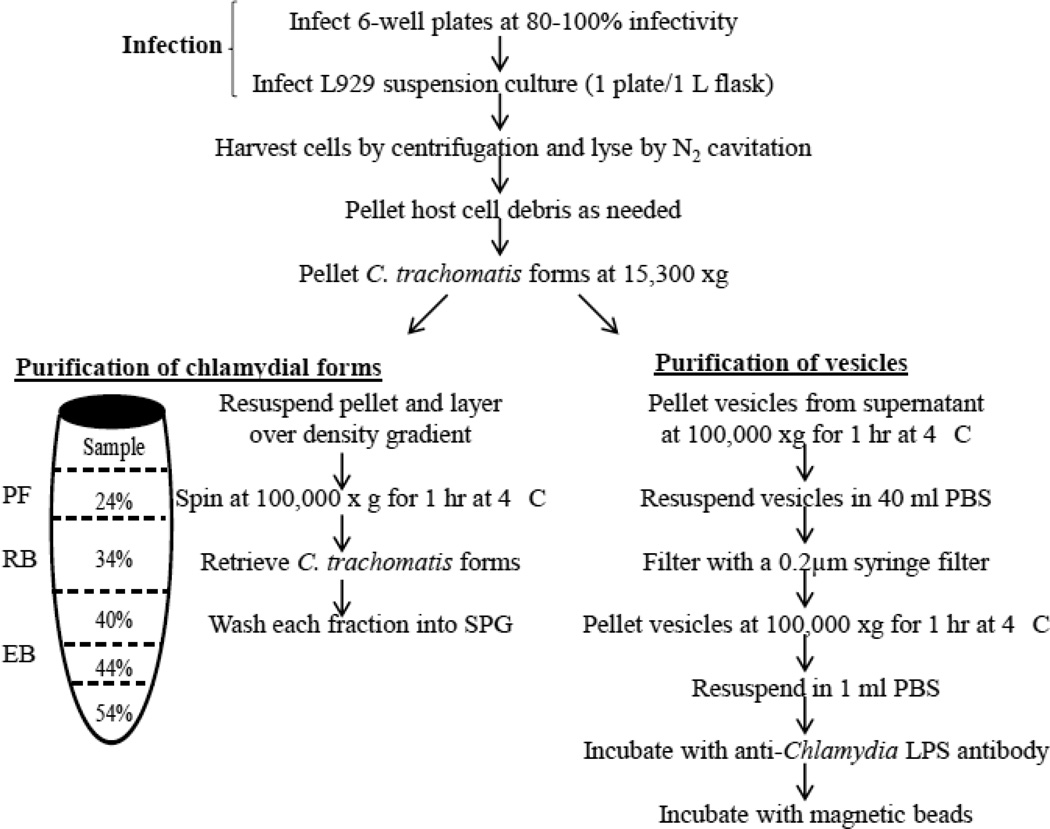
The procedure developed to isolate vesicles and C. trachomatis forms is depicted in Figure 1. Cells were lysed by nitrogen cavitation. Nitrogen cavitation is a technique that is based on rapid nitrogen decompression of a cell suspension from a pressure vessel (Gottlieb and Adachi, 2000). This lysis technique is advantageous because of the gentle lysis conditions and the ability to maintain 0 °C temperatures throughout the lysis procedure. A volume of 4 L of infected L929 cells were pelleted at 3,830 xg for 10 min at 4 °C, resuspended in 40 ml ice-cold SPG and placed into a 50 ml pre-cooled pressure vessel (Parr cell disruption bomb, model # 4639). For 2.5×108 infected L929 cells, we used 125 psi for 5 minute intervals and repeated as needed until greater than 90% host cell lysis was achieved, as determined by trypan blue staining. In order to preserve bacterial integrity, samples were pelleted at 3,830 xg for 10 min at 4 °C prior to each nitrogen re-exposure. The total lysate was cleared of host debris by centrifugation at 3,830 xg for 10 min at 4 °C three times, followed by centrifugation at 5,510 xg for 10 min at 4 °C. The chlamydiae-containing supernatant was centrifuged at 15,300 xg for 1 hr at 4 °C to separate chlamydial particles (pellet) and vesicles (supernatant). Subsequently, the bacterial pellet was resuspended in 5 ml SPG, overlaid and centrifuged through a discontinuous gradient consisting of 6 ml 24%, 7 ml 35%, 8 ml 40%, 5 ml 44% and 3 ml 54% OptiPrep™ density gradient medium (Sigma) diluted with SPG. Samples were centrifuged in a Beckman SW 28 rotor at 100,000 ×g for 1 h at 4 °C. Bands of turbidity at the interfaces of 24–34%, 34–40% and 40–44% OptiPrep™ solution were collected, washed into SPG and sedimented by centrifugation at 100,000 ×g for 1 h at 4 °C. Samples were collected for transmission electron microscopy (TEM) and the resulting pellet was suspended in 1 ml SPG for IFU and immunoblot analysis. Vesicles were pelleted from the supernatant in a Beckman SW 28 rotor at 100,000 ×g for 1 h at 4 °C. The resulting vesicle pellet was resuspended in 10 ml PBS and half of the sample was filtered through a 0.2 µm low protein binding syringe filter (Pall Life Sciences). The two vesicle fractions were pelleted as described previously and gently washed with 2 ml PBS. After samples were taken for electron microscopy, the remaining pellets were resuspended in 0.25 ml PBS per 1 L suspension culture and aliquots were stored at 4 °C or −80 °C for future analysis.
Figure 1.
Schematic of procedure for suspension cell infection with C. trachomatis serovar F/Cal-I-13, purification of chlamydial forms and vesicle isolation. Multiple concentrations of OptiPrep™ density gradient medium used in this study are as indicated. Abbreviations: EB, elementary body; RB, reticulate body; PF, persistent form; SPG, sucrose-phosphate-glutamate buffer; PBS, phosphate buffered saline.
