Abstract
Throughout in utero development, the placenta plays a key role in controlling growth and development. The placenta acts not only as a gatekeeper of nutrient and waste exchange between mother and developing fetus, but also as a regulator of the intrauterine environment. Its functions can be influenced by the environment encountered throughout pregnancy, thereby altering the appropriate genetic programming needed to allow for appropriate fetal growth. Epigenetic alterations related to environmental exposures have been linked to aberrant fetal growth. DNA methylation, which is the best known DNA epigenetic modification, may provide an attractive mechanism linking environmental cues to placental pathology, with consequences for fetal growth and adult life. Alteration of the methylation patterns of genes expressed in the placenta has recently been found to modify gene expression and subsequently impair function of the placenta. Although there is strong evidence to demonstrate that the environment can affect the pattern of DNA methylation of the placenta during fetal development, a direct association between environmental conditions, methylation alterations and gene expression is difficult to confirm. DNA methylation in the placenta has mainly been investigated in the context of imprinted and non-imprinted genes transcription. Several environmental factors have also been assessed in regard to their association with changes to the epigenetic motives of embryonic and extraembryonic tissues and their impact on pregnancy outcome. In this review, we briefly present the available evidence regarding the role of DNA methylation patterns of the placenta on aberrant fetal growth.
Keywords: DNA methylation, epigenetics, fetal growth, H19, human placenta, IGF2, imprinted genes, IUGR
1. Introduction
Epigenetics constitutes an important mechanism capable of regulating gene transcription, linking early life’s events to adult morbidity. It entails heritable changes in chromatin that alter gene expression without altering the DNA sequence (1,2). It is the best-characterized epigenetic modification. Evidence suggests that DNA methylation is closely involved in the regulation of gene expression and that DNA methylation patterns can be distorted during the pathogenetic process of a disease (3). Findings of previous reports suggest that DNA methylation is altered during development and by environmental stress (4,5). However, the mechanisms by which these epigenetic effects are exerted remain to be clarified. In this review, we briefly present the available evidence regarding the role of DNA methylation patterns of the placenta on aberrant fetal growth.
2. DNA methylation
DNA methylation, which is accomplished by DNA- methyltransferases, occurs on the cytosine residues of CG (also designated CpG) dinucleotides. Enzymes known as DNA methyltransferases (DNMTs) catalyse the addition of a methyl group to the cytosine ring to form methyl cytosine, using S-adenosylmethionine as a methyl donor (6). DNA methyltransferase-1 (DNMT1) is the predominant mammalian DNA methylating enzyme responsible for the restoration of hemi-methylated sites to full methylation, termed maintenance methylation, which occurs after DNA replication. DNMT3A and DNMT3B are mainly involved in the methylation of new sites, known as de novo methylation (7). DNMT3L is postulated to play a regulatory role in DNA methylation without DNA methyltransferase activity in itself. In humans and other mammals, DNA modification occurs predominantly on cytosines that precede a guanosine in the DNA sequence (6). These dinucleotides can be clustered in small stretches of DNA, termed CpG islands, which are often associated with promoter regions. In 98% of the genome, CpGs are present approximately once per 80 dinucleotides. By contrast, CpG islands, which comprise 1–2% of the genome, are approximately 200 base pairs (bp) to several kb in length and have a frequency of CpGs approximately five times greater than the genome as a whole (8,9). Most CpG sites outside the CpG islands are methylated, suggesting a role in the global maintenance of the genome, while most CpG islands in gene promoters are unmethylated, which allows active gene transcription (6,10). When a CpG becomes methylated in a cell, it remains methylated in all its descendants (11). Generally, when a given stretch of cytosines in a CpG island located in the promoter region of a gene is methylated, that gene is silenced by methylation; such a CpG island would be termed ‘hypermethylated’. Conversely, when a given stretch of cytosines in a CpG island located in the promoter region of a gene is not methylated, that gene is not silenced by methylation; the CpG island in this case would be ‘hypomethylated’ (12). Methylation of promoters inhibits their recognition by transcription factors and RNA polymerase, as methylated cytosines preferentially bind to a protein known as methyl cytosine binding protein, or MeCP. When a promoter region normally recognized by an activating transcription factor, is methylated, its transcription is inhibited (9).
