Abstract
The aims of the present study were to investigate the genetic characteristics of enterovirus 71 (EV71) and coxsackievirus A16 (CVA16) strains in China and to evaluate the relationship between the genotypes of CVA16 and EV71 and their geographical distribution. A total of 399 stool specimens were collected from children with symptoms of hand, foot and mouth disease (HFMD) in Zhejiang Province. The presence of enteroviruses was determined using reverse transcription-semi-nested PCR targeted to the VP1 gene of all human enteroviruses and DNA sequencing. EV71 and CVA16, the major etiological agents of HFMD, were detected in 38.4% (38/99) and 35.4% (35/99) of HEV-A species-positive cases, respectively. Based on the phylogenetic analysis of the VP1 gene, EV71 strains identified in this study belong to subgenotype C4, and CVA16 strains herein were classified into clusters B2a and B2b within the genotype B2. Taking into consideration other published data, we conclude that the genetic characteristics of enteroviruses in China reflect the pattern of the endemic circulation of the subgenotype C4 to EV71 and clusters B2a and B2b within genotype B2 to CVA16, which have been continuously circulating in China since 1997. This observation indicates that the genetic characteristics of enteroviruses in China seem to depend on their special geographical and climatical features allowing them to be sustained with little external effect.
Keywords: phylogenetic analysis, genotying, enterovirus 71, coxsackievirus A16
Introduction
Hand, foot and mouth disease (HFMD) is generally a benign and common self-limiting childhood illness characterized by fever and vesicular eruption on the hands and feet and in the mouth. This disease can be caused by several enterovirus serotypes, such as coxsackievirus A2, A4, A5, A8, A10, A16 and enterovirus 71 (EV71) (1,2). Of these, coxsackievirus A16 (CVA16) and human EV71 are the major etiologic agents of HFMD. The clinical features of HFMD caused by these two viruses are indistinguishable, but EV71 infection is associated to a greater extent with severe neurological disease and fatalities (3–5). In contrast, CVA16-associated HFMD has a milder outcome, with much lower incidence of neurological disease (6).
EV71 has been classified into three genotypes (A,B,C) and divided into 10 subgenotypes (A, B1-B5, C1-C5) based on the complete VP1 gene over a 40-year period and different geographical distribution (7–9). Although a small number of studies have focused on the molecular characteristics of CVA16, CVA16 has been classified into two genotypes (A and B) based on the VP1 gene. The B genotype can be divided into B1 and B2 subtypes, and subtype B2 is known to contain clusters B2a and B2b (10–13). Whereas identification of the genotypes or subgenotypes of CVA16 and EV71 has been used to reveal the origins of virus in HFMD outbreaks, (9,10) the relationship between the genotype of CVA16 and EV71 and their geographical distribution has not been previously investigated.
In this study, we analyzed the genetic characteristics of the partial VP1 gene of CVA16 and EV71 strains in China using phylogenetic analysis, and we specifically aimed to evaluate the relationship between genotype and geographical epidemics.
Materials and methods
Clinical specimens, sample processing, RNA extraction
A total of 399 stool specimens were obtained with informed consent from children with signs of HFMD, including a brief febrile illness and typical vesicular rashes on the palms and soles in Zhejiang Province between May and August 2010. We collected these specimens from the Pediatrics Department of The First Affiliated Hospital, School of Medicine, Zhejiang University (Hangzhou, China). Stool samples were immediately stored at −80˚C until further analysis. Before commencement of this study, the project was approved by the Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University.
A 10% stool suspension was made by adding 0.5 g of stool (0.5 ml for fluid stools) to 5 ml of 1% phosphate-buffered saline. The suspension was centrifuged at 12.000 × g for 10 min and filtered, and then subsequently processed. Vial RNA extraction was carried out using a high Pure Viral RNA kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's procedures and stored at −80˚C until further analysis.
