Abstract
The purpose of this study was to explore the functional role and mechanism of miR-155 in the development of renal cell carcinoma (RCC). miR-155 expression was quantified in renal cancers, matched adjacent non-tumor tissues and renal cell lines using quantitative real-time PCR (RT-PCR). Cell proliferation, apoptosis and migratory activity were measured following suppression of miR-155 expression by antisense oligonucleotides. miR-155 targets were scanned using target prediction programs. Following the inhibition of miR-155, target gene expression was detected by western blotting. The expression of miR-155 was upregulated in clear cell RCC (ccRCC) tissue and renal cancer cell lines. The suppression of miR-155 inhibited cell proliferation and migratory activity and induced apoptosis in renal cancer cells. The suppressor gene suppressor of cytokine signaling (SOCS-1) and BACH1 were predicted as potential target genes by bioinformatics analysis. The suppression of miR-155 inhibited BACH1 protein expression. miR-155 may function as an oncogene by targeting BACH1. Thus, the inhibition of miR-155 may be an effective way to treat RCC.
Keywords: microRNA-155, renal cancer, BACH1
Introduction
Renal cell carcinoma (RCC) accounts for 2–3% of all malignant diseases in adults and is the seventh most common cancer in males and the ninth most common cancer in females (1). In the US, 58,240 new cases and 13,040 mortalities are estimated to have occurred in 2010 (2). However, treatments for renal cancer, particularly for metastatic tumors, remain unsatisfactory. The prognosis of patients with advanced metastatic RCC is poor. Less than 10% of the patients are reported to have approximately 5-year survival rates (3). Therefore, it is necessary to develop more effective therapy for renal cancers.
microRNAs (miRNAs) are approximately 22-nucleotide noncoding RNAs that function as negative regulators of gene expression. They may repress translation or induce mRNA cleavage by binding to the 3′ untranslated region (UTR) of target mRNA (4). Between 74 and 92% of known human genes are estimated to be under the control of miRNAs (5). In recent years, increasing evidence implicates the involvement of miRNAs in carcinogenesis. Deregulated expression of miRNAs has been shown in various types of human malignancies, some of which function as tumor-suppressor genes or oncogenes (6–8). Previous miRNA profiling studies revealed that the level of miRNA-155 (miR-155) is upregulated in various types of human malignancy, including different forms of lymphoma and carcinomas of the nasopharynx, breast and colon (9–13). Functional analysis of miR-155 indicates that it functions as an oncogenic miRNA in several types of cancer. Ectopic expression of miR-155 was shown to promote proliferation and facilitate invasion and migration of breast cancer cells (9,14). Overexpression of miR-155 was also found to promote development of lymphoma and hepatocellular carcinoma (12,15). Furthermore, elevated expression of miR-155 was associated with poor prognosis of nasopharyngeal and colorectal cancer patients (10,13). On the other hand, the miR-155 level was also shown to be downregulated in melanoma cells, possibly playing a tumor-suppressor role (16). Several studies have demonstrated the upregulation of miR-155 levels in renal cancers (17–19). However, the function of miR-155 in renal cancers has not been reported.
In this study, we examined the expression of miR-155 in clear cell RCC (ccRCC) using RT-PCR, and investigated cell proliferation, apoptosis and migration following miR-155 knockdown in renal cancer cells. Furthermore, we explored the mechanism of miR-155 involvement in the development of renal cancer by identifying possible target genes. Our study may present an attractive, novel target for further studies of the therapeutics of renal cancer.
Materials and methods
Tissue samples and cell lines
RCC samples and normal adjacent tissue samples were obtained from 16 patients with a mean age of 61.5 years (50–75 years). All samples were frozen in liquid nitrogen immediately after tumors were resected. Prior written and informed consent was obtained from every patient, and the study was approved by the Ethics Review Board of Second Xiangya Hospital, Central South University.
Human RCC cell lines ACHN and KC and the normal renal tubule epithelium cell line HK2 were purchased from the Cell Bank of Xiangya Medical College, Central South University. The ACHN, KC and HK2 cells were cultured in DMEM medium (Gibco, Invitrogen, Carlsbad, CA, USA) complemented with 10% fetal bovine serum (Gibco). All cell lines were cultured in a humidified incubator (5% CO2) at 37˚C.