3. DNA methylation in the developing embryo and placenta
Methylation of gene promoters is probably one of the foremost mechanisms responsible for cell differentiation during embryogenesis: the transcription of unwanted genes is eliminated by methylation of their promoters (13). As oocytes and spermatozoa are more differentiated than the pluripotent cells of the early embryo, the DNA of morula (16-cell embryo, third day post-conception) undergoes global demethylation. CpGs are demethylated on a large scale, thus reactivating the near-totality of the genome (a few genes escape this demethylation, e.g., the genes subject to genomic imprinting). Subsequently, as cells start differentiating, the gene promoters involved in this differentiation become methylated according to a strict sequence depending on each cell type (14).
On fertilisation a rapid paternal-specific asymmetric loss of methylation is observed (15,16). This process occurs in the absence of transcription or DNA replication and is termed active demethylation. Thereafter, there is a step-wise decline in methylation until the morula stage (17,18). The initiation of the de novo methylation occurs after the fifth cell cycle and coincides with the time of the first differentiative event. The establishment of the first two cell lineages results in a significant asymmetry. The inner cell mass (ICM), which gives rise to all the tissues of the adult, becomes hypermethylated, while the trophectoderm (TE), which forms most of the structure of the placenta, is hypomethylated (17,18). This differential methylation is maintained and reflected in highly methylated somatic tissues and the distinctively hypomethylated extra-embryonic tissues of the placenta. This epigenetic inequality with higher overall DNA methylation levels in the embryo compared with the placenta is maintained throughout gestation (18).
4. Imprinting
Despite the genome-wide decline in DNA methylation, certain sequences remain refractory to the general demethylation during preimplantation development. Imprinted genes escape this epigenetic reprogramming (15). They are protected from demethylation because it is crucial that the parental imprints are preserved in the developing embryo (19).
Genomic imprinting refers to silencing of one parental allele in the zygotes of gametes leading to monoallelic expression of these genes in the offspring. During the process of imprinting, the male and female germ line confer a gender-specific mark (imprint) on certain chromosomal regions (20). Only one allele of the imprinted genes, the maternal or paternal, can be active and expressed. Each cluster is controlled by an imprinting control region (ICR) that usually contains a stably maintained or developmentally changing Differentially Methylated Region (DMR) (21,22). Genomic imprinting arose during the mammalian evolution (approximately 150 million years ago) and may be associated with the evolution of intrauterine development that requires the formation of a placenta (20).
The prevailing hypothesis on the evolution of genomic imprinting is the ‘conflict hypothesis’ theory. This theory suggests that paternally expressed genes strongly favor using maternal resources to benefit offspring, while maternally expressed genes attempt to preserve such maternal resources and thus, are in direct conflict with one another. Many imprinted genes are involved in fetal development and growth, and some affect behaviour (20,23). Imprinting appears to be particularly important for placental development (24,25). Knockout studies of several paternally or maternally imprinted genes result in intrauterine growth restriction (IUGR) and smaller placental size or the overgrowth and hyperplasia of the placenta, respectively (25–27). Certain maternal genes are required for proper development of the embryo, whereas extraembryonic tissues depend on the presence of active paternal genes. Approximately 60 genes have been shown to be imprinted in humans, two thirds of which are paternally expressed (maternally imprinted) and one third maternally expressed (paternally imprinted) (28).
