Abstract
Lung cancer remains the leading cause of cancer-related death worldwide for both men and women, and non-small cell lung cancer (NSCLC) accounts for approximately 80% of all cases. Despite improvements in early diagnosis and newly developed therapies, the 5-year survival rate for NSCLC patients remains low (15%). Therapy in NSCLC has reached a plateau. Understanding genomic medicine may provide insight into the oncogenesis of lung cancer and open the door to molecular diagnosis, new biomarkers and a more accurate prognosis of lung cancer. It is well known that almost half of the genes regulated by microRNAs (miRNAs) are located in cancer-associated genomic regions. In the present study, we discuss the potential of miRNAs to function as suppressors and biomarkers for chemoresistance and prognosis of lung cancer.
Keywords: mirans, oncogenes, inhibitors, biomarker, lung cancer
1. Introduction
Lung cancer remains the leading cause of cancer-related death worldwide, and non-small cell lung cancer (NSCLC) accounts for approximately 80% of all cases (1,2). Although novel therapies targeting early diagnosis have been developed, the 5-year survival rate for NSCLC patients remains at a low 15% (3). Therapy in NSCLC has reached a plateau. This unfavourable outcome is due to our relatively limited understanding of tumor pathogenesis and gene expression profiles. Understanding genomic medicine may therefore aid investigations into the oncogenesis of lung cancer, and may also yield prospects in accurate molecular diagnosis, new biomarkers and risk stratification of lung cancer.
The study of C. elegans has facilitated an important discovery: the existence of non-coding microRNAs (miRNAs). These tiny fragments of RNA (approximately 22 nucleotides long) regulate gene expression by hybridizing to complementary sequences in the 3′ untranslated region (3′UTR) of target messenger RNA (mRNA). These RNAs thereby repress the translation of mRNA and silence gene expression by either cleaving target mRNAs or inhibiting their translation (5). In their study, Bartel et al reported that these small non-coding, endogenous, single-stranded RNAs regulate gene expression, especially at the post-transcriptional level (6). miRNAs play a significant role in a wide variety of pathways by regulating gene expression at the post-transcriptional level. It is well known that almost half of the genes are regulated by miRNAs, which are located in cancer-associated genomic regions or fragile genomic sites (4). Weiss et al reported that miRNA-128b directly regulates epidermal growth factor receptor (EGFR); the EGFR mutation is known to correlate with the oncogenesis of lung cancer. Emerging evidence suggests that miRNAs may control lung cancer development and play a critical role in its oncogenesis and pathogenesis (7–15).
miRNAs are short nucleotide strands that are more stable than several proteins and mRNA, which are less prone to enzymatic degradation by RNAses. Furthermore, miRNAs can be measured from formalin-fixed paraffin-embedded samples and bodily fluids, such as blood samples, which can be obtained easily and using non-invasive methods (16). Mitchell et al showed that miRNAs are also present in human plasma in a markedly stable form that is protected from endogenous RNase activity. Tumor-derived miRNAs in the serum or plasma are an important method for the blood-based detection of human cancer (17). Since miRNAs play a substantial role in lung cancer and since detection samples can easily be obtained, miRNAs become a promising means of comprehending the oncogenesis and pathogenesis of lung cancer.
In this review, we briefly describe the role of miRNAs in lung cancer pathogenesis, as predictors of chemoresistance, as biomarkers for risk stratification and as tools for prognostic assessment in lung cancer.