Cell transfections
ACHN cells were transfected with antisense-miR-155 oligonucleotide (As-miR-155), a negative control (miR-CON) (Shanghai GenePharma Co, Ltd. China) or an empty vector (blank) using Lipofectamine 2000™ (Invitrogen) following 24 h of cell seeding. Briefly, Lipofectamine 2000 was mixed with the Opti-MEMI medium (Gibco) and incubated for 5 min. Meanwhile, As-miR-155 or miR-CON was diluted in Opti-MEMI medium. Then, the diluted As-miR-155 or miR-CON and Lipofectamine 2000 were mixed and incubated for 20 min at room temperature. The resultant mixture was added to each well. At 48 h after transfection, the cells were harvested. Experiments were performed in triplicate.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assays
Cell viability was determined by using the MTT method. Briefly, 1×104 ACHN cells were seeded onto 96-well plates. Cells were incubated for 24 h in 37˚C in a 5% CO2 tissue culture incubator. MTT reagent (50 μl; 5 mg/ml) was added to each well and cells were incubated for 4 h. Then the formazan precipitate was dissolved in 150 μl DMSO and the absorbance rate was measured in a microplate reader at a wavelength of 570 nm. All experiments were repeated in triplicate.
Cytofluorometric apoptosis assay
Cells were collected and transferred to 60-mm dishes. The cell apoptosis ratio was analyzed 48 h post-transfection using the Annexin V-FITC Apoptosis Detection kit (BD Biosciences, San Diego, CA), according to the manufacturer's instructions. Annexin V-FITC and propidium iodide double staining was used to evaluate the number of apoptotic cells. Annexin V-positive and propidium iodide-negative cells were designated as apoptotic cells. The tests were performed in triplicate.
Cell migration assay
The invasive potential of cells was measured in 6-well Matrigel-coated invasion chambers (Corning Life Science, Acton, MA). ACHN cells transfected with As-miR-155 or miR-CON were harvested and re-suspended in serum-free DMEM medium. Then aliquots (1×105 cells) of the prepared cell suspension were added into the upper chamber and the lower chamber was filled with 1 ml of DMEM containing fetal bovine serum. Cells were fixed with ice-cold methanol and stained with toluidine blue after they were incubated for 48 h. Cells on the upper side of the membrane were removed by using a cotton swab. Then the invasive cells were counted by a light microscope at ×1000 magnification. All experiments were repeated in triplicate.
Quantitative real-time PCR
Total RNAs were extracted from the fresh tissues or cells with TRIzol reagent (Invitrogen). Real-time PCR assays were performed using an miRNeasy Mini kit (Qiagen, USA) and miRNA Q-PCR Detection kit (GeneCopoeia, USA). Quantitative PCR was performed on a 7500 Fast Real-time PCR System (Applied Biosystems). PCR was performed using an initial step of denaturation at 95˚C for 5 min, 40 cycles of amplification, denaturation at 95˚C for 10 sec, annealing at 60˚C for 30 sec, elongation at 72˚C for 10 sec. All reactions were performed in a 20-μl volume in triplicate. The oligonucleotide primers were as follows: has-miR-155 forward (5′-TAATGCTAA TCGTGATAGGG-3′) and has-miR-155 reverse (5′-TTTGGCACTAGCACATT-3′); U6RNA forward (5′-CTCGCTTCGGCAGCACA-3′) and U6RNA reverse (5′-AACGCTTCACGAATTTGCGT-3′). The miRNA expression was determined using the 2−ΔCt method (20).
Western blotting
ACHN cells were harvested 42 h following transfection of As-miR-155 or miR-CON using the RIPA buffer (ProMab Biotechnology, USA). Equal amounts of the denatured protein sample were separated by SDS-PAGE and were then transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were probed with antibodies against BACH1 (1:400 dilution, cat# sc-14700, Santa Cruz Biotechnology, Santa Cruz, CA, USA), SOCS-1 (1:400 dilution cat# sc-9021, Santa Cruz) or GAPDH (1:1,000 dilution, Mab-2005079, ProMab). GAPDH was used as a loading control. The membrane was incubated with horseradish peroxidase-conjugated secondary antibodies (cat# sc-2030 Santa Cruz), and visualized by an enhanced chemiluminescence detection kit (Pierce, Thermo Scientific, USA) and exposed to film.