5. DNA methylation in the placenta
Throughout in utero development, the placenta plays an important role in controlling growth and development through the transfer of nutrients and waste, and in protecting the fetus from insults (29). Findings of recent studies have shown that placental genetic and epigenetic profiles may serve as markers of the intrauterine and extrauterine environment (30–32). Embryonic and fetal growth depends on genetic, epigenetic and environmental factors, and the process is the result of the interaction between these factors. Approximately 7–9% of live-born infants have a birth weight below the 10th percentile. Intrauterine growth restriction describes a decrease in the fetal growth rate that prevents an infant from obtaining his or her complete growth potential (33). IUGR infants are small for gestational age (SGA) if their birth weight measures <10 to 3% using standard growth curves (34,35). Therefore, the terms IUGR and SGA are related but not synonymous. The IUGR is a pathological condition, whereas SGA may reflect a normal pattern in a given population. The placenta forms the interface between the fetal and maternal circulations. For this reason, fetal disease, maternal disease, primary placental disease, and extrinsic factors could all interfere with the efficiency of nutrient and waste exchange and result in growth restriction (36,37). Fetal growth restriction is a physical sign rather than a single disease. Improper placental function accounts for the majority of IUGR cases.
Epigenetic modification in the placenta may provide an attractive mechanism linking environmental cues to placental pathology, with consequences for fetal growth and adult life. Accumulating evidence suggests that the maternal nutritional status is capable of altering the epigenetic state of the fetal genome and imprinted gene expression. Epigenetic alterations in early embryos may be carried forward to subsequent developmental stages (38). The placenta has been reported to present high variability in overall DNA methylation compared to other tissues (39), probably in response to its role in mediating the conflicting demands of mother and fetus (40). Methylation patterns of several genes (imprinted and non-imprinted) in the placenta have been investigated in an attempt to elucidate the exact role of epigenetic modifications on fetal growth. Administration of a DNA methyltransferase inhibitor to pregnant rats at different gestational ages resulted in significantly smaller placentas and histological evaluation showed the labyrinthine part of the placenta to be severely reduced (41). In a similar study, a lack of the labyrinth layer was observed with a strong proliferative activity of the cells in the basal layer or complete disruption of the placental structure (42). Furthermore, administration of the same agent in human choriocarcinoma-derived cell lines, resulted in disrupted trophoblast migration (43).
DNA methylation and gene transcription
Much of the recent research on placental epigenetics has focused on imprinted genes that are known to affect growth, such as insulin-like growth factor 2 (IGF2). IGF2 and H19 are two oppositely expressed imprinted genes located adjacent to each other at 11p15.5 that share the same transcription regulatory epigenetic mechanisms and have an important role in feto-placental development. The DMR upstream of H19, which harbors sequences known to bind to the zinc finger protein CTCF, if methylated on the paternal chromosome prevents binding with CTCF and allows the IGF2 promoter to assess enhancers located downstream of H19, thereby expressing IGF2 (Fig. 1). On the maternal chromosome the non-methylated H19 DMR is bound to CTCF, thus insulating the IGF2 promoter from the 3′ enhancers and allowing the H19 promoter unimpeded access to the enhancers. Maternal H19 is subsequently transcribed (44). IGF2 is highly expressed in normal mouse and human placenta and affects the functional capacity of the placenta to transfer nutrients to the fetus as well as placental size (45,46). It is expressed in most tissues only from the paternal allele, with the maternal allele being transcriptionally silent. The maternally expressed H19 gene itself does not code a protein, but the RNA has growth-suppressing functions, potentially through inhibiting the translation of IGF2 RNA (47).
Figure 1.
Schematic representation of the imprinted regulation of the IGF2/H19 cluster. Maternal and paternal chromosomes are indicated. Vertical black bars are exons from the expressed allele; red vertical bars are exons from the silenced allele. The arrows show the direction of transcription (white arrows show active transcription from IGF2 P0, P2, P3, P4 and the H19 promoter; black arrow shows the non-imprinted P1 IGF2 promoter). Ovals represent the enhancers. The long arrowed lines indicate enhancer activity. Three known Differentially Methylated Regions (DMR) are presented as sets of triangles. Black triangles are methylated DMRs and white triangles are unmethylated regions. DMR upstream of H19, which harbors sequences known to bind to the zinc finger protein CTCF, if methylated on the paternal chromosome prevents binding with CTCF, allowing the IGF2 promoter to assess enhancers located downstream of H19, thereby expressing IGF2. On the maternal chromosome, the non-methylated H19 DMR is bound to CTCF, thus insulating the IGF2 promoter from the 3′ enhancers and allowing the H19 promoter unimpeded access to the enhancers. Maternal H19 is subsequently transcribed.
