Abstract
Epidemiological studies indicate that infections by certain types of human papillomaviruses (HPVs) are causally linked to the development of cervical cancer. It is also known that HPV infections alone do not cause progression to cervical cancer, as additional genetic changes such as loss of distinct chromosomal regions, inactivation of tumor-suppressor genes and activation of oncogenes must also occur in order for malignant transformation to take place. In the present study, 78 patients diagnosed with cervical cancer and 36 cervical cancer-free cases (control) were analyzed for high-risk HPV genotypes (16 and 18) by polymerase chain reaction (PCR). Loss of heterozygosity (LOH) of the retinoblastoma gene (Rb) at two polymorphic intronic sites (intron 1 and 17) and the p53 polymorphism in codon 72 were detected by RFLP and allele-specific PCR, respectively. HPV 16 and 18 were found at frequencies of 93.6 and 8.3% in the cervical cancer and control samples, respectively. LOH was detected in 63% of patients in intron 1 and/or intron 17. p53 allele frequency for Arg/Arg was 43.6% (34/78), for Arg/Pro 37.2% (29/78) and for Pro/Pro 19.2% (15/78). The relative risk (RR) of LOH and Arg/Arg alone was 1.7 and 1.1, respectively, while the combined RR for Rb LOH and p53 Arg/Arg was 2.5. The present study showed a significant association of the chromosomal allelic loss of Rb in Sudanese cervical cancer patients, while no such association was observed with other parameters, such as clinical stage and degree of differentiation; hence, it cannot be a determinant of tumor behavior in cervical carcinoma. Although the p53 arginine allele is itself an important risk factor for cervical cancer, the combined risk with LOH of Rb, which appears to be greater, might indicate a possible epistatic effect of the two genes/polymorphisms.
Keywords: cervical cancer, human papillomavirus, Sudan, p53, retinoblastoma gene
Introduction
Strong epidemiologic evidence indicates that infections by certain types of human papillomaviruses (HPVs) are causally linked to cervical cancer development (1). Among the high risk HPV types, HPV 16 and 18 are recognized as the main causes of invasive cervical cancer and its precursor lesions. Little is known, however, about HPV prevalence in patients with cervical cancer and healthy women in Sudan, a country with a high incidence of cervical cancer.
In cervical cancer patients, alterations in many tumor-suppressor genes have been reported (2). Among them, the two well-characterized tumor-suppressor genes, the retinoblastoma gene (Rb) on chromosome 13q and the p53 on chromosome 17p, are both frequently inactivated in a broad range of human cancer types (3).
Data concerning mutations of the Rb gene are controversial with respect to cervical carcinoma. Most commonly, pRb is inactivated by mutation of its regulators. pRb is known to be inactivated by virally encoded oncoproteins such as E7 in high-risk papillomaviruses (4,5). However, the genomic changes associated both with mutations of the Rb gene and HPV infection have not been fully established, mainly due to the limited number and low informative capability of the markers used.
A single nucleotide polymorphism in the p53 gene resulting in the substitution of arginine (Arg) by proline (Pro) at codon 72 was identified and shown to alter the primary structure of the p53 protein (6). Biochemical and functional differences between the two p53 forms have been identified (7). In light of the structural differences, it was hypothesized and subsequently demonstrated experimentally that the Arg form of the p53 protein was in fact more susceptible to binding and degradation by the HPV-E6 oncoprotein than the Pro form (8). In the same study it was found that women with invasive cervical cancer were more likely to be homozygous for Arg at codon 72 compared to controls, suggesting that the Arg/Arg genotype may confer greater susceptibility to cervical cancer. Numerous subsequent investigations of the possibility of an increased risk of cervical cancer associated with the Arg/Arg genotype have been conducted on various populations with controversial findings (9–14).
In summary, the two genetic alterations we analyzed in this study have emerged as significant factors in the pathogenesis and progression of many types of tumors and are therefore likely to provide relevant information to assess the risk of cervical cancer (15).