Statistical analyses
The results were provided as the mean ± SD and analyzed using SPSS 17.0 software. Statistical analysis was performed using an independent samples t-test and one-way AVOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of miR-155 is upregulated in renal cancer tissues
To identify miR-155 expression in clinical samples, we performed RT-PCR and found that miR-155 expression in renal cancer tissues (n=16) was significantly higher than that in adjacent normal tissues (Fig. 1A). We further investigated whether miR-155 expression in clinical tissues correlates with pathological stages and Fuhrman grades. Higher expression of miR-155 was observed in cases with higher stages (II+III) in comparison to cases in stage I (Fig. 1B). However, no statistically significant correlation was observed between expression of miR-155 and Fuhrman grades (data not shown).
Figure 1.
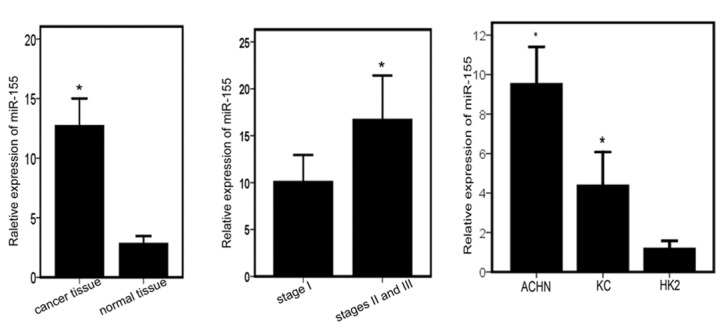
Levels of miR-155 are increased in renal cancer tissues and are associated with advanced pathological stage. (A) Real-time PCR analysis of relative miR-155 expression levels in renal cancer tissues and adjacent normal tissues (*P<0.01). (B) miR-155 expression in different pathological stages of renal cancer tissues (*P<0.05). Patients were staged according to the AJCC TNM Classification, 2002. (C) Relative expression levels of miR-155 in human RCC cell lines (ACHN and KC) and a normal renal cell line (HK2) (*P<0.05).
To choose an appropriate renal cancer cell model for in vitro study, we also determined the expression level of miR-155 in primary RCC cell lines (ACHN and KC) and a normal renal cell line (HK2). The expression levels of miR-155 in the RCC cell lines were upregulated when compared to that in the normal renal cell line and the expression level of miR-155 in the ACHN was similar to levels in the renal cancer tissues (Fig. 1C).
Downregulation of miR-155 expression suppresses proliferation and migration of renal cancer cells
To analyze the function of miR-155 in RCC cell lines, we knocked down the expression of miR-155 in a primary RCC cell line (ACHN) by transfection with As-miR-155. As shown in Fig. 2A, RT-PCR results indicated that miR-155 expression was significantly inhibited. The MTT assay was performed to determine the viability of ACHN cells. A significant decrease in cell viability was observed at 24 and 48 h following As-miR-155 transfection (Fig. 2B), compared to cells transfected with miR-CON. To ascertain whether suppression of miR-155 affects invasion and metastasis of renal cancer cells, a Trans-well migration assay was performed. The number of migrating cells in the miR-155 knockdown group was 14.74±1.06, which was significantly decreased compared with the control (blank- or miR-CON-transfected) groups (41.22±1.65 and 42.17±1.38, respectively). Renal cancer cell migratory activity was significantly decreased in the ACHN cells transfected with As-miR-155 (Fig. 2C and D). Altogether, these results suggest that inhibition of miR-155 suppresses the tumorigenicity of renal cancer cells in vitro.
Figure 2.