2. Defects in the microRNA processing and lung cancer
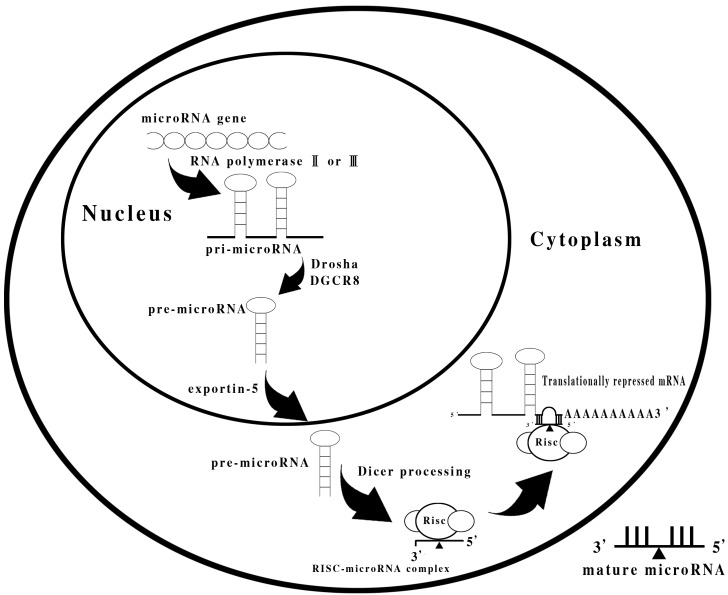
miRNA genes are located within the introns or exons of protein-coding genes, as well as in intergenic areas. miRNA genes are transcribed by RNA polymerase II or III into primary miRNA transcripts (pri-miRNAs), which are then cleaved into B70 nucleotide-long precursor miRNAs (pre-miRNAs) by the nuclear microprocessor complex formed by the RNase III Drosha and DiGeorge syndrome critical region gene 8 (DGCR8). Pre-miRNAs are transported by the exportin-5 into the cytoplasm, where they are cleaved to generate the final products of 22 nucleotides by Dicer. In brief, miRNAs are processed by the type III double-stranded RNase Dicer, and function in an RNA-based mechanism of gene silencing (18). A summary of the miRNA processing is shown in Fig. 1.
Figure 1.
Summary of the miRNA processing.
Global repression of miRNA biogenesis by suppression of the key components of miRNA processing machinery, such as Drosha, DGCR8, DICER1, TRBP and XPO5, promotes cellular transformation and tumorigenesis (19–22). In 2007, Kumar et al found that miRNA processing-impaired cells formed tumors with accelerated kinetics in animals. These tumors were more invasive than control tumors. Furthermore, deletion of DICER1 enhanced tumor development in a K-Ras-induced mouse model of lung cancer (19). TAR RNA-binding protein 2 (TARBP2) encodes an integral component of a DICER1-containing complex. Melo et al showed that the presence of TARBP2 frameshift mutations causes reduced TRBP protein expression and a defect in the processing of miRNAs. The reintroduction of TRBP in the deficient cells restores the efficient production of miRNAs and inhibits tumor growth. Notably, TRBP impairment is associated with destabilization of the DICER1 protein (20). Hill et al reported that DICER1 mutations are associated with familial pleuropulmonary blastoma (21). Recently, Melo et al demonstrated a genetic defect in exportin-5 traps precursor miRNAs in the nucleus of cancer cells. This observed genetic defect is responsible for nuclear retention of pre-miRNAs, thereby reducing miRNA processing. The restoration of XPO5 function reverses the impaired export of pre-miRNAs and has tumor suppressor characteristics (22). Therefore, impairment of these biosynthesis checkpoint control mechanisms of mature miRNAs in cancer cells may lead to an abnormal expression profile of these small non-coding RNAs, thus enhancing the tumorigenic process.
Accordingly, it is suggested that miRNAs have an intrinsic function in tumor inhibition, and their down-regulation eventually accelerates oncogenesis.
