Abstract
Androgens are essential for the development of the prostate and prostate cancer. We examined androgen-regulated gene expression in the human prostate. Samples from benign and malignant prostate tissue and samples containing prostate tissue obtained from prostate cancer patients three days after surgical castration were further processed as probes for a GeneChip array. The comparison of gene expression profiles in castrated samples and in benign or malignant prostate tissue samples revealed androgen-regulated genes. We further evaluated the genes which were differentially expressed in benign and malignant prostate samples. The androgen-regulated expression of dual specificity phosphatase 1 (DUSP1) was confirmed in the LNCaP prostate cancer cell line, as the expression of DUSP1 increased with androgen treatment over the course of time. The expression of the genes CRISP3, PCA3, OR51E2, HOXC6, AGR3, AMACR and SLC14A1 was affected by castration in addition to differential expression in the benign and malignant prostate. These sample results require further investigation for the role of AGR3 and SLC14A1 in prostate cancer as these associations have not been reported previously.
Keywords: gene expression, prostate cancer, benign prostatic hyperplasia, castration
Introduction
Androgen action is essential for the development of the prostate and also for the development of prostate cancer. Androgen deprivation therapy is a fundamental treatment for metastasised prostate cancer. The activity of androgens is mediated mainly via the androgen receptor (AR), although other androgen responsive activation mechanisms may exist, especially in hormone-refractory prostate cancer (1). Androgen-regulated gene expression has been investigated in numerous studies using prostate cancer cell lines. However, to the best of our knowledge, only a few studies have used human tissues to evaluate androgen-regulated genes. One study reported gene expression changes following castration using prostatectomy samples after three months of androgen deprivation therapy (combination of anti-androgen and chemical castration) (2). In another study, a comparison was made of the expression profiles of untreated and androgen-independent prostate cancer (3). Mostaghel et al (4) studied gene expression changes in human prostate tissue at different time points up to nine months after castration for the treatment of localised prostate cancer. The same group further examined the effect of the 5-α reductase inhibitor dutasteride on prostate gene expression (5). The aim of the present study was to identify androgen-regulated genes in the human prostate. Gene expression analysis was performed on human prostate tissue samples from benign and malignant prostate tissue and from prostate biopsy samples obtained three days after surgical castration.
Materials and methods
Patient samples
Prostate samples were collected from three patients undergoing radical prostatectomy for the treatment of prostate cancer. Prostate biopsy samples were obtained from another three patients three days after surgical castration performed as treatment for prostate cancer. These male individuals had newly diagnosed prostate cancer with no previous hormonal treatments. The biopsies were extracted using a biopsy gun technique and 18 G needles. Six biopsies were obtained and a single prophylactic dose of ciprofloxacin (500 mg) was administered. Written informed consent was obtained from every patient giving tissue samples for the study. The study was approved by the Ethics Council of the Northern Ostrobothnia Hospital District. RNA from the samples was isolated using the QuickPrep Total RNA Extraction kit (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's instructions. The RNA from the samples was individually labelled and used for the GeneChip array. Three samples were microdissected and histologically confirmed as benign prostate tissue from radical prostatectomy specimens, and three samples were microdissected and histologically confirmed as Gleason 3+3 prostate cancer tissue from radical prostatectomy specimens and three samples were from biopsies taken following surgical castration performed as a therapeutic procedure for prostate cancer.
Cell culture
The prostate cancer cell line LNCaP (CRL-1740) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). LNCaP cell cultures were maintained in RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10 mM HEPES, 1 mM sodium pyruvate and 2.5 g/l D-glucose or DMEM (Sigma-Aldrich) supplemented with 4,500 mg/l glucose, L-glutamine and 1% penicillin-streptomycin (Invitrogen-Gibco, Carlsbad, CA, USA). The cell cultures were supplemented with 10% foetal bovine serum (FBS) (HyClone, Logan, UT, USA) at 37°C in a humidified atmosphere of 5% CO2. FBS was substituted with charcoal-treated FBS in the hormone-induction experiments. The LNCaP cells were plated at 1×106 cells/plate 72 h prior to the experiments. The cells were treated with 10 nM R1881 (PerkinElmer, Boston, MA, USA) or an equal volume of ethanol for 0, 6, 24, or 48 h. Following incubation, the cells were collected, washed with phosphate-buffered saline and used directly for the isolation of RNA.