2.5 Immunoenrichment and immunoblot analysis
For vesicle immunoenrichment, 500 µl vesicle fractions were combined with 10 ml chlamydial LPS monoclonal antibody supernatant obtained from the culture of hybridoma clone C1A6 (Herrera et al., 2003) and incubated with rotation overnight at 4 °C. The resultant antibody-vesicle mixture was combined with 2.5×108 MagnaBind Protein A Beads (Thermo Scientific) and incubated at 4 °C overnight, rotating. The mixture was washed six times with 1ml PBS to remove unbound material. Protein-antibody-bead complexes were combined with Laemmli sample buffer (BioRad) (1:1, v:v) supplemented with 5% 2-mercaptoethanol and 10 mM dithiothreitol, followed by incubation at 100 °C for 10 min. Samples were then separated by SDS-PAGE on a 4–20% mini-Protean TGX gel (Bio-Rad), transferred to an immobilon-PVDF membrane (Millipore) and subjected to immunoblot analysis as described (Hua et al., 2009). To probe proteins of interest, we used: (i) primary monoclonal antibodies specific to chlamydial MOMP (Wang et al., 2006), small cysteine-rich outer membrane protein OmcA (B12K) (Zhang et al., 1987), chlamydial protease-like activity factor (CPAF) (100a) (Zhong et al., 2001) and Pgp3 (2H4) (Li et al., 2008); (ii) mouse polyclonal antibodies to CT159, CT166 (a kind gift of Dr. Zhong, University of Texas Health Sciences Center at San Antonio, TX) and bacterial RpoB (8RB13) (Neoclone) (Hua et al., 2009); and (iii) antibodies to host components, including TGN38 (Sigma), LAMP-2 (abcam), calnexin (abcam), cadherin (abcam) and EEA-1 (abcam). Immunoblots were developed using a horseradish peroxidase-conjugated secondary antibody and the Super Signal Chemiluminescent Detection kit (Pierce).
2.6 ATP transport assay
The transport assay involving uptake of radio-labeled ATP by the purified C. trachomatis persistent form was measured using the filtration and wash method previously described (Winkler, 1976). Transport assays were initiated by the addition of mock infected samples or purified chlamydiae to assay buffer (SPG containing 50 µM [33P]ATP) and incubating at 37 °C. Samples (100 µl) were removed at regular intervals over a time course, filtered through a Millipore 0.45 µm HAPW02500 nitrocellulose filter and washed with 8 ml SPG. As an additional control, transport assays were also conducted in the presence of excess unlabeled ATP at 20-times the determined Km for C. trachomatis nucleoside phosphate transporter 1 (the primary ATP transporter). Filters were analyzed by liquid scintillation spectrometry (Beckman Coulter LS6500). Individual transport assays were calculated based on total protein of the isolated sample. Transport assay data were normalized to the maximum transport for each purification (100% transport) and expressed as a percentage.
2.7 Electron microscopy
For ultrastructural analysis, infected cells, purified vesicles or OptiPrep™ purified C. trachomatis forms were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde (Polysciences Inc., Warrington, PA) in 200 mM phosphate buffer and processed as described previously (Beatty, 2006). Ultrathin sections were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc., Bannockburn, IL). Samples were viewed on a JEOL 1200 EX II transmission electron microscope (JEOL USA Inc., Peabody, MA).
3.0 Results and discussion
3.1 Large-scale infection of the prevalent C. trachomatis urogenital strain serovar F/Cal-IC/13 in L929 suspension culture
Significant challenges in the study of the obligate intracellular bacterium C. trachomatis include the difficulty of propagation and the ability to obtain sufficient quantities of bacteria to characterize (Hatch, 1996). We employed a two-step infection process that allows for an efficient and effective C. trachomatis genital strain F/Cal-I-13 infection of L929 cells in suspension (Fig. 1). Essentially, several liters of suspension culture were infected with C. trachomatis. After removal of culture medium and gentle lysis, infected cells were separated into three populations: a pellet of host debris and organelles, a pellet of C. trachomatis particles, and a suspension of soluble protein and vesicles. The latter two sub-populations were then isolated by employing differential centrifugation either with or without a density gradient as indicated in Figure 1. A byproduct of this method is a highly concentrated host protein preparation obtained at the time of harvest that can be utilized for analysis of the host proteome during infection.
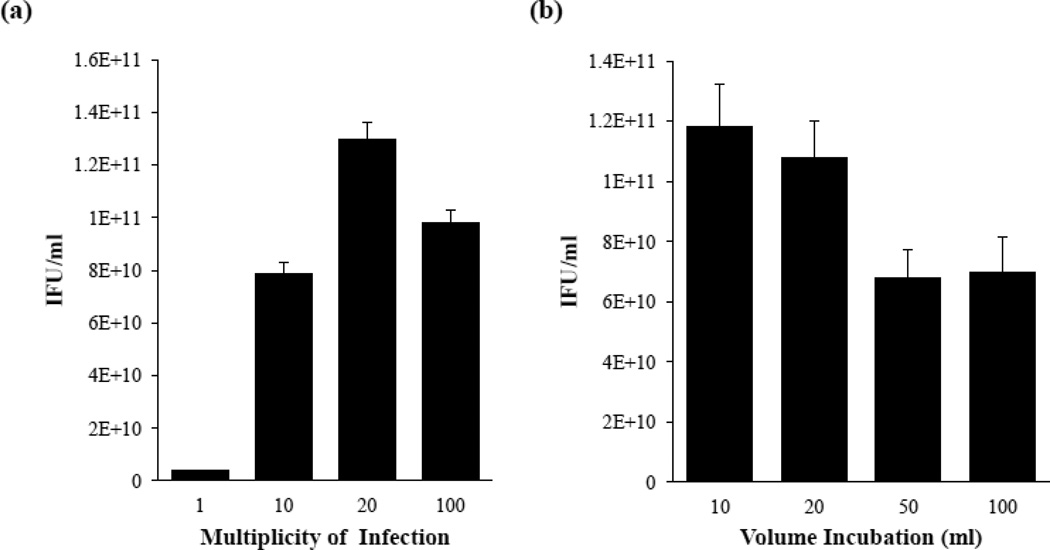
Parameters of the suspension culture infection were optimized for C. trachomatis F/Cal-I-13 from previous reports of suspension culture infections with invasive C. trachomatis LGV biovars (Schachter and Wyrick, 1994; Garrity, 2010) by varying both the MOI and volume of cell-containing buffer (SPG) during the infection process (Fig. 2a and 2b). First, 2.5 ×108 L929 cells collected from suspension were resuspended in a 10 ml volume and co-incubated with increasing concentrations of C. trachomatis F/Cal-I-13. As shown in Figure 2a, a significantly higher yield of EB was obtained with a MOI of 20 as evaluated by IFU measurement (p <0.05). The higher MOI of 100 resulted in a lower yield of EB, likely due to cytotoxicity caused by over-infection (Belland et al., 2001). Next, we examined the impact of cell volume on infection by co-incubating a constant amount of EBs with 2.5×108 L929 cells in 10, 20, 50 and 100 ml of SPG. Figure 2b demonstrates that infections carried out in a total volume of 10 ml or 20 ml produced a significantly higher yield of EB progeny under our test conditions (p <0.05). Smaller volumes likely correlate with a greater yield of EBs due to increased bacterial-host cell contact. Using optimized conditions, we were able to achieve an average yield of 5.5×1012 purified C. trachomatis (as determined by IFU measurement) from 1×109 L929 cells in suspension. The recovery of a high number of IFU demonstrates the ability of suspension culture infections to support C. trachomatis F/Cal-I-13 growth and multiplication. Neither chemical pretreatment with DEAE-dextran nor centrifugation assistance are needed for optimal infectivity. This is a practical approach for obtaining a high yield of C. trachomatis as it is extremely time-cost efficient in comparison to the traditional monolayer infection procedure. The ability to generate large-scale infections has implications in many chlamydial fields including antigen preparation, transport kinetics, biochemistry of the host and bacteria during infection or any application requiring large quantities of bacteria.