Materials and methods
Seventy-eight cervical cancer tissues and matched peripheral blood samples were collected from patients with cervical cancer. Patients were randomly selected regardless of age, ethnicity or duration of the disease. Also, 36 formalin-fixed paraffin-embedded tissues (PETs) of non-cancerous samples were used as control for the presence of HPV. The control for the p53 study was previously published data of 253 Sudanese individuals from different ethnic groups (17). DNA was extracted from fresh tissues using TRIzol reagent protocol, while DNA from PETs was extracted using the JETQUICK kit (Sigma-Aldrich). Histologically, the samples were diagnosed for two types of cellular cancer, squamous cell carcinoma (SCC) and adenocarcinoma (ADCA). The grades and stages were determined.
Detection of HPV-DNA sequence
The following primers were used: for HPV 16 (D, 5′-TTTTGGGTTACACATTTACAA-3′ and R, 5′-TGTCTGCTTTTATACTAACCG-3′); for HPV 18 (D, 5′-GACACCTTAATGAAAAACGACG-3′ and R, 5′-CGT CGTTGGAGTCGTTCCTG-3′). Polymerase chain reaction (PCR) was performed in a total volume of 25 μl. The Master mix contained 1 μl of 10 mM dNTP mix (2.5 mM dATP, 2.5 mM dGTP, 2.5 mM dCTP and 2.5 mM dTTP), 1.5 μl of 25 mM MgCl2, 2.5 μl of 10X PCR buffer [10 mM Tris-HCl (pH 8.3), 50 mM KCl], 1 μl of Taq polymerase (~1 U), 1.5 μl forward primer and 1.5 μl reverse primer. The final volume was set to 25 μl per reaction mix with ddH2O. The amplification was carried out by an initial denaturation stage at 95°C for 5 min, then 35 cycles of 95°C for 1 min and 50°C for 1 min, 55°C for 1 min and a final extension for 10 min at 72°C followed by a final elongation stage at 72°C for 5 min.
PCR for p53 polymorphisms (Arg/Pro)
PCR was carried out for amplification of codon 72 in the p53 gene using the following sets of proline primers: D, 5′-GCCAGAGGC TGCTCCCCC-3′ and R, 5′-CGTGCAAGTCACAGACTT-3′; and arginine primers: D, 5′-TCCCCCTTGCCGTCCCAA-3′ and R, 5′-CTGGTGCAGGGGCCACGC-3′. The cycling profile, as well as agarose gel electrophoresis, followed a previous protocol by Soulitzis et al (16).
PCR-RFLP genotyping for Rb intron 1 and 17
Restriction fragment length polymorphism analysis (RFLP) was utilized in this study to screen for loss of heterozygosity (LOH) in two introns in the Rb gene (intron 1 and 17) using the primers: for intron 1 D, 5′-CAGGACAGCGGCCCGGAG-3′ and R, 5′-CTGCAGACGCTCCGCCGT-3′; and for intron 17 D, 5′-TCCCACCTCAGCCTCCTTAG-3′ and R, 5′-GTAGGCCAAGAGTGGCAGCT-3′. The PCR protocol for amplification of the Rb intron 1 and 17 was as follows: initial denaturation was at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 1 min and annealing at 58°C for the primers of intron 1 and 65°C for the primers of intron 17 for 1 min. One additional cycle for the elongation step at 72°C for 5 min completed the procedure. Five microliters of each PCR product was added to 4 μl of loading dye and loaded onto a 2% agarose gel. DNA molecular weight marker of 1 kb ladder was loaded onto the same gel. The gel was examined under UV light to visualize the DNA. The size of the bands was measured according to the migration of the DNA ladder. Five microliters of the intronic polymorphism products was digested overnight each with the appropriate restriction enzymes – BamH1 for intron 1 and Xba1 for intron 17 (Fermentas Life Sciences). The resulting fragments were separated using 2% agarose gel electrophoresis. The BamHI and Xba1 enzymes digest PCR products with mutated alleles.