Specific regions of differential DNA methylation are regarded as critical for the correct allelic expression of IGF2/H19. Complete loss of methylation of the H19 promoter is reported at all stages of placental development (48). Hypomethylation of IGF2 and H19 promoters as well as the ICR of those genes has been reported in placentas derived from pregnancies complicated with fetal growth restriction (49–51). Since ICR controls the expression of both H19 and IGF2 genes, which are known growth modulators, aberrant methylation in that region may be a potential link between epigenetic modifications and abnormal fetal and placental growth.
Hypomethylation of the IGF2 and H19 promoters would indicate lower transcription levels of these genes in placentas from pregnancies complicated with fetal growth restriction. Underexpression of IGF2 is a repeated event in growth-restricted placentas and it is postulated to be associated with reduced diffusional capacity of the placenta, which in turn affects fetal growth (52). However, hypomethylation of the IGF2 promoter is in contrast to the reduced transcription levels observed in placentas derived from pregnancies with abnormal fetal growth. This discrepancy suggests that there is no direct correlation between methylation and imprinted gene expression in the placenta, and other mechanisms may be involved in this sequence of molecular events. According to recent studies, methylation of the IGF2/H19 promoters is not prerequisite for the regulation of the imprinting domain that controls transcription of the two genes in human placenta (49,53). Moreover, hypomethylation of the same regions does not have an impact on the expression pattern of IGF2 and H19 (53).
Reduced methylation levels of the region that controls the imprinting of IGF2/H19 (ICR) have been reported in placentas derived from pregnancies with poor fetal growth and those complicated with preeclampsia (49–51,54) (Table I). ICR is hypomethylated leading to the repression of IGF2 expression in approximately one-third of patients with Silver Russell syndrome (SRS), a syndrome associated with pre- and post-natal growth deficiency (57). However, where ICR is hypermethylated it leads to an increase in IGF2 expression in some cases of pre- and post-natal overgrowth diagnosed as Beckwith-Wiedemann syndrome (BWS) (58). Methylation at ICR has been shown in a number of studies to be particularly responsive to environmental factors such as culture media (59), environmental toxins (60,61), and prenatal ethanol exposure (61). The reduced methylation levels associated with IUGR may reflect an adaptive process serving to adjust placental and fetal growth in response to poor placental perfusion.
Table I.
Methylation patterns of genes expressed in the placenta of pregnancies that delivered a growth-restricted or small for gestational age neonate.a
| Gene | Imprinting | Tissue | Methylation | Expression | Author/Year (Refs.) |
|---|---|---|---|---|---|
| IGF2/H19 (ICR) | Yes | IUGR and control placentas | Hypomethylation | Decreased (IGF2) | Bourque et al, 2010 (54) |
| IGF2/H19 (ICR) | Yes | IUGR + PET and control placentas | No difference | ||
| CDKN1C (ICR) | Yes | IUGR and control placentas | No difference | No difference | |
| H19 | Yes | IUGR and control placentas | No difference | ||
| CDKN1C | Yes | IUGR and control placentas | No difference | ||
| PEG10 | Yes | IUGR and control placentas | No difference | ||
| PLAGL1 | Yes | IUGR and control placentas | No difference | ||
| SNRPN | Yes | IUGR and control placentas | No difference | Increased | |
| MEST | Yes | IUGR and control placentas | No difference | Increased | |
| PHLDA2 | Yes | IUGR and control placentas | Increased | McMinn et al, 2006 (55) | |
| MEST | Yes | IUGR and control placentas | No difference | Decreased | |
| SERPINA3 | No | IUGR and control placentas | Hypomethylation | Increased | Chelbi et al, 2007 (56) |
| SERPINA3 | No | IUGR+ preeclampsia and control placentas | Hypomethylation | Increased | |
| IGF2 | Yes | SGA placentas and neonatal blood | Hypomethylation | Guo et al, 2008 (49) | |
| H19 | Yes | SGA placentas and neonatal blood | Hypomethylation | ||
| IGF2 | Yes | SGA and control placentas | - | Decreased | |
| H19 | Yes | SGA and control placentas | - | No difference | |
| H19 | Yes | IUGR and control placentas | Hypomethylation | Increased | Koukoura et al, 2011 (51) |
| IGF2/H19 (ICR) | Yes | IUGR and control placentas | No difference | ||
| IGF2 | Yes | IUGR and control placentas | Hypomethylation | Decreased | Koukoura et al, 2001 (50) |
Methylation levels between different samples were assessed and the findings presented (methylation and expression) refer to the values observed in the pathologic tissues in comparison to the controls.