GeneChip protocol
Experimental procedures for GeneChip were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual following the microarray experiment guidelines. In brief, using 8 μg of total RNA as a template, double-stranded DNA was synthesised using the One-Cycle cDNA Synthesis kit (Affymetrix, Santa Clara, CA, USA) and T7-(dT)24 primers. The DNA was purified using a GeneChip Sample Cleanup Module (Qiagen, Venlo, The Netherlands). In vitro transcription was performed to produce biotin-labeled cRNA using an in vitro transcription labeling kit (Affymetrix), according to the manufacturer's instructions. Biotinylated cRNA was cleaned with a GeneChip Sample Cleanup Module (Qiagen), fragmented to 35–200 nucleotides and hybridised to Affymetrix Human Genome U133 Plus 2 arrays that contained ~55,000 human transcripts. After being washed, the array was stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR, USA). The staining signal was amplified using biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA, USA) and a second staining was performed with streptavidin-phycoerythrin before the array was scanned on a GeneChip Scanner 3000. The expression data were analysed using Affymetrix GeneChip Operating System Software. Signal intensities of all probe sets were scaled to the target value of 500.
Quantitative reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from LNCaP cells for quantitative RT-PCR measurements was isolated with TRIzol reagent (Invitrogen-Gibco) according to the manufacturer's instructions. Using the First-Strand cDNA synthesis kit (Amersham Biosciences) the first-strand cDNA was synthesised with 1 μg of RNA and pd(N)6 random hex deoxynucleotides according to the manufacturer's instructions. The mRNA levels for LNCaP cells were measured by quantitative RT-PCR analysis (ABI 7700, Applied Biosystems, Foster City, CA, USA) as described previously (6). The forward and reverse primers for for the detection of dual specificity phosphatase 1 (DUSP1) mRNA were 5′-TCCTTCTTCGCTTTCAACGC-3′ and 5′-ACGATGGTGCTGAAGCGC-3′, respectively. Amplicons were detected using the fluorogenic probe 5′-FAM-CACA TCGCCGGCTCTGTCAACG-TAMRA-3′. The primers and probe for the 18 S amplicon were 5′-TGGTTGCAAAGCTGA AACTTAAAG-3′ (forward), 5′-AGTCAAATTAAGCCGCA GGC-3′ (reverse) and 5′-VIC-CCTGGTGGTGCCCTTCCG TCA-TAMRA-3′, respectively.
Analysis of gene expression profiles
The expression profiles from the GeneChip array were analysed using Chipster software (http://chipster.csc.fi). The GeneChips were normalised using the robust multiarray average (RMA) method and gene expression intensity estimates were received in log2-transformed values. The pathways involved were identified using Chipster software utilising data from ConsensusPathDB (7). The data discussed in this study have been deposited in the NCBI's Gene Expression Omnibus (8) and are accessible through GEO Series accession number GSE32982 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32982).
Statistical analysis
Statistical analysis of the levels of DUSP1 mRNA in LNCaP cells was performed using SPSS version 15.0 (SPSS, Chicago, IL, USA). The Student's t-test was used for comparison between the two groups. Values were presented as the means ± SD.
Results
Genes differentially expressed in prostate cancer and benign prostate tissue, and genes differentially expressed in benign prostate tissue or prostate cancer and castrated prostate cancer are listed in Table I. These genes contain several well-known androgen-regulated genes and prostate cancer-associated genes, including MSMB, SORD, CRISP3 and PCGEM1. The consensus pathways involved between benign and malignant tissue and between benign tissue and castrated prostate cancer are shown in Table II.
Table I.