Figure 2.
Dose and volume optimization of suspension culture infection with C. trachomatis serovar F/Cal-I-13. (a) Suspension cultures were incubated with increasing amounts of bacteria. (b) Bacteria and host cells were incubated in varying volumes of infection buffer (SPG) to determine optimal infection conditions. Endpoint infection assays determining the IFU of progeny on monolayers of L929 cells resulting from each infection were used as an indication of quality of infection for both experiments. Each result is indicated as the mean value of at least three independent experiments ± the standard deviation (SD). Significant difference was analyzed by Student’s t-test. A p-value <0.05 was used as the cut-off for statistical significance.
3.2 Isolation of C. trachomatis productive and persistent forms and ATP transport assays
We have streamlined and optimized an effective method for the purification of C. trachomatis forms (Schachter and Wyrick, 1994; Scidmore, 2005; Wang et al., 2011) employing gentle lysis by nitrogen cavitation followed by a modified OptiPrep™ gradient centrifugation protocol in conjunction with suspension culturing techniques (Fig. 1). Cell disruption by nitrogen cavitation is based on the rapid decompression of a cell suspension from a pressure vessel (Gottlieb and Adachi, 2000), ensuring that vesicular and organelle structures are well preserved. After gentle lysis and removal of host debris, the C. trachomatis forms were collected and overlaid onto a density gradient consisting of 24%, 34%, 40%, 44% and 54% OptiPrep™-SPG. Figure 1 outlines the sedimentation of each form, including enlarged aberrant RB persistent forms from ampicillin-exposed C. trachomatis infected L929 cells.
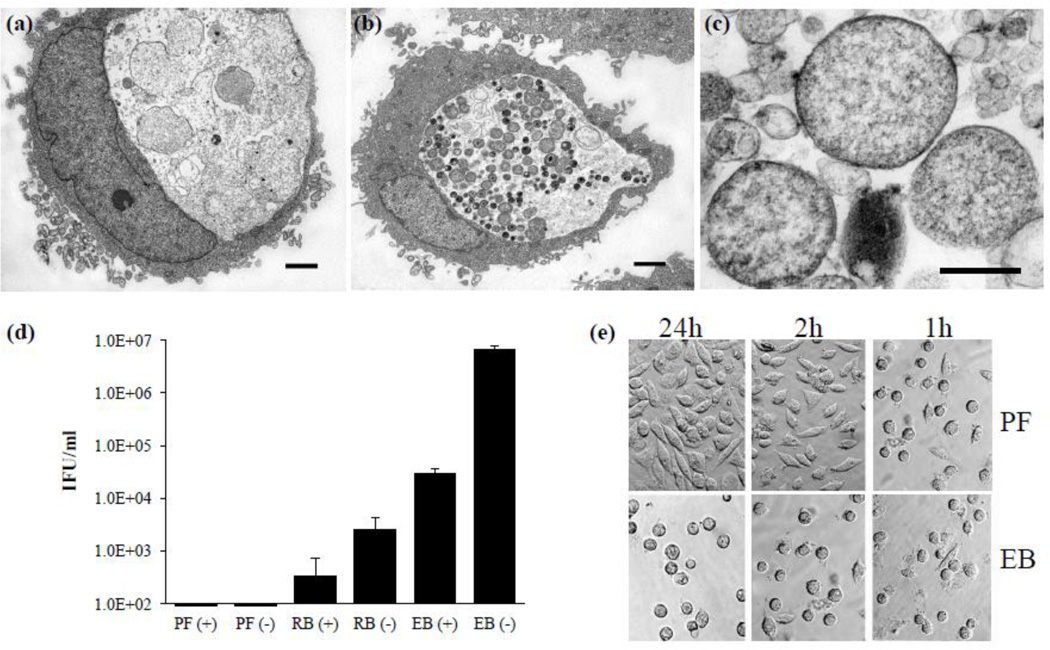
Previous reports indicated the heterogeneity of intracellularly persistent forms generated during ampicillin or IFN-γ exposure (Matsumoto and Manire, 1970; Wyrick, 2010; Wang et al., 2011). We confirmed these isolated persistent forms also exhibited heterogeneity (Fig. 3). TEM conducted on the sedimented samples reveals that the persistent forms partition at the 24–34% interface (Fig. 3c). The size and density of the isolated persistent forms is consistent with the infected L929 cells exposed to ampicillin in Figure 3a. In contrast, ampicillin-unexposed control infections appeared to have more homogeneous RBs or EBs (Fig. 3b) and yielded a population of RB at 34–40% and a distinct layer of purified EB at the 40–44% interface (data not shown). Each interface appears to be relatively separate from other chlamydial forms. Accordingly, based on IFU measurements conducted with ampicillin-exposed and unexposed control purifications, the majority of EBs sediment to the 40–44% interface (Fig. 3d). Although a very small amount of contaminating EBs are found in the RB fraction, we are unable to detect EBs in the persistent form fraction. As further evidence of the isolation of distinct populations of chlamydial forms, we compared the ability of isolated persistent forms vs. EBs to generate a normal infection in a suspension culture as indicated by the ability of L929 cells to adhere to cover slips. Cell adherence is a good indicator of infection as infectious EBs of C. trachomatis genital strains can release cytotoxic and host modulatory factors that disrupt the cytoskeleton of host cells, thus preventing host cell attachment to coverslips post-infection (Moulder et al., 1976; Belland et al., 2001; Carabeo et al., 2002; Clifton et al., 2004; Kumar and Valdivia, 2008). As demonstrated by Figure 3e, only those cells incubated with the non-infectious persistent form were able to adhere to coverslips. Accordingly, cells incubated with EBs remained rounded and failed to adhere to coverslips. These results further confirm the validity of this culture method and demonstrate that the persistent form fraction is properly identified and does not contain any detectable infectious forms (EBs).