ICR, imprinting control center; IUGR, intrauterine growth restriction; SGA, small for gestational age; PET, preeclampsia.
Hypomethylation of the imprinted gene promoters is not a universal finding in cases where fetal growth is compromised. In their study, Lambertini et al demonstrated a slight tendency towards hypermethylation of the DMRs of all known imprinted genes identified to be expressed in growth-restricted placentas (62). These authors suggested that differential methylation changes in growth-restricted placentas occur throughout the genomic regions, encompassing genes actively expressed in the placenta. Analysis of other imprinted genes from the placentas of pregnancies complicated with IUGR, revealed a lack of altered DNA methylation at their imprinting centers (55), although they demonstrated differences in their transcription levels (Table I).
The contradictory results that stem from different studies regarding imprinted gene methylation patterns in the placenta highlight the already reported DNA methylation variation at the imprinted genes. Inter-individual, tissue-specific variation in DNA-methylation level is widespread in the human genome, with implications on phenotypic variation and disease (63). Several genes have been described to exhibit this polymorphic pattern of DNA methylation in the human placenta (64). Therefore, the exact epigenetic defects in the human placenta, which control imprinted gene expression and affect fetal development, remain to be determined.
The hypothesis that variation in the DNA methylation profile of human term placenta can serve as a marker of growth has been confirmed by Banister et al who demonstrated a pattern of methylation of 22 critical loci in human term placentas. Specific methylation alterations of these genes were highly predictive of IUGR or SGA (65). A significant association has also been shown between the differential methylation of the glucocorticoid receptor gene in the placenta and Large for Gestational Growth (LGA) infants (31). Placental gene serine protease inhibitor A3 (SERPINA3), whose expression is known to be affected by placental pathologies such as preeclampsia, has been shown to exhibit hypomethylation of its promoters in IUGR placentas. Hypomethylation coincided with increased transcription levels of the same gene in placentas derived from IUGR pregnancies as well as preeclamptic IUGR cases. The hypomethylated CpGs were found to be located at putative binding sites for developmental and stress response (hypoxia and inflammation) factors (56).
Recent studies have demonstrated significant associations between infant growth, in utero exposures and repetitive element methylation in placental tissue (66). These DNA repetitive elements are made up of interspersed and tandem repeats and comprise at least half of the human genome (67). Interspersed repeats are composed of long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). A significant correlation was found between methylation levels and the birthweight percentile. A 10% methylation increase in LINE-1 mean levels caused the birthweight percentile to significantly increase by 9.7. Similarly, a 10% methylation increase in AluYb8 mean levels caused the birthweight percentile to significantly increase by 14.5. Furthermore, mean AluYb8 levels differed significantly due to maternal tobacco use during pregnancy; whereas, mean LINE-1 levels only significantly differed due to maternal alcohol use during pregnancy. Authors of these studies concluded that the alterations may reflect underlying functional epigenetic alterations to genes important in placental growth and development.