Differentially expressed genes in benign prostate tissue samples, prostate cancer samples and samples obtained following surgical castration. a
| Fold overexpression | ||||
|---|---|---|---|---|
|
|
||||
| Symbol | Description | P-value | Cancer vs. benign | Benign/cancer vs. castrated |
| CRISP3 | Cysteine-rich secretory protein 3 | <0.001 | 5.78 | −3.08 |
| FOS | FBJ murine osteosarcoma viral oncogene homologue | <0.001 | 4.76 | |
| PCGEM1 | Prostate-specific transcript 1 (non-protein coding) | <0.001 | 4.42 | |
| MYBPC1 | Myosin binding protein C, slow type | <0.001 | 4.30 | |
| PCA3 | Prostate cancer antigen 3 (non-protein coding) | <0.001 | 4.18 | 3.74 |
| OR51E2 | Olfactory receptor, family 51, subfamily E, member 2 | <0.001 | 4.01 | 3.29 |
| ANPEP | Alanyl (membrane) aminopeptidase | <0.001 | 3.84 | |
| DPP4 | Dipeptidyl-peptidase 4 | 0.005 | 3.79 | |
| GCNT2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) | 0.001 | 3.73 | |
| HOXC6 | Homeobox C6 | 0.003 | 3.69 | −2.62 |
| AGR3 | Anterior gradient homologue 3 (Xenopus laevis) | 0.003 | 3.63 | 3.00 |
| RGS1 | Regulator of G-protein signalling 1 | 0.002 | 3.55 | |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 0.002 | 3.50 | |
| DUSP1 | Dual specificity phosphatase 1 | 0.011 | 3.48 | |
| AMACR | α-methylacyl-CoA racemase | 0.008 | 3.40 | 3.00 |
| LOC728606 | Hypothetical LOC728606 | 0.015 | 3.36 | |
| DLX1 | Distal-less homeobox 1 | 0.011 | 3.30 | |
| VEGFA | Vascular endothelial growth factor A | 0.004 | 3.30 | |
| MSMB | Microseminoprotein, β- | 0.006 | 3.21 | |
| TDO2 | Tryptophan 2,3-dioxygenase | 0.025 | 3.19 | |
| NCAPD3 | Non-SMC condensin II complex, subunit D3 | 0.008 | 3.14 | |
| EGR1 | Early growth response 1 | 0.027 | 3.14 | |
| CD38 | CD38 molecule | 0.008 | 3.13 | |
| C15orf48 | Chromosome 15 open reading frame 48 | 0.022 | 3.11 | |
| RASD1 | RAS, dexamethasone-induced 1 | 0.030 | 3.09 | |
| AGR2 | Anterior gradient homologue 2 (Xenopus laevis) | 0.023 | 3.07 | |
| SORD | Sorbitol dehydrogenase | 0.012 | 3.02 | |
| C12orf56 | Chromosome 12 open reading frame 56 | 0.037 | 3.01 | |
| SFTPA2 | Surfactant protein A2 | 0.037 | 2.99 | |
| ST6GALNAC1 | ST6 (α-N-acetyl-neuraminyl-2,3-β-galactosyl-1,3)-N-acetylgalactosaminide α-2,6-sialyltransferase 1 | 0.038 | 2.98 | |
| ACADL | Acyl-CoA dehydrogenase, long chain | 0.013 | 2.98 | |
| EGR3 | Early growth response 3 | 0.044 | 2.94 | |
| ERG | V-ets erythroblastosis virus E26 oncogene homologue (avian) | 0.045 | 2.90 | |
| FOSB | FBJ murine osteosarcoma viral oncogene homologue B | 0.016 | 2.90 | |
| SCD | Stearoyl-CoA desaturase (δ-9-desaturase) | 0.024 | 2.80 | |
| GNMT | Glycine N-methyltransferase | 0.028 | 2.76 | |
| CCK | Cholecystokinin | 0.028 | 2.75 | |
| PEBP4 | Phosphatidylethanolamine-binding protein 4 | 0.031 | 2.72 | |
| ATF3 | Activating transcription factor 3 | 0.031 | 2.71 | |
| HLA-DQA1 | Major histocompatibility complex, class II, DQ α 1 | 0.032 | 2.70 | |
| SELE | Selectin E | 0.035 | 2.66 | |
| PENK | Proenkephalin | 0.043 | 2.61 | |
| CD177 | CD177 molecule | 0.048 | 2.58 | |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 (non-protein coding) | 0.032 | −2.69 | |
| ASPN | Asporin | 0.029 | −2.74 | |
| PSPH | Phosphoserine phosphatase | 0.027 | −2.77 | |
| SLC14A1 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | 0.048 | −2.88 | −3.05 |
| HBB | Haemoglobin, β | 0.016 | −2.89 | |
| SCGB1A1 | Secretoglobin, family 1A, member 1(uteroglobin) | 0.045 | −2.91 | |
| KRT14 | Keratin 14 | 0.032 | −2.99 | |
| NEFH | Neurofilament, heavy polypeptide | 0.022 | −3.10 | |
| TMEM45B | Transmembrane protein 45B | 0.007 | −3.16 | |
| GPM6A | Glycoprotein M6A | 0.027 | −3.17 | |
| RLN1 | Relaxin 1 | 0.016 | −3.21 | |
| GREM1 | Gremlin 1 | 0.004 | −3.29 | |
| MME | Membrane metallo-endopeptidase | 0.004 | −3.57 | |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | 0.003 | −3.67 | |
| CYP3A5 | Cytochrome P450, family 3, subfamily A, polypeptide 5 | 0.001 | −3.92 | |
| WIF1 | WNT inhibitory factor 1 | <0.001 | −4.14 | |
Gene symbols and gene descriptions are presented.