3. microRNAs as tumor inhibitors or oncogenes in lung cancer
More and more studies report that the mutation or aberrant expression of many miRNAs has often been found in cancer patients, leading to the study of miRNAs as regulators of oncogenes and tumor inhibitor genes. Hammond has shown that miRNAs, whose expression is increased in tumors, may function as a novel class of oncogenes or tumor inhibitor genes (15). In cell studies, Johnson et al reported that all synthetic pre-let-7 molecules reduced the cell number in transfected A549 lung cancer cells compared to a negative control miRNA, and inhibition of let-7 with an anti-let-7 molecule results in a 100% increase in the number of A549 cells compared to a control transfection and other tested anti-miRNA (anti-oncomirs). These authors concluded that cell growth is inhibited by the overexpression of let-7 miRNA in the A549 cell line, which functions as a tumor suppressor (23). Kumar et al not only showed that in lung cancer cells the ectopic expression of let-7g induced both cell cycle arrest and cell death, but also demonstrated that in an autochthonous model of NSCLC in the mouse, let-7g expression substantially reduced lung tumor burden (24). Esquela-Kerscher et al found that lung adenocarcinoma is capable of being repressed by administering let-7 intranasally (25). Briefly, it is suggested that the miRNA let-7 family functions as a tumor inhibitor in cell and animal studies.
Aside from the let-7 family, Davidson et al noted that miR-218 expression was down-regulated in 85% (33/39) of NSCLC tumor compared to paired normal lung; statistically significant decreases were observed in both squamous cell carcinomas (p<0.0001) and, to a lesser extent, in adenocarcinomas (p=0.001). This down-regulation of miR-218 was found to be associated with a history of cigarette smoking; thus, it was reported that miR-218 is a strong candidate tumor suppressing miRNA potentially involved in lung cancer (26). Wang et al reported that the expression levels of miR-451 were found to be significantly correlated with tumor differentiation, pathological stage and lymph node metastasis (27). Moreover, low miR-451 expression levels were also correlated with shorter overall survival of NSCLC patients (p<0.001). Furthermore, these authors observed that miRNA-451 functions as a tumor inhibitor in human NSCLC by targeting ras-related protein 14 (RAB14) (27).
In other respects, several miRNAs functioning as oncogenes, known as oncomirs, usually promote tumor progression by negatively inhibiting tumor suppressor genes. Liu et al recently reported that the engineered knockdown of miR-31 substantially repressed lung cancer cell growth and tumorigenicity in a dose-dependent manner (28). They also showed that miRNA-31 functions as an oncogenic miRNA by repressing specific tumor suppressors, such as large tumor suppressor 2 (LATS2) and PP2A regulatory subunit B α isoform (PPP2R2A) (28).
4. microRNAs as biomarkers for chemoresistance or chemosensitivity in NSCLC
Several studies have demonstrated that aberrant miRNA expression, resulting in altered function of the target mRNA, which affects the expression of the target proteins and fundamentally silences the target gene (29), can be related to chemotherapy resistance. Mechanisms of drug resistance are often associated with changes in relevant proteins, such as PTEN, PDCD4, P-glycoprotein (P-gp) and multidrug resistance 1 (MDR1).
Current investigations support the hypothesis that the over- or under-expression of any miRNA is directly linked to a patient’s response to chemotherapeutic agents. Specific examples of miRNAs related to chemoresistance have been discussed in the literature. For example, Ranade et al recently reported that the miRNAs significantly associated with chemoresistance in SCLC were miR-92a-2* (p=0.010), miR-147 (p=0.018) and miR-574-5p (p=0.039). Among these miRNAs, higher tumor miR-92a-2* levels are associated with chemoresistance and with decreased survival in patients with SCLC. Therefore, tumor miR-92a-2* may be applied in the screening of patients with SCLC at risk for de novo chemoresistance (30). These miRNA biomarkers may assist in treatment stratification and may also predict chemoresistance for SCLC.