Reverse transcription-semi-nested PCR and sequencing
Reverse transcription-semi-nested PCR amplification was performed using a set of primers for the VP1 gene as described previously (14). Briefly, the 992-bp fragment encompassing the VP3 and VP1 gene was firstly amplified with degenerate primers (AN32, AN33, AN34, AN35 and 224, 222). First round PCR condition was subjected to an initial cycle for reverse transcription at 50˚C for 30 min and the following cycling conditions: 94˚C for 10 min, 40 cycles of 94˚C for 30 sec, 42˚C for 30 sec and 60˚C for 1 min followed by a final extension at 72˚C for 4 min. Nested PCR was performed using 1 μl of the first-round product as template, with the primers AN88 (5′-TACTGGACCACCTGGNGGNAYRWACAT-3′) and AN89 (5′-CCAGCACTGACAGCAGYNGARAYNGG-3′). Nested PCR was subjected to an initial cycle of 95˚C for 4 min and 30 cycles (95˚C for 30 sec, 60˚C for 20 sec and 72˚C for 15 sec), followed by a final incubation at 72˚C for 4 min. RT-semi-nested PCR resulted in a 350- to 400-bp fragment of the VP1 gene. The water was included as negative control for the first and second round PCR. All reactions were performed in a Bio-Rad Thermal cycler (Bio-Rad, Hercules, CA, USA) with thin-walled reaction tubes. Five microliters of the nested PCR products was run on 1% agarose gel made with Tris-boric acid-EDTA (TBE) buffered and strained with ethidium bromide (0.5 μg/ml). For sequencing, PCR products of the appropriate size that produced visible bands upon UV illumination were purified from the gel using QIAquick gel extraction kit (Qiagen) and directly sequenced by an automated DNA sequence analyzer (Applied Biosystem, Carlsbad, CA, USA). Sequences obtained were compared pairwise with the enterovirus sequences available in GeneBank (http://www.ncbi.nlm.nih.gov/blast).
Phylogenetic analysis
Clustal W program was applied for multiple sequence alignment. The dendrograms were constructed by the neighbor-joining method in the MEGA program. The reliability of neighbor-joining tree was estimated by bootstrap analysis with 1.000 pseudoreplicate data sets. Genetic distances were calculated with the Kimura 2-parameter model nucleotide substitution (15).
Results
To determine the prevalence of HFMD in Zhejiang Province, 399 stool specimens from children with typical symptoms of fever and vesicular eruption on the palms and soles were obtained and examined with reverse transcription-semi-nested PCR approach adopted by Nix et al (14). We found that 35% (139/399) of the specimens were positive for enterovirus. Of the children with positive specimens, the ages ranged from 11 months to 6 years, with a mean age being 2.3 years. There were 75 boys and 64 girls, for a male to female ratio of 1.2–1. RT-semi-nested PCR products targeting the 350–400 bp (Fig. 1) fragment of the VP1 gene of 139 enterovirus-positive samples were directly sequenced and 99 sequences were obtained. All 99 sequences were assigned to 7 serotypes within the HEV-A species by comparing them with all enterovirus sequences in the GenBank database. EV71 and CVA16 were the frequently detected serotypes, accounting for 38.4% (38/99) and 35.4% (35/99) respectively, in the HEV-A species-positive cases. Other serotypes were responsible for the remaining 26.2%.
Figure 1.
Ethidium bromide-stained 1% agarose gel shows the results of VP1 RT-semi-nested PCR amplification of the RNA extracted from stool specimens. DL2000 DNA marker was included in lane M. Lane 1 to lane 24 represent RT-semi-nested the PCR-amplified 350–400 bp fragments of representative stool specimens ranging from 170EV-ZJ2010 to 207EV-ZJ2010.
To determine the genetic characteristics of EV71 and CVA16 strains circulating in their geographic location, phylogenetic analysis of these strains was based on the alignment of partial VP1 gene sequences. A total of 61 EV71 strains were used for phylogenetic analysis of the VP1 gene including the 38 EV71 strains identified in this study, 3 EV71 strains from 3 provinces in mainland China, and 20 international EV71 strains that represented all 11 known genotypes or subgenotypes (A, B1-B5, C1-C5) available from the GenBank (Fig. 2). All 38 EV71 strains from Zhejiang Province belonged to the subgenotype C4, which was similar to EV71 sequences isolated from 3 provinces in mainland China.