P-values for differential expression were produced using Chipster software. Only genes with P-values <0.05 are presented. Negative values in the fold overexpression column indicate that the gene was underexpressed in cancer compared with benign samples or underexpressed in benign or cancer samples compared with castrated samples.
Table II.
Pathways potentially active in the transition from benign to malignant prostate tissue and during surgical castration based on gene-expression differences.a
| P-value | |||
|---|---|---|---|
|
|
|||
| Pathway | Database | Benign tissue vs. cancer | Castrated tissue vs. benign tissue |
| AP-1_transcription_factor_network | PID | 0.007 | <0.001 |
| Beta1_integrin_cell_surface_interactions | PID | <0.001 | |
| Bevacizumab_Pathway | SMPDB | 0.006 | |
| Cholesterol_biosynthesis | Wikipathways | <0.001 | |
| Cholesterol_biosynthesis | Reactome | 0.002 | |
| cholesterol_biosynthesis_I | HumanCyc | 0.003 | |
| cholesterol_biosynthesis_II_(via_24,25-dihydrolanosterol) | HumanCyc | 0.003 | |
| cholesterol_biosynthesis_III_(via_desmosterol) | HumanCyc | 0.003 | |
| Collagen_adhesion_via_alpha_2_beta_1_glycoprotein | Reactome | 0.001 | <0.001 |
| ECM-receptor_interaction_-_Homo_sapiens_(human) | KEGG | <0.001 | |
| Fatty_Acyl-CoA_Biosynthesis | Reactome | 0.007 | |
| Focal_Adhesion | Wikipathways | 0.004 | |
| Focal_adhesion_-_Homo_sapiens_(human) | KEGG | 0.003 | |
| GPCR_signalling-cholera_toxin | INOH | 0.004 | 0.003 |
| GPCR_signalling-G_alpha_i | INOH | 0.006 | |
| GPCR_signalling-pertussis_toxin | INOH | 0.006 | |
| HIF-1-alpha_transcription_factor_network | PID | 0.008 | |
| Immunoregulatory_interactions_between_a_ Lymphoid_and_a_non-Lymphoid_cell | Reactome | <0.001 | <0.001 |
| Integrin | INOH | 0.006 | |
| Integrins_in_angiogenesis | PID | 0.004 | |
| Ketogenesis | HumanCyc | 0.006 | |
| Neurophilin_interactions_with_VEGF_and_VEGFR | Reactome | 0.005 | |
| Platelet_degranulation_ | Reactome | 0.006 | |
| Prostaglandin_Synthesis_and_Regulation | Wikipathways | 0.007 | |
| Protein_digestion_and_absorption_-_Homo_ sapiens_(human) | KEGG | 0.002 | |
| Response_to_elevated_platelet_cytosolic_Ca2+ | Reactome | 0.007 | |
| Signalling_by_PDGF | Reactome | <0.001 | |
| Signalling_by_VEGF | Reactome | 0.008 | |
| Smooth_Muscle_Contraction | Reactome | 0.003 | |
| Steroid_Biosynthesis | SMPDB | <0.001 | |
| Steroid_biosynthesis_-_Homo_sapiens_(human) | KEGG | 0.002 | |
| Superpathway_of_cholesterol_biosynthesis | HumanCyc | <0.001 | |
| Syndecan-1-mediated_signalling_events | PID | 0.001 | |
| Valine_degradation_I | HumanCyc | 0.009 | |
| Vatalanib_Pathway | SMPDB | 0.006 | |
| VEGF_ligand-receptor_interactions | Reactome | 0.008 | |
The active pathways identified and the respective P-values were produced using Chipster software. Databases used for the identification of the pathways are presented.
PID, pathway interaction database; SMPDB, small molecule pathway database; KEGG, Kyoto encyclopedia of genes and genomes; INOH, integrating network objects with hierarchies.