Figure 3.
The persistent form purification. Representative transmission electron micrographs of infected L929 cells (a) exposed or (b) unexposed to ampicillin and (c) the purified persistent form fraction containing aberrant RBs with a less electron dense appearance. Based on multiple cross-sections purified persistent form size is estimated at approximately 0.7–2.0 µm. Scale bars in (a) and (b) represent 2 µm and 0.5 µm in (c). (d) Endpoint infection assays determining IFUs on monolayers of L929 cells in each isolated density gradient fraction. Values represent mean IFU ± SD of three independent chlamydial preparations. (e) Representative images of the cell adherence assays demonstrating the inability of the persistent form to produce infection and inhibit cover slip adherence. Abbreviations: PF (+), persistent fraction purified from an infection exposed to ampicillin; PF (−) persistent fraction purified from an infection with no ampicillin exposure; RB (+), reticulate body fraction purified from an infection exposed to ampicillin; RB (−), reticulate body fraction purified from an infection with no ampicillin exposure; EB (+), elementary body fraction purified from an infection exposed to ampicillin; EB (−), elementary body fraction purified from an infection with no ampicillin exposure.
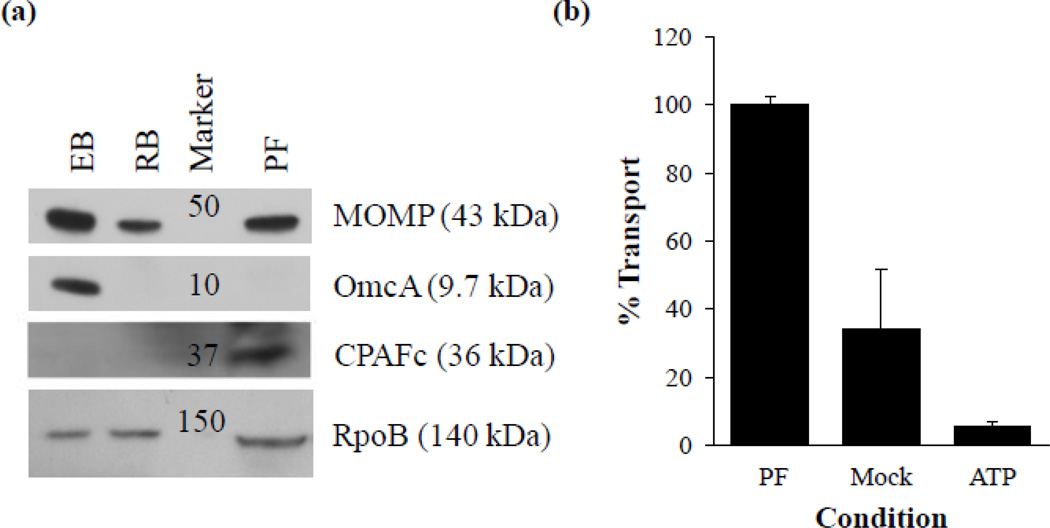
Western blot analysis of the chlamydial fractions isolated with this procedure clearly demonstrates the appropriate presence of stage specific markers (Fig. 4a). As expected, the essential major outer membrane protein, MOMP, is present in all chlamydial isolations, but the EB specific membrane protein OmcA is detectable only in the EB fraction of this purification. Chlamydial protease-like activity factor (CPAF), an effecter produced and secreted during normal RB growth, but retained under persistence inducing conditions (Wang et al., 2011) was detectable in the persistent fraction only. The presence of the bacterial cytoplasmic protein RpoB (a subunit of RNA polymerase) in all chlamydial fractions confirms that these bacteria are intact and have not lost their cytoplasmic contents during isolation (Fig. 4a). Taken together, these data demonstrate the relative purity of each density gradient fraction and the isolation of a persistent form fraction.
Figure 4.
Characteristics of the persistent form fraction. (a) Western blot analysis of isolated chlamydial forms with monoclonal antibodies specific to chlamydial MOMP, OmcA, CPAF and RpoB. Sample loading normalized based on total protein. All samples prepared in Laemmli sample buffer (1:1, v:v) supplemented with 5% 2-Mercaptoethanol, 10 mM dithiothreitol and heated for 10 min at 100 °C. (b) ATP transport assays conducted with isolated persistent forms. ATP indicates control transport assays containing 20-times the determined Km for C. trachomatis nucleoside phosphate transporter 1. All data normalized by adjusting maximum transport to 100%. Experiments were performed in triplicate on three independent chlamydial preparations. Abbreviations: EB, elementary body; RB, reticulate body; PF, persistent form; ATP, adenosine-5'-triphosphate.
We next conducted ATP transport assays with isolated persistent forms. Chlamydia posses the rare ability to scavenge ATP from the host cell cytosol via two nucleoside phosphate transporters termed NTP 1 and NTP 2 (Hatch et al., 1982; Tjaden et al., 1999). Although both are able to concentrate exogenous ATP to the bacterial cytoplasm, NTP 1 is an antiporter that primarily scavenges ATP as an energy source while NTP 2 is a symporter that serves as a general nucleotide transport system with a relatively low affinity for ATP (Tjaden et al., 1999). As detailed in Figure 4b C. trachomatis persistent forms isolated from the 24–34% interface were able to transport ATP above mock-infected and negative controls. These data demonstrate that the purification process detailed here is able to produce purified bacteria with intact membranes and suggests that the persistent form is able to transport ATP from the host. These data are in agreement with the finding of Gerard et al. who report NTP 1 transcript present in not only RB, but also in the persistent form (Gerard et al., 2002). Further work is warranted to confirm and detail the contribution of energy scavenging during the persistent state of chlamydial growth. Since it is critical to conduct biochemical assays as quickly as possible after the removal of bacteria from the host, the ability to isolate large quantities of even the most fragile chlamydial forms in less than four hours makes this protocol exquisitely suited for further transport and biochemical assays.