Results
Histopathology
Histopathology revealed that 82.1% (64/78) of the cervical cancer cases were SCC and 17.9% (14/78) were ADCA. The cases were characterized by different grades: 50% (39/78) were moderately differentiated, 25.6% (20/78) were well differentiated and 24.4% (19/78) were poorly differentiated. Stage II was noted in 51.3% (40/78) of the tumors followed by 24.4% (19/78) of stage III. Stage I and IV accounted for 19.2% (15/78) and 5.1% (4/78) of the tumors, respectively. HPV was detected in 93.6% (73/78) of cancer samples while in non-cancerous samples these genotypes appeared in only 8.3% (3/36) of cases. Sixty tumor samples harbored HPV genotype 18. Single infection with this genotype was found in 40 samples (51.3%), while mixed infection was detected in 20 samples (25.6%). The HPV genotype 16 was found in 33 tumor samples. Single infection with this genotype appeared in 13 patients (16.7%), while 20 samples (25.6%) showed mixed infection. Genotype analysis of the p53 codon 72 showed Arg/Arg in 43.6% (34/78), Arg/Pro in 37.2% (29/78) and Pro/Pro in 19.2% (15/78) of tumor samples. The P-values, confidence interval (CI) and odd ratios (OD) were calculated for homozygous and heterozygote Arg allele. P-value was 0.015 and the OR was 2.4 (CI, 1.12–5.33) for Arg/Arg while the heterozygous P-value was 0.78 and the OR was 0.91 (CI, 0.43–1.96). The relative risk for Arg/Arg was 1.1.
p53 genotype distributions and allele frequencies of cervical cancer patients were compared with data from a healthy Sudanese control population published in 2003 (17) (Fig. 1).
Figure 1.
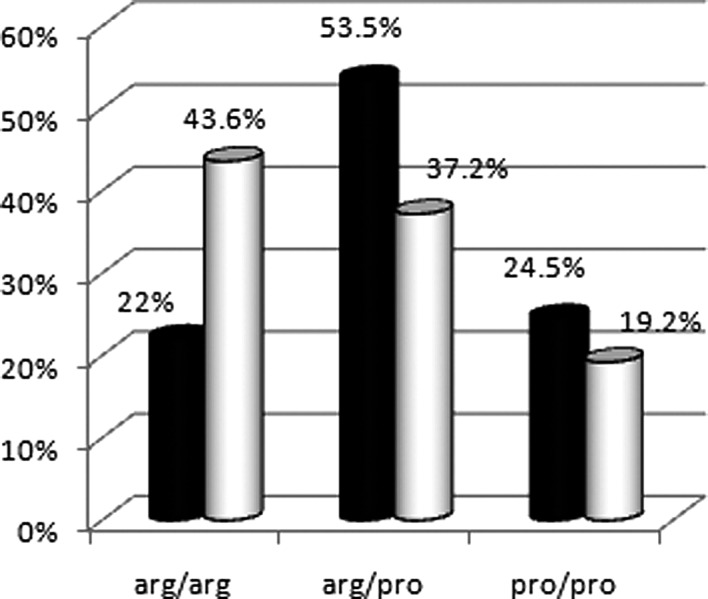
Frequency of Arg/Pro polymorphisms in cervical cancer patients (white cylinders) and controls (black cylinders).
LOH analysis of Rb intron 1 and 17 using RFLP
LOH was detected in 25.6% (20/78) of patients in intron 1 and in 37.2% (29/78) of patients in intron 17. LOH was found in 62.8% of patients in intron 1 and/or 17. The relative risk for LOH was 1.7.
Analysis of Rb RFLP with the p53 polymorphism
The genotypes of the p53 gene for patients wih LOH in Rb were 43.9, 43.9 and 12.2% for Arg/Arg, Arg/Pro and Pro/Pro, respectively, with a significant difference for the Arg allele among this group (P=0.012). Possible epistatic effect of the Rb RFLPs with the p53 Arg/Pro polymorphism was calculated and the relative risk was found to be 2.5.
Histopathology of patients with LOH in the Rb gene
SCC was noted in 80.5% of patients with LOH in the Rb gene. Histopathology revealed that 51.2% of the cases were moderately differentiated, 26.8% were poorly differentiated and 22% were well differentiated. Stage II was predominant among the tumors with LOH in the Rb gene, which was found in 61% of tumors, followed by stage III in 24.4%, stage I in 9.8% and finally stage IV in 4.9% of tumors. Table I summarizes the molecular abnormalities found in patients compared with the healthy control subjects.
Table I.