The androgen-regulated expression of DUSP1 in LNCaP prostate cancer cell lines was also evaluated (Fig. 1). The treatment of LNCaP cells with synthetic androgen R1881 led to an increased expression of DUSP1 mRNA, with a peak of 2.6-fold expression following 48 h of treatment compared with androgen-depleted conditions.
Figure 1.
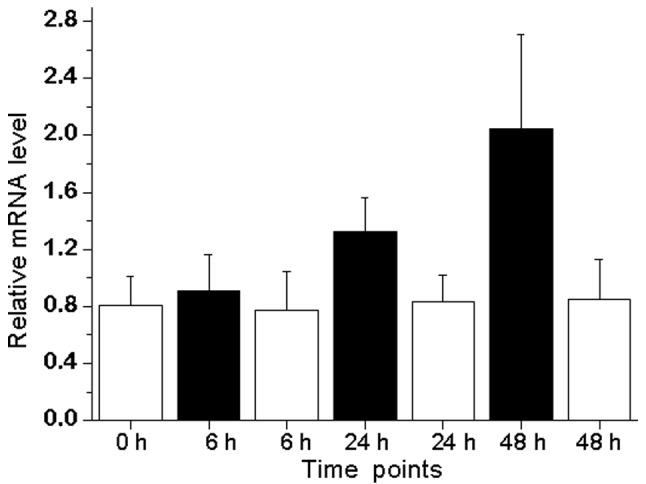
Response of DUSP1 mRNA in LNCaP cells following treatment with 10 nM synthetic androgen R1881 (solid bars). Statistically significant induction of DUSP1 mRNA was observed after 24 and 48 h compared with the control after 24 and 48 h with P=0.04 and P<0.001, respectively. The values are the means ± standard deviations of four individual samples. DUSP1, dual specificity phosphatase 1.
Discussion
To identify androgen-regulated genes in the human prostate, we used human prostate tissue mRNAs in a GeneChip array. The main limitation of widely used human prostate cancer cell lines such as LNCaP, PC-3 and DU-145 is that they do not represent the prostate per se, but are isolated from a prostate cancer metastasis. We evaluated the gene expression profiles from microdissected tissue samples from freshly prepared radical prostatectomy samples and from transrectal prostate biopsies obtained three days after surgical castration performed as a treatment for prostate cancer.
Certain known androgen-regulated genes were identified, including MSMB (9) and SORD (10). Furthermore, a number of previously identified prostate cancer-associated genes, such as CRISP3 (11), PCGEM1 (12), PCA3 (13) and OR51E2 (also known as PSGR) (14) were also differentially expressed.
Cholesterol biosynthesis has several correlations with prostate cancer. Low serum cholesterol levels have been correlated with a lower risk of high-grade prostate cancer (15). Furthermore, the use of cholesterol-lowering drugs, such as statins, is associated with a lower risk of advanced prostate cancer (16). Increased cholesterol synthesis may serve as a precursor for intratumoural androgen synthesis in castration-resistant prostate cancer (17).
Focal adhesion pathways are involved in gene-expression changes between benign and malignant prostate tissue. Cell adhesion is a significant process in cancer (18). The steroid biosynthesis pathway was found to be active between the castrated and benign tissue, as expected.
DUSP1 (also known as mitogen-activated protein kinase phosphatase 1, MKP1) is androgen-regulated in the rat prostate (19). DUSP1 expression was detected in prostate cancer with a decreased expression in poorly differentiated carcinomas. Moreover, DUSP1 expression was downregulated in androgen-depleted clinical prostate cancer samples and the expression of DUSP1 was inversely correlated with apoptosis (20). In prostate cancer specimens, the expression of DUSP1 was low in hormone-refractory prostate cancer, whereas it was high in benign prostatic hyperplasia samples and in non-hormone-treated prostate cancer (21). The results of our study confirm those of previous results as DUSP1 expression was 3.48 times lower in castrated samples compared with benign or malignant prostate samples (Table I). We also provide data for the androgen-mediated induction of DUSP1 in androgen-sensitive LNCaP prostate cancer cell lines (Fig. 1). Taken together, it appears that DUSP1 is not important in hormone-refractory prostate cancer, as increased apoptosis detected in androgen-depleted prostate cancer by Magi-Galluzzi et al (20) may be explained by the low expression of DUSP1 under androgen-depleted conditions and high apoptotic indices under the same conditions in hormone-sensitive prostate cancer. The low expression of DUSP1 in hormone-refractory prostate cancer (21) may be explained by the androgen-dependent expression of DUSP1 shown in our study and previously by Leav et al (19), as castration is an ongoing process in hormone-refractory prostate cancer.