Figure 2.
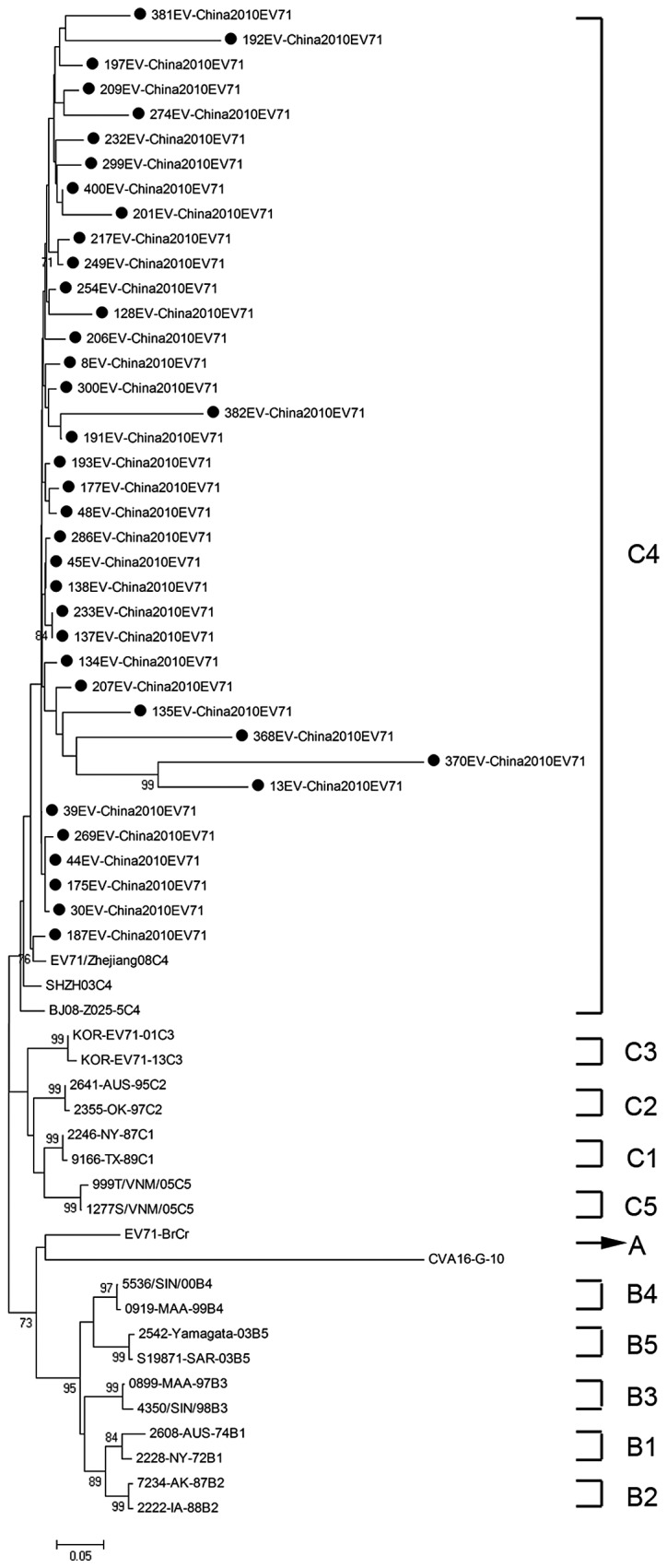
Phylogenetic tree showing the relationship between the Zhejiang EV71 strains (filled circles), and other EV71 sequences available from the GeneBank database. Genotypes are shown at the right of the tree, and the bootstrap values (percentage of 1.000 pseudoreplicates) are shown at the nodes of major clades. The scale at the bottom indicates the measurement of the relative phylogenetic distance. The prototype CAV16 strain (G-10) was used as an out-group.