Comparison between the healthy control subjects and the cervical cancer patients regarding three parameters: presence of HPV, p53 Arg/Pro polymorphism and LOH in the Rb gene (intron 1 and 17).
| Molecular abnormality | Cervical cancer patients (%) | Healthy controls (%) |
|---|---|---|
| HPV infection | ||
| Frequency | 93.6 | 8.3 |
| Types | ||
| 18 | 51.3 | |
| 16 | 16.7 | |
| 16 plus 18 | 25.6 | |
| p53 | ||
| Arg/Arg | 43.6 | 22.0 |
| Arg/Pro | 37.2 | 37.2 |
| Pro/Pro | 19.2 | 53.5 |
| Rb gene (13q14) LOH | ||
| Intron 1 | 25.6 | 3.0 |
| Intron 17 | 37.2 | 3.2 |
Discussion
The so-called ‘high-risk’ HPVs infect the anogenital tract epithelium and are associated with the appearance of cervical dysplasia in almost all cases of cervical cancer (18). In the present study, the frequency of high-risk types HPV 16 and 18 is considered the etiological factor in Sudanese cervical cancer patients with predominance of HPV 18 (P=0.02). The tumor-suppressor protein p53 plays a critical role in cell cycle control and apoptosis and response to genome aberrations and environmental factors, including tumor viruses. Somatic mutations in p53 have been found to exist in more than 50% of human cancers (19). One of the most common variant associated with cancer development is the codon 72 single nucleotide polymorphism (SNP), which results in the substitution of proline for arginine. Codon 72 arginine homozygote in p53 was reported to increase the risk of human papillomavirus-associated cervical cancer (20). In this study we investigated the association of the p53 codon 72 with cervical cancer and the association between cancer and Arg/Arg was found to be significant (P=0.01 and OR=2.4) although not as strongly. The Rb as well as the p53 gene are thought to be the major target genes contributing to the malignant transformation of the cervical epithelium in relation to the role of HPV infection. We detected LOH of the Rb regions in 62.8% of our samples in intron 1 and/or intron 17 which is higher than the 14% reported by Kim et al (21) and the 29% reported by Park et al (22). The same markers showed 55% LOH in informative cases of esophageal squamous cell carcinoma specimens from a high-risk population in northern China (23). Although it has been suggested that the LOH of the chromosome 13 may be associated with a more aggressive tumor behavior in other types of cancer, in our cases the LOH of the Rb gene was not significantly associated with conventional clinicopathological parameters, including clinical stage and degree of differentiation, but our data showed a significant association between LOH and histological type of the tumor. Infections by the high-risk human papillomavirus (HPV) genotypes 16 and 18 are significantly involved in cervical cancer in Sudanese patients, and HPV 18 is the predominant one.
The p53 Arg/Arg polymorphism may be an important determinant of the risk for cervical cancer, but it does not appear to be sufficient for carcinogenesis. Furthermore, the Arg72-containing allele is preferentially mutated and retained in various human tumors, suggesting that polymorphic residue within p53 modifies mutant behavior and in the presence of LOH Rb acts as an additive factor increasing the risk of cervical cancer.
The LOH of the Rb gene was not significantly associated with other parameters, such as clinical stage and degree of differentiation and hence it cannot be a determinant of the tumor behavior in cervical carcinoma. Although the significance of the chromosomal allelic loss of Rb in this study may suggest that it is a molecular marker for cervical cancer, further supportive evidence is required.
Acknowledgements
The authors wish to acknowledge the collaboration of the staff of the Khartoum Teaching Hospital and to thank the patients who participated in this study. This study received partial financial support from the International Center of Genetic Engineering and Biotechnology (ICGEB).