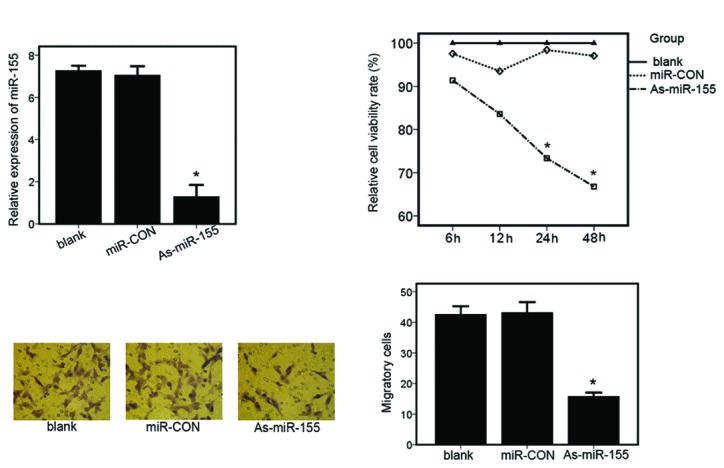
Suppression of miR-155 expression reduces renal cancer cell proliferation and migration. (A) Relative expression of miR-155 in ACHN cells transfected with the controls (blank- or miR-CON-transfected) or As-miR-155. Expression of miR-155 was detected by real-time PCR (*P<0.01). (B) Cellular viability assay in ACHN cells transfected with controls (blank- or miR-CON-transfected) or As-miR-155 (*P<0.01). (C) Transwell-migration assay in ACHN cells transfected with the controls (blank or miR-CON transfected) or As-miR-155 (*P<0.01). (D) Values shown were calculated from at least three independent experiments, including that shown in panel C.
Suppression of miR-155 expression induces apoptosis of renal cancer cells
To detect the effect of miR-155 inhibition on apoptosis of renal cancer cells, a cytofluorometric apoptosis assay was performed. At 48 h post-transfection, we determined apoptosis in ACHN cells transfected with controls (blank- or miR-CON-transfected) and As-miR-155 by flow cytometric analysis of Annexin-V-FITC-PI-stained cells (Fig. 3A). It was found that the average apoptotic cell fractions were significantly increased upon the inhibition of miR-155 expression compared with the miR-CON- or blank-transfected cells (Fig. 3B).
Figure 3.
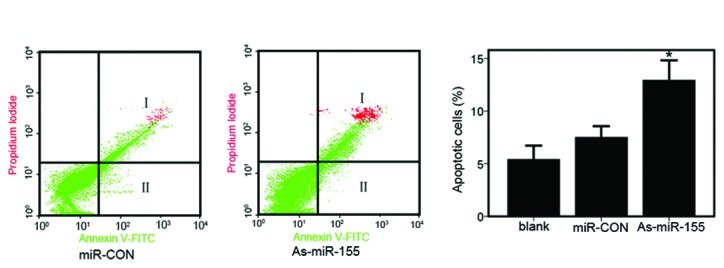
Downregulation of miR-155 expression induces renal cancer cell apoptosis. (A) Apoptosis assay in ACHN cells following transfection with miR-CON or As-miR-155. I, Apoptotic cells (%); II, early apoptotic cells (%). (B) Representation of average apoptotic cell fractions (both early and late apoptotic cells) in each group (*P<0.01).
BACH1 may be a target gene of miR-155
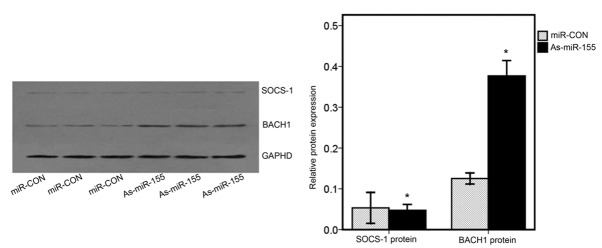
Three target scan algorithms (miRanda, PicTar and Targetscan) were used to predict targets of miR-155. Two putative target genes, BACH1 and SOCS-1, which are involved in oncogenesis (9,21) were selected for further investigation. To determine whether BACH1 and SOCS-1 are targets of miR-155, ACHN cells were transfected with miR-CON or As-miR-155. Western blot analyses were performed at 48 h post-transfection. The western blotting results indicated that BACH1 levels in ACHN cells transfected with As-miR-155 were significantly increased when compared with that in cells transfected with miR-CON (Fig. 4). However, the levels of SOCS-1 in ACHN cells transfected with As-miR-155 were not significantly affected when compared with that in the cells transfected with miR-CON (Fig. 4). These results suggest that BACH1 may be a target gene of miR-155.
Figure 4.
miR-155 negatively regulates BACH1. (A) Western blot analysis was used to detect the expression levels of endogenous BACH1 and SOCS-1 in ACHN cells transfected with miR-CON or As-miR-155. GAPHD was used as a loading control. (B) Values shown were calculated from at least three independent experiments, including that shown in panel A.