3.3 C. trachomatis membrane vesicle purification
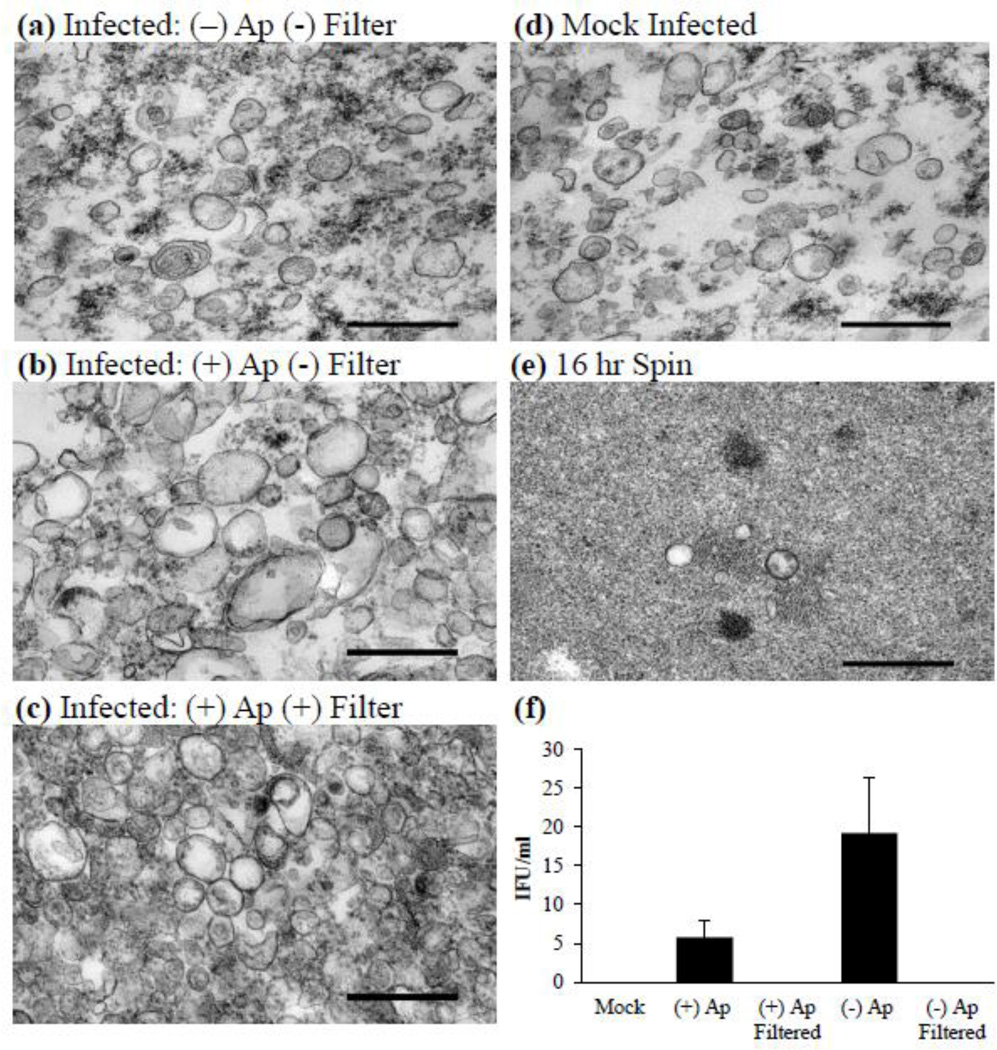
We previously confirmed the presence of vesicles in association with C. trachomatis infection in several cell culture models, including HeLa 229, L929, and human primary endocervical cell culture (Wang et al., 2011). We observed that in these infection models vesicle production seemingly increased during persistence-inducing conditions (Wang et al., 2011). Some of these vesicles appear to be bacterial in nature, supporting the hypothesis that chlamydiae are able to produce membrane vesicles during the normal growth cycle and that this population of vesicles is modulated upon stress such as exposure to ampicillin or IFN-γ (Giles et al., 2006; Wang et al., 2011). As a first step to better understanding the role of vesicles during infection, we developed a purification protocol for isolation the total vesicle population during infection (Fig. 1). The vesicle purification procedure was designed such that both bacteria and vesicles could be harvested from a single infected culture. We chose to employ the gentle host cell lysis method of nitrogen cavitation in order to avoid the generation of non-native or mechanically generated vesicles and decrease bacterial lysis. After removal of contaminating C. trachomatis by centrifugation as detailed in section 2.4, vesicles were separated from the majority of free protein by ultracentrifugation at 100,000 ×g for one hour, filtered through a 0.2 µM filter and gently washed. Figure 5a–e contains representative TEM samples obtained from vesicle pellets and demonstrates the population of vesicles isolated from infections not exposed to ampicillin (Fig. 5a), vesicles isolated from infections exposed to ampicillin with and without 0.2 µm filtration (Fig. 5b and c), and those isolated from mock infected cultures (Fig. 5d). To confirm that the majority of vesicles were collected during the initial high speed spin, the resultant supernatant was centrifuged for an additional 16 hours. Clearly depicted in figure 5e, there are very few vesicles remaining in the supernatant after the initial vesicle collection protocol. Purification of vesicles from non-ampicillin-exposed or mock-infected samples yielded a population similar in appearance by TEM. It was confirmed that filtered vesicle populations were free of contaminating EB particles by IFU assessment (Fig. 5f). Thus, we can be confident that chlamydial proteins detected in vesicle samples are present due to an association with vesicles and not co-purified intact EBs. Since the vesicle population is representative of both bacterial and host-derived vesicles present during infection at the time of harvest, it can be used to study the dynamics of both populations.
Figure 5.
Vesicle purification. (a–e) Representative transmission electron micrographs of various vesicle purifications from infected cultures that are (a) non-ampicillin-exposed, non-filtered; (b) ampicillin-exposed, non-filtered; (c) ampicillin-exposed, filtered; (d) mock infected; or (e) pellet collected from the vesicle isolation supernatant after 16 hr of centrifugation. Experiments performed with the initial fraction (15,300 ×g vesicle supernatant) confirmed that vesicles remain intact after 16 hr of centrifugation. (f) An endpoint assay determining the IFU present in each fraction was conducted to determine possible C. trachomatis elementary body contamination. Values represent mean IFU ± SD from three independent vesicle preparations. Scale bars represent 0.5 µm. Abbreviations: (+) Ap, vesicles isolated from an infection exposed to ampicillin; (−) Ap, vesicles isolated from an infection not exposed to ampicillin.