Previous investigations emphasized marked similarities between the proliferative, migratory and invasive properties of placental cells and those of cancer cells. Alterations in the expression of tumour suppressor gene expression profiles have been identified in placentas from preeclamptic pregnancies (68). A distinct pattern of tumour-associated methylation, linking a coordinated series of epigenetic silencing events, similar to those associated with some tumours, in the distinct, features of normal human placental invasion and function has been observed (69). A genome-wide methylation analysis revealed reduced methylation levels of trophoblastic tissues derived from chorionic villous sampling during the first trimester of pregnancy. The highly proliferative and invasive nature of early placenta may explain this relative hypomethylation as a requirement for an intensively active transcriptional state. Trophoblasts and cancer cells may use common epigenetic modifications to facilitate their proliferative, migratory and invasive properties (70). However, no data are currently available that may indicate a correlation between the epigenetic modification of tumor-associated genes and fetal growth. In a study where the methylation status of genes regulating vitamin D bioavailability and activity in the placenta was investigated, the CYP24A1 gene was methylated in human placenta, purified cytotrophoblasts, and primary and cultured chorionic villus sampling tissue, whereas vitamin D receptor (VDR) and CYP27B1 genes were non-methylated. All three genes were hypermethylated in choriocarcinoma cell lines, emphasizing the role of vitamin D deregulation in this type of cancer. The promoter methylation of the CYP24A1 gene, directly downregulated basal promoter activity and abolished vitamin D-mediated feedback activation. This event resulted in maximizing active vitamin D bioavailability at the fetomaternal interface suggesting a role in pregnancy progression (71).
Environmental impact on DNA methylation in the placenta
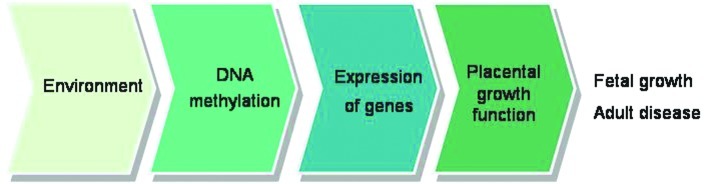
There is a critical window, at some stage in intrauterine life, during which balanced homeostasis is essential for normal fetal growth and development. Adverse effects during that period alter the structure and function of distinct cells, organ systems or homeostatic pathways, thereby ‘programming’ the individual for an increased risk of developing diseases in adult life. Placental phenotype is responsive to environmental conditions and may help predict the risk of adult disease programmed in utero. The placenta responds to and is potentially marked in an epigenetic context by environmental insults, suggesting that the placental epigenome serves, not only as a record of in utero exposure, but also as a mediator and/or modulator of disease pathogenesis.
Accumulating evidence suggests that the maternal nutritional status is capable of altering the epigenetic state of the fetal genome and imprinted gene expression. Ethanol-exposed midgestation placentas and embryos were severely growth retarded when compared with the controls. The relationship between placental weight and ethanol treatment suggested that this was partially dependent on DNA methylation at the CCCTC-binding factor (CTCF) site on the paternal allele in placentas (61). Preimplantation embryo culture has been shown to affect the methylation and expression of imprinted genes in several animal models. One particularly favoured explanation for the association between the environmental impact in early life and long-term physiological functions lies with the epigenetic modification of gene expression (71). Although there is strong evidence to demonstrate that the environment affects the pattern of DNA methylation during fetal development, the direct association between environmental conditions, methylation alterations and gene expression is difficult to verify (Fig. 2).
Figure 2.
A potential link between environmental factors and aberrant fetal growth in the context of methylation modifications in the placenta. Environmental factors such as assisted reproduction techniques, alcohol, chemical agents, tobacco and malnutrition are capable of altering the epigenetic profile of several imprinted and non-imprinted genes expressed in the placenta. This event may influence the proper structure or function of the trophoblast and in turn affect fetal growth; however, the exact sequence of events requires further elucidation.
6. Conclusion
Numerous links have been made between infant growth restriction and specific epigenetic alterations, including changes to the gene imprinting status and to DNA methylation. Fetal growth is affected by the proper function of many imprinted and non-imprinted genes which are subject to epigenetic control through methylation of their promoters. DNA methylation has a critical role in placenta development, and alterations to its methylation pattern can lead to adverse placental morphology and birth outcome. However, since DNA methylation represents a delicate molecular mechanism that is easily affected by various factors, data that associate methylation patterns with placental pathology or abnormal fetal growth, should be interpreted with caution.