Discussion
miR-155 is an miRNA involved in various biological processes including haematopoiesis, inflammation and immunity (10). Recent studies have demonstrated that miR-155 is one of the most prominent miRNAs implicated in the genesis and progression of human cancer. In our study, we determined the upregulation of miR-155 expression in renal cancer tissues. Moreover, it was found that the suppression of miR-155 expression induced apoptosis and inhibited proliferation and migration in renal cancer cells. This is the first report to reveal that miR-155 may function as an oncogenic miRNA in renal cancers, although some evidence has demonstrated that miR-155 is upregulated and acts as an oncogene in several types of human cancer, such as breast, colon cancer and lymphoma (9,11,22). In addition, miR-155 was also reported to be downregulated and to function as a tumor suppressor in melanoma cell lines (16). Similar evidence was also found for miR-126. miR-126 in lung cancer cells acts as an inhibitor of motility only, without any effects on proliferation (23), whereas in breast cancers, miR-126 is an inhibitor of cell proliferation (24). Taken together, the roles of miRNA in cancers are indicated to be tissue- and tumor-specific. The identification of the oncogenic role of miR-155 in renal cancer is necessary, and inhibition of miR-155 may serve as a new anti-miRNA therapeutic in renal cancers. Two anti-miR-122 oligonucleotides have been proven to be specific, stable and non-toxic in two primate studies (25,26).
Although the prospect of miRNA-based therapy is interesting, there have not been enough studies on the mechanisms of the function of miR-155 in renal cancers. BACH1 and SOCS-1 are potential targets of miR-155 predicted by all three target scan algorithms: miRanda, PicTar and Targetscan. In our study, BACH1 was identified to be a target of miR-155 in renal cancer by western blotting, since its protein expression level increased significantly following inhibition of miR-155 in ACHN cells.
BACH1 belongs to a member of the basic leucine zipper transcription factor family and is a key player in the regulation of oxidative stress (27). Recent studies show that BACH1 is a broadly expressed transcriptional repressor, which negatively regulates genes involved in cell cycle progression, apoptosis, and the hypoxia response (21). An important one of these genes is heme-oxygenase-1 (HO-1), which may be important in induction of the tumorigenic pathway (21,28). The expression of HO-1 is significantly upregulated in various types of cancer and overexpressed HO-1 is found to promote tumor growth and metastasis and inhibit the apoptosis of tumor cells (29,30). Furthermore, BACH1 is identified to suppress growth and survival of acute myeloid leukemia (AML) cells by downregulation of HO-1 expression and may enhance the cytotoxic effects of certain anti-cancer drugs in AML cells (31). Although the mechanisms by which BACH1 plays a role in renal cancer development have not been defined, there is a study showing that BACH1 functions as a repressor of HO-1 in human cells. Moreover, it is reported that the expression of HO-1 is upregulated in human renal cancer tissues following activation of the Ras-ERK-Nrf2 pathway and the overexpression of HO-1 inhibits apoptosis of renal cancer cells (32). These data combined with our findings indicate that BACH1 may function as a tumor suppressor by downregulating HO-1, which plays a key role in the Ras-ERK-Nrf2 pathway in renal cancer.
Our results suggest that miR-155 may play an oncogenic role by inhibiting the expression of BACH1 in renal cancers. However, BACH1 is not necessarily the only target that mediates function of miR-155 in renal cancers. Several targets of miR-155 have been identified to be regulated by miR-155 in certain types of cancer (11,33,34). For example, Forkhead boxO3A (FOXO3a) and SOCS-1 were identified as targets of miR-155 in breast cancer (9,33). Inositol polyphosphate-5-phosphatase (SHIP1) and SMAD family member 5 (SMAD5) are targets of miR-155 in lymphomas (11,34). Hypoxia-inducible factor-1a (HIF-1a) is one of the putative miR-155 targets identified by using both PicTar and miRanda algorithms. HIF-1a encodes a subunit of HIF-1 and regulates many hypoxia-inducible genes that are involved in cell proliferation, angiogenesis and iron metabolism (35). Moreover, factors inhibiting HIF-1 promote renal cancer cell survival by protecting cells from HIF-1-mediated apoptosis (36). In our study, SOCS-1 was found not to be a target of miR-155 in renal cancer cells. However, SOCS-1 has been identified to be a target of miR-155 in breast cancer cells (9). These findings suggest that miRNA targeting is cell type-specific, which was also previously reported by other investigators (37,38). More studies related to targets of miR-155 in renal cancers will be helpful for elucidating the mechanisms of miR-155 functioning in renal cancers. In conclusion, miR-155 may play an oncogenic role in renal cancers by targeting BACH1, which functions as a tumor suppressor. Therapy targeting miR-155 inhibition may be an effective method for the treatment of renal cancers.