A number of miRNAs have also been shown to be differentially expressed in docetaxel-resistant NSCLC, such as specifically increased levels of miR-98, -192 and -424, and decreased levels of miR-194, -200b and -212 (31). Recently, Xie et al found that cell-free miRNAs in the supernatant of effusions may aid in the diagnosis of malignancy and predict chemosensitivity to docetaxel (32). Ceppi et al found that miR-200c overexpression restored the sensitivity of NCI-H1299 cells to cisplatin and cetuximab (37). Zhu et al found that miR-497 was down-regulated in the multidrug-resistant human lung cancer cell line A549/cisplatin (CDDP), and the down-regulation of miR-497 was concurrent with the up-regulation of the BCL2 protein. Using an in vitro drug sensitivity assay, these authors demonstrated that an overexpression of miR-497 sensitized A549/CDDP cells to anticancer drugs. The luciferase activity of BCL2 3′UTR region-based reporter constructed in A549/CDDP cells suggested that BCL2 was the direct target gene of miR-497. Enforced miR-497 expression reduced the BCL2 protein level and sensitized A549/CDDP cells to VCR- and CDDP-induced apoptosis, respectively (38).
The various studies regarding miRNAs as biomarkers for chemoresistance in NSCLC and SCLC are shown in Table I. These examples clearly demonstrate the role of miRNAs in chemo-sensitivity/-resistance, and show that manipulating miRNAs may be beneficial in modulating cancer chemoresistance and chemosensitivity. Therefore, miRNAs serve as the most accurate predictive biomarkers in establishing personalized medicine currently available in the market, providing the most precise prognosis for treatment response and outcomes. miRNAs should be used, first, as biomarkers to predict drug resistance or sensitivity in order to establish the most appropriate personalized treatment, and, second, as possible drug targets to reverse resistance. We also anticipate that the therapeutic development of miRNA mimics or antagomirs may reveal novel therapies and improve prognosis for NSCLC.
Table I.
microRNAs as biomarkers for chemoresistance in NSCLC and SCLC.
| miRNA | Cancer type | miRNA intracellular levels | Protein target | Drug | Biomarker or confirmed in resistance | Ref. |
|---|---|---|---|---|---|---|
| −21 | NSCLC | Increased | PTEN | N/A | B | 32 |
| −21, −23b | NSCLC | Increased | TKR | Erlotinib, sunitinib, vandetanib | R | 34,35 |
| −92a−2, −147, −574−5p | SCLC | Increased | N/A | Platinum | R | 30 |
| −98, −192, −424 | NSCLC | Increased | N/A | Docetaxel | B | 31 |
| −194, −200b, −212 | NSCLC | Decreased | N/A | Docetaxel | B | 32 |
| −221, −222 | NSCLC | Increased | Kit, p27, PTEN, TIMP3 | TRAIL | B | 36 |
| −424 | NSCLC | Decreased | N/A | Erlotinib, vandetanib | R | 34 |
NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer; B, biomarker; R, resistance.
5. EGFR signaling pathway and miRNAs
The potent role of EGF in transforming normal cells into tumors, with specific implications for lung cancer, had already been observed by the early 1980s. Recently, EGFR signaling and EGFR mutations became a controversial topic of NSCLC studies. In pace with the recent studies of miRNAs and the EGFR signaling pathway, it has gradually become evident that a relationship exists between miRNAs and the EGFR signaling pathway. miRNAs are emerging as important modulators in cell pathways. Certain miRNAs down-regulate lung cancer through the EGFR signaling pathway. For example, Dacic et al demonstrated that miR-155 was up-regulated only in the EGFR/KRAS-negative group, miR-25 was up-regulated only in the EGFR-positive group and miR-495 was up-regulated only in the KRAS-positive adenocarcinomas. By contrast, let-7g was down-regulated in all three groups, with more significant down-regulation in the EGFR/KRAS-negative adenocarcinomas (39). The study by these authors showed that among lung adenocarcinomas, some miRNAs showed unique expression patterns, which were strongly correlated with the mutation type, suggesting different carcinogenic pathways for these tumors. These miRNAs can be further explored for their diagnostic role and as biomarkers for targeted therapies (39).