References
- 1.Nakao M. Epigenetics: interaction of DNA methylation and chromatin. Gene. 2001;278:25–31. doi: 10.1016/s0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 2.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 3.Van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007;64:1531–1538. doi: 10.1007/s00018-007-6526-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 5.Anway MD, Skinner MK. Epigenetic programming of the germ line: effects of endocrine disruptors on the development of transgenerational disease. Reprod Biomed Online. 2008;16:23–25. doi: 10.1016/s1472-6483(10)60553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 7.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 8.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 9.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 12.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62:78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oligny LL. Human molecular embryogenesis: an overview. Pediatr Dev Pathol. 2001;4:324–343. doi: 10.1007/s10024001-0033-2. [DOI] [PubMed] [Google Scholar]
- 14.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 15.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 16.Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14:93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- 17.Dean W, Santos F, Stojkovic M, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 20.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 21.Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 24.Constancia M, Hemberger M, Hughes J, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 25.Frank D, Fortino W, Clark L, et al. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci USA. 2002;99:7490–7495. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Kobayashi T, Kanayama N. p57(Kip2) regulates the proper development of labyrinthine and spongiotrophoblasts. Mol Hum Reprod. 2000;6:1019–1025. doi: 10.1093/molehr/6.11.1019. [DOI] [PubMed] [Google Scholar]
- 28.Glaser RL, Ramsay JP, Morison IM. The imprinted gene and parent-of-origin effect database now includes parental origin of de novo mutations. Nucleic Acids Res. 2006;34(Database issue):D29–D31. doi: 10.1093/nar/gkj101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins JC, Marsit CJ, Padbury JF, Sharma SS. Endocrine disruptors, environmental oxygen, epigenetics and pregnancy. Front Biosci (Elite Ed) 2011;3:690–700. doi: 10.2741/e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci USA. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filiberto AC, Maccani MA, Koestler D, et al. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6:566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 33.Resnik R. Intrauterine growth restriction. Obstet Gynecol. 2002;99:490–496. doi: 10.1016/s0029-7844(01)01780-x. [DOI] [PubMed] [Google Scholar]
- 34.Brodsky D, Christou H. Current concepts in intrauterine growth restriction. J Intensive Care Med. 2004;19:307–319. doi: 10.1177/0885066604269663. [DOI] [PubMed] [Google Scholar]
- 35.Lubchenco LO, Hansman C, Dressler M, Boyd E. Intrauterine growth as estimated from liveborn birth-weight data at 24 to 42 weeks of gestation. Pediatrics. 1963;32:793–800. [PubMed] [Google Scholar]
- 36.Snijders RJ, Sherrod C, Gosden CM, Nicolaides KH. Fetal growth retardation: associated malformations and chromosomal abnormalities. Am J Obstet Gynecol. 1993;168:547–555. doi: 10.1016/0002-9378(93)90491-z. [DOI] [PubMed] [Google Scholar]
- 37.Odegard RA, Vatten LJ, Nilsen ST, Salvesen KA, Austgulen R. Preeclampsia and fetal growth. Obstet Gynecol. 2000;96:950–955. [PubMed] [Google Scholar]
- 38.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Houseman EA, Christensen BC, Yeh RF, et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics. 2008;9:365. doi: 10.1186/1471-2105-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- 41.Vlahovic M, Bulic-Jakus F, Juric-Lekic G, Fucic A, Maric S, Serman D. Changes in the placenta and in the rat embryo caused by the demethylating agent 5-azacytidine. Int J Dev Biol. 1999;43:843–846. [PubMed] [Google Scholar]