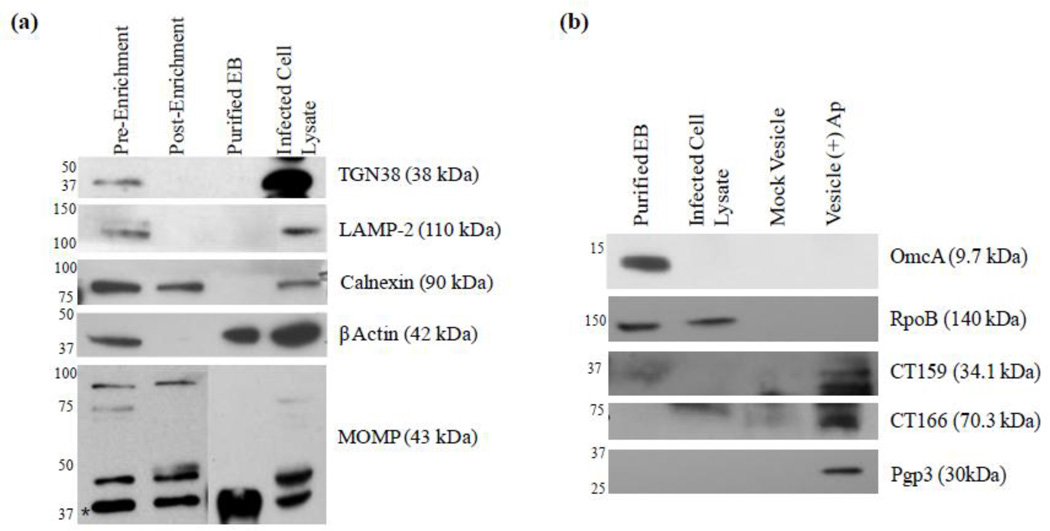
To enrich C. trachomatis-derived vesicles from the total vesicle population, we developed an immunoenrichment protocol using anti-chlamydial LPS antibody and magnetic beads. As detailed in Section 2.5, the vesicle preparation was incubated with anti-chlamydial LPS antibody and the vesicle-antibody population was separated from the total vesicle mixture by magnetic beads. After washing, the resulting vesicle-antibody-bead complex was resuspended in an equivalent volume of buffer such that the host vesicle concentration should decrease while the bacterial vesicle concentration remains relatively unchanged. Western blot analysis of various host vesicle markers was employed to demonstrate bacterial membrane vesicle enrichment. Figure 6a demonstrates an overall decrease in contaminating host vesicle associated proteins, including the lysosome marker LAMP-2, the trans-golgi network protein TGN38 and the ER membrane marker calnexin. β-actin was also greatly reduced. It is noteworthy that only a mild decrease in calnexin is observed in comparison to the other host markers, suggesting that there could be association of bacterial vesicles with the ER. These results correlate with those of Giles and Wyrick in which they observed a co-localization of selective chlamydial major antigens (MOMP and LPS) with ER markers, but not markers of mitochondria, Golgi, lysosomes or endosomes (Giles and Wyrick, 2008). It is unlikely that the decrease in host vesicle markers is due to a loss of total vesicles during the enrichment procedure as the overall amount of chlamydial MOMP detected in the pre- and post-enrichment samples does not significantly change (Fig. 6a). Host vesicle associated proteins not detected in the total vesicle population include the cell plasma membrane marker cadherin and the early endosome marker EEA-1 (data not shown). Taken together, these data demonstrate the immune-enrichment of bacterial vesicles and suggest the ER as a target and/or a possible route of delivery for at least a sub-population of C. trachomatis vesicles. The immune-enrichment procedure employed here to enrich a population of bacteria-derived vesicles could be used further to negatively or positively select sub-populations of host and bacterial vesicles respectively.
Figure 6.
Analysis of vesicle populations by Western blot. (a) Relative enrichment of chlamydial LPS-containing vesicles results in a decrease of host derived vesicles. (b) Purity of the vesicle fraction and the association of putative virulence factors with the chlamydial vesicle population. Vesicle samples prepared in Laemmli sample buffer (1:1, v:v) supplemented with 5% 2-Mercaptoethanol, 10 mM dithiothreitol and heated for 10 min at 100 °C. Abbreviation: EB, elementary body.
In order to further define the bacterial vesicle population, we next examined if selective bacterial proteins that are known to be either outer membrane components or putative secreted cytotoxic proteins are present in the immune-enriched population by Western blot analysis (Fig. 6b). We can infer from the results that these C. trachomatis vesicles are likely derived from persistent or RB forms as the EB-specific cysteine-rich outer membrane protein OmcA was not detected. Additionally, a lack of the bacterial cytoplasmic protein RpoB further confirms that the purified vesicles are free of C. trachomatis particles and do not contain RpoB (Fig. 6b). In addition to MOMP (Fig. 6a), the hypothetical protein CT159, as well as the putative cytotoxic protein CT166, were also confirmed to be associated with the vesicle preparation (Fig. 6b). Both of these proteins could serve as cytotoxic effectors during infection as CT159 is predicted to contain a conserved phospholipase D superfamily catalytic domain and CT166 has been shown to display cytotoxic activity by targeting Rac1 during infection (Belland et al., 2001; Thalmann et al., 2010). CT166 protein is produced during the mid- and late-stages of the chlamydial developmental cycle and is thought to play a role early in infection as evidenced by morphological changes in host cells when treated with high C. trachomatis MOIs (Belland et al., 2001; Thalmann et al., 2010). Although we did not observe CT166 in purified EBs, likely due to a difference in loading concentration, antibody selection, and probing methods, the presence of this protein in association with vesicles, coupled to the report of high enzymatic activity by Thalmann et al. (Thalmann et al., 2010), suggests a role for CT166 in addition to that of initial bacterial entry. Of particular interest are the apparent alternate forms of MOMP detected in the vesicle and whole cell lysate populations (as probed by monoclonal antibody) (Fig. 6a, lower panel). While a relatively strong band at approximately 40 kDa (indicated with an asterisk in Fig. 6a), similar to the predicted molecular weight of MOMP, was present in all populations, multiple distinct bands of larger sizes ranging from approximately 45 to 80 kDa were detected only in the vesicle and/or whole lysate populations (Fig. 6a). This difference in migration could be due to either MOMP oligomerization or post-translational modification as previously reported (Swanson and Kuo, 1994; Sun et al., 2007) and suggests a potential conformation difference of MOMP between vesicles and the C. trachomatis membrane. Many bacterial outer membrane vesicles display modified LPS that has been shown to be highly immunogenic and elicit an enhanced immune response (Andersen et al., 1997; Arigita et al., 2005). Whether or not MOMP structural differences contribute to alternate immunogenicity is not yet known. Figure 6b also demonstrates the presence of the plasmid encoded effector Pgp3 in association with C. trachomatis vesicles. Pgp3 is an immunodominant antigen and found in association with the chlamydial outer membrane complex, but is primarily located in the host cell cytoplasm (Li et al., 2008; Chen et al., 2010). These observations strongly support the hypothesis that membrane vesicles are generated by C. trachomatis as a means of carrying and delivering outer membrane components (LPS and MOMP) and virulence proteins (Pgp3, CT159 and CT166) necessary for pathogenesis. Although we cannot determine the exact destination of chlamydial vesicles in this study, it is probable that there are distinct subpopulations present at multiple cellular compartments: within the inclusion, host cytoplasm, or trafficked through cellular machinery alongside host vesicles. These multiple distinct populations of chlamydial membrane vesicles appear to serve as carriers and delivery systems for cytotoxic or host modulatory proteins. As the vesiculation process may vary under different growth conditions, the properties of vesicles under normal growth and ampicillin–exposed or stress conditions likely differ. Further characterization of chlamydial membrane vesicle components is ongoing and will further illuminate the complexity of host-pathogen interactions and their role in disease pathogenesis.