Seike et al reported a significant correlation between phosphorylated-EGFR (p-EGFR) and miR-21 levels in lung carcinoma cell lines, suggesting that EGFR signaling is a pathway positively regulating miR-21 expression. The aberrantly increased expression of miR-21, which is further enhanced by the activated EGFR signaling pathway, plays a significant role in lung carcinogenesis in never-smokers, as well as in smokers, and is a potential therapeutic target in both EGFR-mutant and wild-type cases (40). Webster et al found that miR-7 down-regulates EGFR mRNA and protein expression in cancer cell lines (lung, breast and glioblastoma), inducing cell cycle arrest and cell death (41). Furthermore, these investigators found that miR-7 attenuated the activation of protein kinase B (Akt) and extracellular signal-regulated kinase 1/2, two critical effectors of EGFR signaling, in different cancer cell lines (41). Cho et al reported miR-145 in the negative regulation of EGFR and NUDT1 expression at both the mRNA and protein levels. Further analysis showed that miR-145 is capable of inhibiting cell proliferation on transfected lung adenocarcinoma cells over three time points (24, 48 and 72 h). Up-regulation of miR-145 appeared to be an important gene regulation mechanism for the proliferation of lung adenocarcinoma cells and it correlated strongly with the down-regulation of EGFR and NUDT1 (42). Despite the progress that has been made, much uncertainty remains regarding the role of miRNA and EGFR in human cancer. We believe that extensive miRNA studies are likely to yield promising evidence explicating the function of miRNA regulation and the EGFR network in lung cancer.
6. microRNAs as risk stratification and prognosis prediction in NSCLC
In an era of personalised medicine, accurate prognosis has become the focus of considerable attention, with studies in miRNA becoming more and more relevant. Investigators have been using miRNA expression signatures not only to classify human cancers, but also to define miRNA markers that may provide favorable prognosis (10). Saito et al investigated the expression of miR-21, miR-17 and miR-155 measured by quantitative RT-PCR in tissues from 317 NSCLC patients (43). They found that more advanced stage tumors expressed significantly higher levels of miR-21 compared to TNM stage I tumors. TNM stage I patients were evaluated separately and elevated levels of miR-21 were found to be associated with higher rates of cancer mortality (HR=2.16; 1.11–4.21) and relapse-free survival, independent of other clinical factors. This finding suggests that an increased miR-21 expression is associated with disease progression and survival in stage I lung cancer, and the expression of miR-21 may contribute to early-stage prognostic biomarkers for lung adenocarcinoma (43).
Boeri et al explored the miRNA expression profiles of lung tumors, normal lung tissues and plasma samples from cases with variable prognosis (44). miRNA expression analyses in plasma samples collected 1–2 years prior to the onset of disease, at the time of CT detection and in disease-free smokers enrolled in the screening trial, resulted in the generation of miRNA signatures with strong predictive, diagnostic and prognostic potential (area under the ROC curve ≥0.85). These results indicate that miRNAs play a role in lung tissues and plasma as molecular predictors of lung cancer development and aggressiveness, and have theoretical and clinical implications for lung cancer management (44). These results suggest that the integration of microarray-based genomic information with existing clinicopathological models may enhance the ability of clinicians to determine the most effective treatment for individual patients. Such a strategy may improve survival rates and reduce treatment-related morbidity in NSCLC patients. Roybal et al showed that the miR-200 family is part of a gene expression signature that predicts poor prognosis in lung cancer patients. Furthermore, they found that forced miR-200 expression suppressed Flt1 levels whose knockdown decreased the growth and metastasis of tumor cells. Therefore, they conclude that miR-200 suppresses lung tumorigenesis by targeting Flt1, which correlates inversely with the duration of survival (45).
miRNA signature is a novel method that may provide fresh insight to old problems. It is difficult to say whether miRNA signature is superior or inferior to gene expression profiling, with respect to risk stratification and prognosis prediction. A recent study by Voortman et al investigated the prognostic and predictive values of miRNA expression for survival using a Cox model, which included every factor used in stratified randomization, clinicopathological prognostic factors and other factors statistically related to miRNA expression. Results by these authors indicated that the miRNA expression patterns examined were neither predictive nor prognostic in a large patient cohort with radically resected NSCLC, randomized to receive adjuvant cisplatin-based chemotherapy vs. follow-up only (46). Therefore, the role of miRNAs in outcome prediction remains controversial and requires further investigation.