We performed a similar analysis of the CVA16 detection in this study. A total of 46 CAV16 strains were used for phylogenetic analysis for the VP1 gene, including the 35 CAV16 strains identified in this study and 11 other CAV16 strains available from the GenBank (Fig. 3). It shows that 33 out of the 35 identified strains herein belong to clusters B2a and B2b which were found to be the prevalent viruses circulating in China and in neighboring countries and regions. However, the remaining 2 CAV16 strains could not be classified and formed a cluster distinct from genotype A and B.
Figure 3.
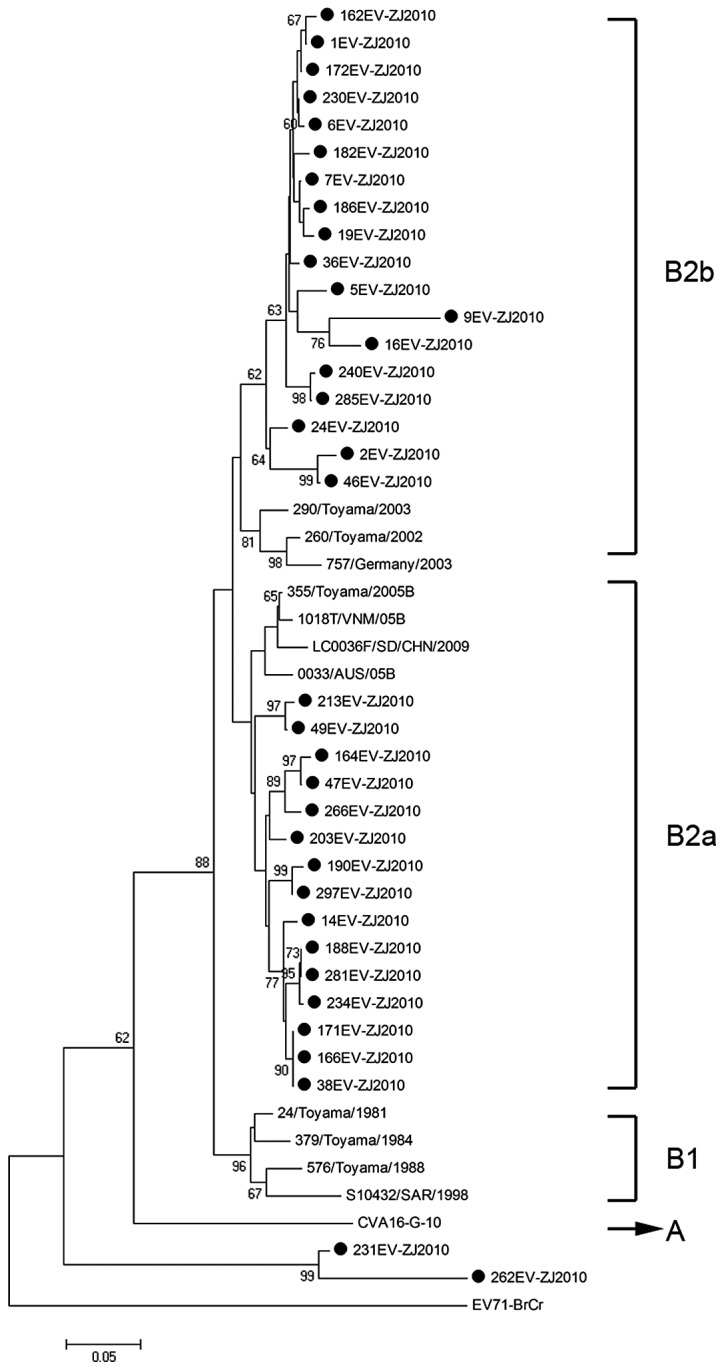
Dendrogram constructed based on the 350–400 bp nucleotides in the VP1 gene of 35 Zhejiang CAV16 strains and selected CAV16 sequences available from GeneBank database. Genotypes are shown at the right of the tree, and the bootstrap values (percentage of 1.000 pseudoreplicates) are shown at the nodes of major clades. The scale at the bottom indicates the measurement of the relative phylogenetic distance. The prototype EV71 strain (BrCr) was used as an out-group.