Of note are the genes that are differentially expressed in benign tissue and cancer, which are also overexpressed or underexpressed following castration, meaning that the gene is regulated by androgens and potentially involved in prostate carcinogenesis. These genes are CRISP3, PCA3, OR51E2, HOXC6, AGR3, AMACR and SLC14A1 (Table I). To the best of our knowledge, the association between prostate cancer and AGR3 or SLC14A1 has yet to be established. A high level of the immunoexpression of AGR3 has been linked with prolonged survival in ovarian cancer (22) and AGR3 expression has previously been detected in breast tumours (23). SLC14A1 was recently described as a urinary bladder cancer susceptibility gene (24).
Certain well-known androgen-regulated genes, such as kallikreins (25,26), are not shown in Table I. This is due to the variation in gene expression levels in different samples leading to increased P-values. Thus genes, such as kallikreins, did not exceed the given threshold level of P<0.05 for genes presented in Table I (data not shown). The number of samples analysed in our study was limited (three samples per group) and the biopsies obtained from castrated patients were random biopsies, thus these samples may represent cancer or benign tissue, or both. Despite these limitations, our data may be valuable for further studies in prostate cancer.
In conclusion, we have described the identification of androgen-regulated genes in the human prostate, some of which are potential new diagnostic or therapeutic targets in prostate cancer. Particular attention should be paid to AGR3 and SLC14A1 for their roles in prostate cancer. Furthermore, these gene expression profiles may be useful in sophisticated gene expression analyses utilising expression profiles from several different sources.
Acknowledgements
We are grateful to Ms. Mirja Mäkeläinen, Ms. Tarja Piispanen and Ms. Marja Tolppanen for technical assistance. Dr Mika Ilves performed the quantitative RT-PCR experiments. Dr Jussi Vuoristo performed the GeneChip arrays. The computations presented in this document were performed by the Centre for Scientific Computing (CSC) environment. CSC is the Finnish IT centre for science and is owned by the Ministry of Education. This study was supported by grants from the Finnish Medical Fund, the Päivikki and Sakari Sohlberg Foundation and the Urologic Research Foundation, Finland.
References
- 1.Bennett NC, Gardiner RA, Hooper JD, Johnson DW, Gobe GC. Molecular cell biology of androgen receptor signalling. Int J Biochem Cell Biol. 2010;42:813–827. doi: 10.1016/j.biocel.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best CJ, Gillespie JW, Yi Y, Chandramouli GV, Perlmutter MA, Gathright Y, Erickson HS, Georgevich L, Tangrea MA, Duray PH, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mostaghel EA, Page ST, Lin DW, Fazli L, Coleman IM, True LD, Knudsen B, Hess DL, Nelson CC, Matsumoto AM, et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007;67:5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 5.Mostaghel EA, Geng L, Holcomb I, Coleman IM, Lucas J, True LD, Nelson PS. Variability in the androgen response of prostate epithelium to 5alpha-reductase inhibition: implications for prostate cancer chemoprevention. Cancer Res. 2010;70:1286–1295. doi: 10.1158/0008-5472.CAN-09-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majalahti-Palviainen T, Hirvinen M, Tervonen V, Ilves M, Ruskoaho H, Vuolteenaho O. Gene structure of a new cardiac peptide hormone: a model for heart-specific gene expression. Endocrinology. 2000;141:731–740. doi: 10.1210/endo.141.2.7312. [DOI] [PubMed] [Google Scholar]
- 7.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar R, Domrachev M, Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlman A, Edsjö A, Halldén C, Persson JL, Fine SW, Lilja H, Gerald W, Bjartell A. Effect of androgen deprivation therapy on the expression of prostate cancer biomarkers MSMB and MSMB-binding protein CRISP3. Prostate Cancer Prostatic Dis. 2010;13:369–375. doi: 10.1038/pcan.2010.25. [DOI] [PubMed] [Google Scholar]