Acknowledgements
We thank Professors Luoyan Yang and Xiaokun Zhao for their valuable assistance.
References
- 1.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Reeves DJ, Liu CY. Treatment of metastatic renal cell carcinoma. Cancer Chemother Pharmacol. 2009;64:11–25. doi: 10.1007/s00280-009-0983-z. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 6.Asangani IA, Rasheed S, Nikolova D, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2236. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 7.Navarro A, Diaz T, Martinez A, et al. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood. 2009;114:2945–2951. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an oncomir in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 10.Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX, Shao JY, Ernberg I. Upregulation of miR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 2011;7:e1001305. doi: 10.1371/journal.ppat.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 14.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RHOA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Majumder S, Nuovo G, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levati L, Pagani E, Romani S, et al. MicroRNA-155 targets the ski gene in human melanoma cell lines. Pigment Cell Melanoma Res. 2011;24:538–550. doi: 10.1111/j.1755-148X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 17.Neal CS, Michael MZ, Rawlings LH, Van der Hoek MB, Gleadle JM. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010;8:64. doi: 10.1186/1741-7015-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White NM, Bao TT, Grigull J, et al. MiRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011;186:1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- 19.Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Ridzon D, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;27:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warnatz JH, Schmidt D, Manke T, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem. 2011;286:23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakirtzi K, Hatziapostolou M, Karagiannides I, Polytarchou C, Jaeger S, Iliopoulos D, Pothoulakis C. Neurotensin signaling activates microRNAs -21 and -155 and akt, promotes tumor growth in mice, and is increased in human colon tumors. Gastroenterology. 2011;141:1749–1761. doi: 10.1053/j.gastro.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Ga O, Nuovo G, Marsh CB, Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373:607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 24.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 26.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada S, Muto A, Ogawa E, Nakanome A, Katoh Y, Ikawa S, Aiba S, Igarashi K, Okuyama R. Bach1-dependent and -independent regulation of HEME oxygenase-1 in keratinocytes. J Biol Chem. 2010;285:23581–23589. doi: 10.1074/jbc.M109.068197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 29.Jozkowicz A, Was H, Dulak J. HEME oxygenase-1 in tumors: is it a false friend? Antioxid Redox Signal. 2007;9:2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Was H, Dulak J, Jozkowicz A. HEME oxygenase-1 in tumor biology and therapy. Curr Drug Targets. 2010;11:1551–1570. doi: 10.2174/1389450111009011551. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki T, Kirino Y, Takeno M, et al. Expression of HEME oxygenase-1 in human leukemic cells and its regulation by transcriptional repressor Bach1. Cancer Sci. 2010;101:1409–1416. doi: 10.1111/j.1349-7006.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta D, Banerjee P, Gasser M, Waaga AG, Pal S. CXCR3-B can mediate growth-inhibitory signals in human renal cancer cells by down-regulating the expression of HEME oxygenase-1. J Biol Chem. 2010;285:33684–36842. doi: 10.1074/jbc.M110.170324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci USA. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 36.Khan MN, Bhattacharyya T, Andrikopoulos P, Esteban M, Barod R, Connor T, Ashcroft M, Maxwell PH, Kiriakidis S. Factor inhibiting HIF (FIH-1) promotes renal cancer cell survival by protecting cells from HIF-1α-mediated apoptosis. Br J Cancer. 2011;104:1151–1159. doi: 10.1038/bjc.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17–92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 38.Lössner C, Meier J, Warnken U, Rogers M, Lichter P, Pscherer A, Schnölzer M. Quantitative proteomics identify novel miR-155 target proteins. PLoS One. 2011;6:22146. doi: 10.1371/journal.pone.0022146. [DOI] [PMC free article] [PubMed] [Google Scholar]