Discussion
In the present study, we investigated the prevalence of HFMD in Zhejiang Province between May and August 2010, and found that EV71 and CVA16 were the major etiological pathogens. Although there were no deaths caused by HFMD in the present study, EV71-related deaths from encephalitis, pulmonary edema and hemorrhage, have been previously noted (4,16,17), and fatal CVA16 infections have also been described in children with myocarditis (3) and in an adult with pneumonitis (18). Therefore, the identification of the major causal pathogen of HFMD is imperative.
Based on a study of molecular characteristics of the VP1 gene by Brown et al, 3 genotypes of EV71 (A, B, C) were identified (9). The EV71 prototype strain (BrCr-CA-70) isolated in California in 1970, is the sole member of the genotype A (4). At present, the B genotype is known to contain 5 subgenotypes (B1-B5), and the C genotype is known to contain another 5 subgenotypes (C1-C5). The genotype B was predominant in the US and Australia, during the period from 1972 to 1968, in Colombia in 1994, and in Malaysia in 1997 (9). Whereas the genotype C was predominant in east Asia, particularly in mainland China and Vietnam (11,19,20). Based on phylogenetic analysis, the EV71 strains identified in this study belong to the subgenotype C4 and show high homology with isolates from 3 representative provinces of the mainland China. Notably, the result was consistent with previous studies (7,20). The C4 subgenotype of EV71 has been in continuous circulation for at least 10 years since the first reported occurrence in ShenZhen city in 1998 (11).
The prototype G-10 strain of CVA16, first isolated in South Africa in 1951, is the sole member of genotype A (10). The genotype B can be further divided into 2 subgenotypes (B1, B2). CVA16 strains isolated in mainland China and the majority of international countries from 1981 to 2000 were all members of subgenotype B1. Since 1997, subgenotype B1 has gradually been replaced by subgenotypes B2, and CVA16 from clusters B2a and B2b has become the predominant virus circulating in mainland China and in neighboring countries to date (13). Based on phylogenetic analysis, the majority of CVA16 strains detected in the present study belong to clusters B2a and B2b, which was consistent with a previous study (13). Briefly, the genotypes of CVA16 identified in their study were similar to those detected in other Chinese provinces and countries. In addition, the other 2 CAV16 strains formed a new cluster distinct from genotype A and B, which indicated that CAV16 circulating in Zhejiang Province of China was genetically diverse and additional surveillance is necessary.
In conclusion, our study reveals that genetic characteristics of enteroviruses in China since 1998, reflect the pattern of endemic circulation of the subgenotype C4 to EV71 and clusters B2a and B2b within the genotype B2 to CVA16. We hypothesis that EV71 and CVA16 strains in China may derive from a respective ancestor associated with special geographic and climatic features allowing it to be sustained with little external effect. Therefore, future investigation of the respective ancestor of EV71 and CVA16 strains in China is warranted in order to take effective measures to prevent an HFMD outbreak.
Acknowledgements
This study was funded by grants from the National Basic Research Program of China (973 program) no. 2007CB513001, China's National Science and Technology Major Project no. 2008ZX10002-009, and a Qiu Shi Professorship from Zhejiang University to C.X. The authors thank Michael Brownstein for his critical reading of our manuscript.