- 10.Szabo Z, Hamalainen J, Loikkanen I, Moilanen AM, Hirvikoski P, Vaisanen T, Paavonen TK, Vaarala MH. Sorbitol dehydrogenase expression is regulated by androgens in the human prostate. Oncol Rep. 2010;23:1233–1239. doi: 10.3892/or_00000755. [DOI] [PubMed] [Google Scholar]
- 11.Bjartell A, Johansson R, Bjork T, Gadaleanu V, Lundwall A, Lilja H, Kjeldsen L, Udby L. Immunohistochemical detection of cysteine-rich secretory protein 3 in tissue and in serum from men with cancer or benign enlargement of the prostate gland. Prostate. 2006;66:591–603. doi: 10.1002/pros.20342. [DOI] [PubMed] [Google Scholar]
- 12.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 14.Xu LL, Stackhouse BG, Florence K, Zhang W, Shanmugam N, Sesterhenn IA, Zou Z, Srikantan V, Augustus M, Roschke V, et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 2000;60:6568–6572. [PubMed] [Google Scholar]
- 15.Platz EA, Till C, Goodman PJ, Parnes HL, Figg WD, Albanes D, Neuhouser ML, Klein EA, Thompson IM, Jr, Kristal AR. Men with low serum cholesterol have a lower risk of high-grade prostate cancer in the placebo arm of the prostate cancer prevention trial. Cancer Epidemiol Biomarkers Prev. 2009;18:2807–2813. doi: 10.1158/1055-9965.EPI-09-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murtola TJ, Tammela TL, Lahtela J, Auvinen A. Cholesterol-lowering drugs and prostate cancer risk: a population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:2226–2232. doi: 10.1158/1055-9965.EPI-07-0599. [DOI] [PubMed] [Google Scholar]
- 17.Leon CG, Locke JA, Adomat HH, Etinger SL, Twiddy AL, Neumann RD, Nelson CC, Guns ES, Wasan KM. Alterations in cholesterol regulation contribute to the production of intratumoral androgens during progression to castration-resistant prostate cancer in a mouse xenograft model. Prostate. 2010;70:390–400. doi: 10.1002/pros.21072. [DOI] [PubMed] [Google Scholar]
- 18.Mol AJ, Geldof AA, Meijer GA, van der Poel HG, van Moorselaar RJ. New experimental markers for early detection of high-risk prostate cancer: role of cell-cell adhesion and cell migration. J Cancer Res Clin Oncol. 2007;133:687–695. doi: 10.1007/s00432-007-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leav I, Galluzzi CM, Ziar J, Stork PJ, Ho SM, Loda M. Mitogen-activated protein kinase and mitogen-activated kinase phosphatase-1 expression in the Noble rat model of sex hormone-induced prostatic dysplasia and carcinoma. Lab Invest. 1996;75:361–370. [PubMed] [Google Scholar]
- 20.Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, Wishnow K, Kaplan I, Stork PJ, Loda M. Mitogen-activated protein kinase phosphatase 1 is overexpressed in prostate cancers and is inversely related to apoptosis. Lab Invest. 1997;76:37–51. [PubMed] [Google Scholar]
- 21.Rauhala HE, Porkka KP, Tolonen TT, Martikainen PM, Tammela TL, Visakorpi T. Dual-specificity phosphatase 1 and serum/glucocorticoid-regulated kinase are downregulated in prostate cancer. Int J Cancer. 2005;117:738–745. doi: 10.1002/ijc.21270. [DOI] [PubMed] [Google Scholar]
- 22.King ER, Tung CS, Tsang YT, Zu Z, Lok GT, Deavers MT, Malpica A, Wolf JK, Lu KH, Birrer MJ, Mok SC, et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am J Surg Pathol. 2011;35:904–912. doi: 10.1097/PAS.0b013e318212ae22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher GC, Patel S, Tyson K, Adam PJ, Schenker M, Loader JA, Daviet L, Legrain P, Parekh R, Harris AL, Terrett JA. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88:579–585. doi: 10.1038/sj.bjc.6600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafnar T, Vermeulen SH, Sulem P, Thorleifsson G, Aben KK, Witjes JA, Grotenhuis AJ, Verhaegh GW, Hulsbergen-van de Kaa CA, Besenbacher S, et al. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum Mol Genet. 2011;20:4268–4281. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henttu P, Vihko P. Prostate-specific antigen and human glandular kallikrein: two kallikreins of the human prostate. Ann Med. 1994;26:157–164. doi: 10.3109/07853899409147884. [DOI] [PubMed] [Google Scholar]
- 26.Young CY, Andrews PE, Tindall DJ. Expression and androgenic regulation of human prostate-specific kallikreins. J Androl. 1995;16:97–99. [PubMed] [Google Scholar]