References
- 1.Zur Hausen H. Papillomavirus infections - a major cause of human cancers. Biochim Biophys Acta. 1996;1288:55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 2.Kloth JN, Kenter GG, Spijker HS, et al. Expression of Smad2 and Smad4 in cervical cancer: absent nuclear Smad4 expression correlates with poor survival. Mod Pathol. 2008;21:866–875. doi: 10.1038/modpathol.2008.62. [DOI] [PubMed] [Google Scholar]
- 3.Bookstein R, Allred DC. Recessive oncogenes. Cancer. 1993;71(Suppl 3):S1179–S1186. doi: 10.1002/1097-0142(19930201)71:3+<1179::aid-cncr2820711442>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Kamil JP, Hume AJ, Jurak I, Münger K, Kalejta RF, Coena DM. Human papillomavirus 16 E7 inactivator of retinoblastoma family proteins complements human cytomegalovirus lacking UL97 protein kinase. Proc Natl Acad Sci USA. 2009;106:16823–16828. doi: 10.1073/pnas.0901521106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heck DV, Yee CL, Howley PM, Münger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 8.Storey A, Thomas M, Kalita A, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 9.Koushik A, Platt RW, Franco EL. p53 codon 72 polymorphism and cervical neoplasia: a meta-analysis review. Cancer Epidemiol Biomarkers Prev. 2004;13:11–22. doi: 10.1158/1055-9965.epi-083-3. [DOI] [PubMed] [Google Scholar]
- 10.Jee SH, Won SY, Yun JE, Lee JE, Park JS, Ji SS. Polymorphism p53 codon-72 and invasive cervical cancer: a meta-analysis. Int J Gynaecol Obstet. 2004;85:301–308. doi: 10.1016/j.ijgo.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Settheetham-Ishida W, Singto Y, Yuenyao P, Tassaneeyakul W, Kanjanavirojkul N, Ishida T. Contribution of epigenetic risk factors but not p53 codon 72 polymorphism to the development of cervical cancer in Northeastern Thailand. Cancer Lett. 2004;210:205–211. doi: 10.1016/j.canlet.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 12.Lee SA, Kim JW, Roh JW, et al. Genetic polymorphisms of GSTM1, p21, p53 and HPV infection with cervical cancer in Korean women. Gynecol Oncol. 2004;93:14–18. doi: 10.1016/j.ygyno.2003.11.045. [DOI] [PubMed] [Google Scholar]
- 13.Comar M, Molin GD, Guaschino S, Campello C. p53 at codon 72 polymorphism, human papillomavirus infection and cervical lesions: a cross-sectional study from northeastern Italy. Eur J Obstet Gynecol Reprod Biol. 2004;114:210–214. doi: 10.1016/j.ejogrb.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Ueda M, Hung YC, Terai Y, Saito J, Nunobiki O, Noda S, Ueki M. Glutathione-S-transferase and p53 polymorphisms in cervical carcinogenesis. Gynecol Oncol. 2005;96:736–740. doi: 10.1016/j.ygyno.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Scambia G, Lovergine S, Masciullo V. RB family members as predictive and prognostic factors in human cancer. Oncogene. 2006;25:5302–5308. doi: 10.1038/sj.onc.1209620. [DOI] [PubMed] [Google Scholar]
- 16.Soulitzis N, Sourvinos G, Sourvinos DN, Spandidos DA. P53 codon 72 polymorphism and its association with bladder cancer. Cancer Lett. 2002;179:175–183. doi: 10.1016/s0304-3835(01)00867-9. [DOI] [PubMed] [Google Scholar]
- 17.Bereir RE, Mohamed HS, Seielstad M, et al. Allele frequency and genotype distribution of polymorphisms within disease-related genes is influenced by ethnic population sub-structuring in Sudan. Genetica. 2003;119:57–63. doi: 10.1023/a:1024486716497. [DOI] [PubMed] [Google Scholar]
- 18.Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 19.Smardová J, Nemajerová A, Trbusek M, Vagunda V, Kovarík J. Rare somatic p53 mutation identified in breast cancer: a case report. Tumor Biol. 2001;22:59–66. doi: 10.1159/000050597. [DOI] [PubMed] [Google Scholar]
- 20.Zehbe I, Voglino G, Wilander E, et al. P53 codon 72 polymorphism and various human papillomavirus 16 E6 genotypes are risk factors for cervical cancer development. Cancer Res. 2001;61:608–611. [PubMed] [Google Scholar]
- 21.Kim JW, Lee CG, Han SM, et al. Loss of heterozygosity of the retinoblastoma and p53 genes in primary cervical carcinomas with human papillomavirus infection. Gynecol Oncol. 1997;67:215–221. doi: 10.1006/gyno.1997.4847. [DOI] [PubMed] [Google Scholar]
- 22.Park JS, Dong SM, Kim HS, et al. Detection of p16 gene alteration in cervical cancer using tissue microdissection and LOH study. Cancer Lett. 1999;136:101–108. doi: 10.1016/s0304-3835(98)00366-8. [DOI] [PubMed] [Google Scholar]
- 23.Xing EP, Yang GY, Wang LD, Shi ST, Yang CS. Loss of heterozygosity of the Rb gene correlates with pRb protein expression and associates with p53 alteration in human esophageal cancer. Clin Cancer Res. 1999;5:1231–1240. [PubMed] [Google Scholar]