- 42.Serman L, Vlahovic M, Sijan M, et al. The impact of 5-azacytidine on placental weight, glycoprotein pattern and proliferating cell nuclear antigen expression in rat placenta. Placenta. 2007;28:803–811. doi: 10.1016/j.placenta.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Rahnama F, Shafiei F, Gluckman PD, Mitchell MD, Lobie PE. Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology. 2006;147:5275–5283. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- 44.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65(Suppl 3):50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- 46.Randhawa R, Cohen P. The role of the insulin-like growth factor system in prenatal growth. Mol Genet Metab. 2005;86:84–90. doi: 10.1016/j.ymgme.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 47.Petry CJ, Ong KK, Barratt BJ, et al. Common polymorphism in H19 associated with birthweight and cord blood IGF-II levels in humans. BMC Genet. 2005;6:22. doi: 10.1186/1471-2156-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jinno Y, Ikeda Y, Yun K, et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat Genet. 1995;10:318–24. doi: 10.1038/ng0795-318. [DOI] [PubMed] [Google Scholar]
- 49.Guo L, Choufani S, Ferreira J, et al. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol. 2008;320:79–91. doi: 10.1016/j.ydbio.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 50.Koukoura O, Sifakis S, Soufla G, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med. 2011;28:481–487. doi: 10.3892/ijmm.2011.754. [DOI] [PubMed] [Google Scholar]
- 51.Koukoura O, Sifakis S, Zaravinos A, et al. Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta. 2011;32:51–57. doi: 10.1016/j.placenta.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 52.Sibley CP, Coan PM, Ferguson-Smith AC, et al. Placental-specific insulin-like growth factor 2 (IGF2) regulates the diffusional exchange characteristics of the mouse placenta. Proc Natl Acad Sci USA. 2004;101:8204–8208. doi: 10.1073/pnas.0402508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabano S, Colapietro P, Cetin I, et al. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics. 2010;5:313–324. doi: 10.4161/epi.5.4.11637. [DOI] [PubMed] [Google Scholar]
- 54.Bourque DK, Avila L, Penaherrera M, von Dadelszen P, Robinson WP. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta. 2010;31:197–202. doi: 10.1016/j.placenta.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 55.McMinn J, Wei M, Schupf N, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Chelbi ST, Mondon F, Jammes H, et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension. 2007;49:76–83. doi: 10.1161/01.HYP.0000250831.52876.cb. [DOI] [PubMed] [Google Scholar]
- 57.Gicquel C, Rossignol S, Cabrol S, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 58.Weksberg R, Shuman C, Smith AC. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137C:12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 59.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 60.Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and IGF2. Biol Reprod. 2004;70:1790–1797. doi: 10.1095/biolreprod.103.025387. [DOI] [PubMed] [Google Scholar]
- 61.Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the H19 imprinting control region. Biol Reprod. 2009;81:618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- 62.Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3:261–269. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monk D, Sanches R, Arnaud P, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 64.Yuen RK, Avila L, Penaherrera MS, et al. Human placental-specific epipolymorphism and its association with adverse pregnancy outcomes. PLoS One. 2009;4:e7389. doi: 10.1371/journal.pone.0007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6:920–927. doi: 10.4161/epi.6.7.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, et al. In Utero Exposures, Infant Growth, and DNA Methylation of Repetitive Element and Developmentally Related Genes in Human Placenta. Environ Health Perspect. 2011 doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity (Edinb) 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 68.Heikkilä A, Tuomisto T, Häkkinen SK, Keski-Nisula L, Heinonen S, Yla-Herttuala S. Tumor suppressor and growth regulatory genes are overexpressed in severe early-onset preeclampsia-an array study on case-specific human preeclamptic placental tissue. Acta Obstet Gynecol Scand. 2005;84:679–689. doi: 10.1111/j.0001-6349.2005.00814.x. [DOI] [PubMed] [Google Scholar]
- 69.Novakovic B, Rakyan V, Ng HK, et al. Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol Hum Reprod. 2008;14:547–554. doi: 10.1093/molehr/gan046. [DOI] [PubMed] [Google Scholar]
- 70.Ferretti C, Bruni L, Dangles-Marie V, Pecking AP, Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 2007;13:121–141. doi: 10.1093/humupd/dml048. [DOI] [PubMed] [Google Scholar]
- 71.Novakovic B, Sibson M, Ng HK, et al. Placenta-specific methylation of the vitamin D 24-hydroxylase gene: implications for feedback autoregulation of active vitamin D levels at the fetomaternal interface. J Biol Chem. 2009;284:14838–14848. doi: 10.1074/jbc.M809542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]