References
- 1.Yamashita T, Ito M, Taniguchi A, Sakae K. Prevalence of coxsackievirus A5, A6, and A10 in patients with herpangina in Aichi Prefecture, 2005. Jpn J Infect Dis. 2005;58:390–391. [PubMed] [Google Scholar]
- 2.Yang F, Du J, Hu Y, et al. Enterovirus coinfection during an outbreak of hand, foot and mouth disease in Shandong, China. Clin Infect Dis. 2011;53:400–401. doi: 10.1093/cid/cir346. [DOI] [PubMed] [Google Scholar]
- 3.McMinn P, Stratov I, Nagarajan L, Davis S. Neurological manifestations of enterovirus 71 infection in children during an outbreak of hand, foot and mouth disease in Western Australia. Clin Infect Dis. 2001;32:236–242. doi: 10.1086/318454. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt NJ, Lennette EH, Ho HH. An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis. 1974;129:304–309. doi: 10.1093/infdis/129.3.304. [DOI] [PubMed] [Google Scholar]
- 5.Shimizu H, Utama A, Yoshii K, et al. Enterovirus 71 from fatal and nonfatal cases of hand, foot and mouth disease epidemics in Malaysia, Japan and Taiwan in 1997–1998. Jpn J Infect Dis. 1999;52:12–15. [PubMed] [Google Scholar]
- 6.Chang LY, Lin TY, Huang YC, et al. Comparison of enterovirus 71 and coxsackie-virus A16 clinical illnesses during the Taiwan enterovirus epidemic, 1998. Pediatr Infect Dis J. 1999;18:1092–1096. doi: 10.1097/00006454-199912000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Tan XJ, Wang HY, et al. An outbreak of hand, foot and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J Clin Virol. 2009;44:262–267. doi: 10.1016/j.jcv.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Huang YP, Lin TL, Kuo CY, et al. The circulation of subgenogroups B5 and C5 of enterovirus 71 in Taiwan from 2006 to 2007. Virus Res. 2008;137:206–212. doi: 10.1016/j.virusres.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Brown BA, Oberste MS, Alexander JP, Jr, Kennett ML, Pallansch MA. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol. 1999;73:9969–9975. doi: 10.1128/jvi.73.12.9969-9975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera D, Yusof MA, Podin Y, et al. Molecular phylogeny of modern coxsackievirus A16. Arch Virol. 2007;152:1201–1208. doi: 10.1007/s00705-006-0934-5. [DOI] [PubMed] [Google Scholar]
- 11.Li L, He Y, Yang H, et al. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J Clin Microbiol. 2005;43:3835–3839. doi: 10.1128/JCM.43.8.3835-3839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwai M, Masaki A, Hasegawa S, et al. Genetic changes of coxsackievirus A16 and enterovirus 71 isolated from hand, foot and mouth disease patients in Toyama, Japan between 1981 and 2007. Jpn J Infect Dis. 2009;62:254–259. [PubMed] [Google Scholar]
- 13.Zong W, He Y, Yu S, et al. Molecular phylogeny of coxsackievirus A16 in Shenzhen, China, from 2005 to 2009. J Clin Microbiol. 2011;49:1659–1661. doi: 10.1128/JCM.00010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nix WA, Oberste MS, Pallansch MA. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J Clin Microbiol. 2006;44:2698–2704. doi: 10.1128/JCM.00542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 16.Chang LY, Huang YC, Lin TY. Fulminant neurogenic pulmonary oedema with hand, foot and mouth disease. Lancet. 1998;352:367–368. doi: 10.1016/S0140-6736(98)24031-1. [DOI] [PubMed] [Google Scholar]
- 17.Lum LC, Wong KT, Lam SK, Chua KB, Goh AY. Neurogenic pulmonary oedema and enterovirus 71 encephalomyelitis. Lancet. 1998;352:1391. doi: 10.1016/s0140-6736(05)60789-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang JR, Tuan YC, Tsai HP, Yan JJ, Liu CC, Su IJ. Change of major genotype of enterovirus 71 in outbreaks of hand, foot and mouth disease in Taiwan between 1998 and 2000. J Clin Microbiol. 2002;40:10–15. doi: 10.1128/JCM.40.1.10-15.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu PV, Thao NT, Perera D, et al. Epidemiologic and virologic investigation of hand, foot and mouth disease, Southern Vietnam, 2005. Emerg Infect Dis. 2007;13:1733–1741. doi: 10.3201/eid1311.070632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhu Z, Yang W, et al. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand, foot and mouth disease in Fuyang City of China. Virol J. 2010;7:94. doi: 10.1186/1743-422X-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]