4.0 Conclusion
We optimized and developed these protocols in order to facilitate the study of intracellular populations that are difficult to isolate. The large-scale propagation of C. trachomatis is critical to its study and facilitates the isolation of distinct populations, such as vesicles and the persistent form, not easily separated due to their obligate intracellular nature. We present an optimized persistent form purification and demonstrate that the persistent form is able to transport radiolabeled ATP, suggesting that these aberrant forms employ some degree of energy parasitism. This method is not only used to isolate fragile persistent forms of Chlamydia, but also allows for the isolation of other sub-cellular fractions such as bacterial and host vesicles. The isolated vesicle population was enriched for a bacterial sub-population of vesicles that contain common bacterial antigens such as LPS and MOMP, as well as the putative cytotoxic or host modulatory proteins Pgp3, CT159, and CT166. Additionally, slight modifications of the protocol could yield numerous other populations of either organelles or subpopulations of host vesicles that are suitable for the study of host adaptation/modulation during infection with intracellular bacteria.
Highlights.
Large scale L929 cell suspension cultures are infected with C. trachomatis strain F.
Persistent forms and other C. trachomatis forms are purified from the host cell.
Persistent forms transport ATP and likely rely upon host ATP as an energy source.
C. trachomatis and host vesicles are purified and vesicles containing LPS are enriched.
C. trachomatis vesicles contain bacterial membrane markers and putative virulence factors.
Acknowledgements
We are grateful to Drs. Kenneth Johnston, Priscilla Wyrick and Wandy Beatty for helpful discussions and critical review of the manuscript and Dr. Guangming Zhong for the kind gift of antibodies. This work was supported, in part, by the Louisiana Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents and by NIH grants AI055869 and AI055869S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen SR, Bjune G, Hoiby EA, Michaelsen TE, Aase A, Rye U, Jantzen E. Outer membrane vesicle vaccines made from short-chain lipopolysaccharide mutants of serogroup B Neisseria meningitidis: effect of the carbohydrate chain length on the immune response. Vaccine. 1997;15:1225–1234. doi: 10.1016/s0264-410x(97)00030-3. [DOI] [PubMed] [Google Scholar]
- Arigita C, Luijkx T, Jiskoot W, Poelen M, Hennink WE, Crommelin DJ, Ley P, Els C, Kersten GF. Well-defined and potent liposomal meningococcal B vaccines adjuvated with LPS derivatives. Vaccine. 2005;23:5091–5098. doi: 10.1016/j.vaccine.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 2006;119:350–359. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Scidmore MA, Crane DD, Hogan DM, Whitmire W, McClarty G, Caldwell HD. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13984–13989. doi: 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- Carabeo RA, Grieshaber SS, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect. Immun. 2002;70:3793–3803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 2005;73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Lei L, Lu C, Galaleldeen A, Hart PJ, Zhong G. Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 2010;192:6017–6024. doi: 10.1128/JB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosinephosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage BL, Cookson BT. Membrane vesicle release in Bacteria, Eukaryotes and Archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012 doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields KA, Hackstadt T. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 2002;18:221–245. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- Garrity GM. Bergey's Manual of Systematic Bacteriology. Place Springer New York, Published; 2010. [Google Scholar]
- Gerard HC, Freise J, Wang Z, Roberts G, Rudy D, Krauss-Opatz B, Kohler L, Zeidler H, Schumacher HR, Whittum-Hudson JA, Hudson AP. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microbes Infect. 2002;4:13–22. doi: 10.1016/s1286-4579(01)01504-0. [DOI] [PubMed] [Google Scholar]
- Giles DK, Whittimore JD, LaRue RW, Raulston JE, Wyrick PB. Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect. 2006;8:1579–1591. doi: 10.1016/j.micinf.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Giles DK, Wyrick PB. Trafficking of chlamydial antigens to the endoplasmic reticulum of infected epithelial cells. Microbes Infect. 2008;10:1494–1503. doi: 10.1016/j.micinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb RA, Adachi S. Nitrogen cavitation for cell disruption to obtain mitochondria from cultured cells. Methods Enzymol. 2000;322:213–221. doi: 10.1016/s0076-6879(00)22022-3. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Intracellular Biology. Pathogenesis, and Immunity. Washington, DC: R. S. Stephens. American Socienty for Microbiology; 1999. Developmental Biology.Chlamydia; pp. 29–67. [Google Scholar]
- Hatch TP, Al-Hossainy E, Silverman JA. Adenine nucleotide and lysine transport in Chlamydia psittaci. J. Bacteriol. 1982;150:662–670. doi: 10.1128/jb.150.2.662-670.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera VL, Shen L, Lopez LV, Didishvili T, Zhang YX, Ruiz-Opazo N. Chlamydia pneumoniae accelerates coronary artery disease progression in transgenic hyperlipidemia-genetic hypertension rat model. Mol. Med. 2003;9:135–142. doi: 10.2119/2003-00009.herrera. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Rao X, Feng X, Luo X, Liang Y, Shen L. Mutagenesis of region 4 of sigma 28 from Chlamydia trachomatis defines determinants for protein-protein and protein-DNA interactions. J. Bacteriol. 2009;191:651–660. doi: 10.1128/JB.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg KR, Horoschak KD, Moulder JW. Toxicity of low and moderate multiplicities of Chlamydia psittici for mouse fibroblasts (L cells) Infect. Immun. 1977;18:531–541. doi: 10.1128/iai.18.2.531-541.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y, Valdivia RH. Reorganization of the host cytoskeleton by the intracellular pathogen Chlamydia trachomatis. Commun. Integr. Biol. 2008;1:175–177. doi: 10.4161/cib.1.2.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambden PR, Pickett MA, Clarke IN. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology. 2006;152:2573–2578. doi: 10.1099/mic.0.29032-0. [DOI] [PubMed] [Google Scholar]
- Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 2008;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Manire GP. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 1970;101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder JW, Hatch TP, Byrne GI, Kellogg KR. Immediate toxicity of high multiplicities of Chlamydia psittaci for mouse fibroblasts (L cells) Infect. Immun. 1976;14:277–289. doi: 10.1128/iai.14.1.277-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralinath M, Kuehn MJ, Roland KL, Curtiss R., 3rd Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect. Immun. 2011;79:887–894. doi: 10.1128/IAI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves W, Asakrah S, Qazi O, Brown KA, Kurtz J, Aucoin DP, McLachlan JB, Roy CJ, Morici LA. A naturally derived outer-membrane vesicle vaccine protects against lethal pulmonary Burkholderia pseudomallei infection. Vaccine. 2011;29:8381–8389. doi: 10.1016/j.vaccine.2011.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya ID, Szwedo JW, Goodwin A, Lupashina TV, Nagarajan UM, Lupashin VV. Chlamydia trachomatis hijacks intra-Golgi COG complex-dependent vesicle trafficking pathway. Cell Microbiol. 2012;14:656–668. doi: 10.1111/j.1462-5822.2012.01747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect. Immun. 2009;77:3284–3293. doi: 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Troncoso G, Ferreiros CM, Criado MT. Evaluation of cross-reactive antigens as determinants of cross-bactericidal activity in pathogenic and commensal Neisseria. Vaccine. 2001;19:3390–3398. doi: 10.1016/s0264-410x(01)00077-9. [DOI] [PubMed] [Google Scholar]
- Schachter J, Wyrick PB. Culture and isolation of Chlamydia trachomatis. Methods Enzymol. 1994;236:377–390. doi: 10.1016/0076-6879(94)36028-6. [DOI] [PubMed] [Google Scholar]
- Scidmore MA. Cultivation and Laboratory Maintenance of Chlamydia trachomatis. Curr. Protoc. Microbiol. 2005 doi: 10.1002/9780471729259.mc11a01s00. Chapter 11, Unit 11A 11. [DOI] [PubMed] [Google Scholar]
- Su EL, Snape MD. A combination recombinant protein and outer membrane vesicle vaccine against serogroup B meningococcal disease. Expert Rev. Vaccines. 2011;10:575–588. doi: 10.1586/erv.11.32. [DOI] [PubMed] [Google Scholar]
- Subtil A. Rerouting of host lipids by bacteria: are you CERTain you need a vesicle? PLoS Pathog. 2011;7:e1002208. doi: 10.1371/journal.ppat.1002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Pal S, Sarcon AK, Kim S, Sugawara E, Nikaido H, Cocco MJ, Peterson EM, De la Maza LM. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 2007;189:6222–6235. doi: 10.1128/JB.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson AF, Kuo CC. Binding of the glycan of the major outer membrane protein of Chlamydia trachomatis to HeLa cells. Infect. Immun. 1994;62:24–28. doi: 10.1128/iai.62.1.24-28.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Matsumoto A, Manire GP, Higashi N. Electron microscopic observations on the structure of the envelopes of mature elementary bodies and developmental reticulate forms of Chlamydia psittaci. J. Bacteriol. 1971;105:355–360. doi: 10.1128/jb.105.1.355-360.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann J, Janik K, May M, Sommer K, Ebeling J, Hofmann F, Genth H, Klos A. Actin re-organization induced by Chlamydia trachomatis serovar D--evidence for a critical role of the effector protein CT166 targeting Rac. PLoS One. 2010;5:e9887. doi: 10.1371/journal.pone.0009887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J, Winkler HH, Schwoppe C, Van Der Laan M, Mohlmann T, Neuhaus HE. Two nucleotide transport proteins in Chlamydia trachomatis one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 1999;181:1196–1202. doi: 10.1128/jb.181.4.1196-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooij C, Apodaca G, Engel J. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect. Immun. 1997;65:758–766. doi: 10.1128/iai.65.2.758-766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooij C, Kalman L, Van I, Nishijima M, Hanada K, Mostov K, Engel JN. Host cell-derived sphingolipids are required for the intracellular growth of Chlamydia trachomatis. Cell Microbiol. 2000;2:627–637. doi: 10.1046/j.1462-5822.2000.00077.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Frohlich KM, Buckner L, Quayle AJ, Luo M, Feng X, Beatty W, Hua Z, Rao X, Lewis ME, Sorrells K, Santiago K, Zhong G, Shen L. Altered protein secretion of Chlamydia trachomatis in persistently infected human endocervical epithelial cells. Microbiology. 2011;157:2759–2771. doi: 10.1099/mic.0.044917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 2006;15:122–134. doi: 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler HH. Rickettsial permeability. An ADP-ATP transport system. J. Biol. Chem. 1976;251:389–396. [PubMed] [Google Scholar]
- Wyrick PB. Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 2010;201(Suppl 2):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Watkins NG, Stewart S, Caldwell HD. The low-molecular-mass, cysteine-rich outer membrane protein of Chlamydia trachomatis possesses both biovar- and species-specific epitopes. Infect Immun. 1987;55:2570–2573. doi: 10.1128/iai.55.11.2570-2573.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial proteaselike activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]