In conclusion, miRNAs have emerged as one of the key players in regulating gene expression. Numerous studies have reported the implications of miRNAs in virtually every carcinogenesis process of lung cancer, including tumor development, apoptosis, invasion and metastasis, as well as anticancer drug resistance. Thus, miRNAs are not only promising biomarkers for chemoresistance that are likely to guide personalized treatments, but they also precisely and directly target agents that may develop personalized medicine and improve prognostics.
References
- 1.Ramalingam S, Pawlish K, Gadgeel S, Demers R, Kalemkerian GP. Lung cancer in young patients: analysis of a Surveillance, Epidemiology, and End Results database. J Clin Oncol. 1998;16:651–657. doi: 10.1200/JCO.1998.16.2.651. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Miller YE. Pathogenesis of lung cancer: 100 year report. Am J Respir Cell Mol Biol. 2005;33:216–223. doi: 10.1165/rcmb.2005-0158OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs: micro-RNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, Varella-Garcia M, Bunn PA, Jr, Haney J, Helfrich BA, Kato H, Hirsch FR, Franklin WA. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 8.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2005;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Eder M, Scherr M. MicroRNA and lung cancer. N Engl J Med. 2005;352:2446–2448. doi: 10.1056/NEJMcibr051201. [DOI] [PubMed] [Google Scholar]
- 13.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 14.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Ba Y, Ligia M. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 19.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 20.Melo SA, Ropero S, Moutinho C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo SA, Moutinho C, Ropero S, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 26.Davidson MR, Larsen JE, Yang IA, et al. MicroRNA-218 is deleted and downregulated in lung squamous cell carcinoma. PLoS One. 2010;5:e12560. doi: 10.1371/journal.pone.0012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. Clin Invest. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Xiao H. miRNAs modulate the drug response of tumor cells. Sci China C Life Sci. 2009;9:797–801. doi: 10.1007/s11427-009-0114-4. [DOI] [PubMed] [Google Scholar]
- 30.Ranade AR, Cherba D, Shridhar S, et al. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in small cell lung cancer patients. J Thorac Oncol. 2010;5:1273–1278. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13:57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie L, Chen X, Wang L, et al. Cell-free miRNAs may indicate diagnosis and docetaxel sensitivity of tumor cells in malignant effusions. BMC Cancer. 2011;11:256. doi: 10.1186/1471-2407-10-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:11–12. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 34.Weiss G, Nelson K, Edwards D, Boisvert S, Sima C. MicroRNA biomarkers associated with vandetanib, sunitinib, and/or erlotinib resistance. AACR/IASLC. 2010 [Google Scholar]
- 35.Nelson K, Sima C, Edwards D, Weiss G. MicroRNA biomarkers associated with sunitinib resistance in non-small cell lung cancer. AACR. 2010 abstract 3048. [Google Scholar]
- 36.Garofalo M, Di LG, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ceppi P, Mudduluru G, Kumarswamy R, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 38.Zhu W, Zhu D, Lu S, et al. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2011 Jan; doi: 10.1007/s12032-010-9797-4. [E-pub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 39.Dacic S, Kelly L, Shuai Y. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23:1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 40.Seike M, Goto A, Okano T, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2009;284:5731–5741. doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 42.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–2206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 43.Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early stage, non small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeri M, Verri C, Conte D. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roybal JD, Zang Y, Ahn YH, et al. miR-200 inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voortman J, Goto A, Mendiboure J, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70:8288–8298. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]