Abstract
Accumulating data reveal that aldehyde dehydrogenase (ALDH) activity is a cancer stem cell marker in several types of human cancers. Whether ALDH also defines cancer stem cells of tongue squamous cell carcinoma is unknown. In the present study, we performed the Aldefluor assay to detect ALDH enzymatic activity, and used flow cytometry to isolate ALDH+ and ALDH− cells based on their ALDH activity in the Tca8113 tongue squamous cell carcinoma cell line. We found that Tca8113 cells harbored 1.3% of ALDH+ cells, which exhibited higher proliferation capacity than their ALDH− counterparts and parental cells. Sorted ALDH+ cells were able to differentiate and generate ALDH− cells in vitro. Moreover, in serum-free medium, ALDH+, but not ALDH− cells, survived and formed tumorspheres. The suppression subtractive hybridization assay revealed 68 overexpressed or underexpressed genes in the ALDH+ subpopulation relative to ALDH− cells, which included a set of known cancer stem cell-related genes. Thus, our data demonstrated that a small subset of Tca8113 cells with high ALDH enzymatic activity display characteristics of cancer stem cells, suggesting that ALDH activity may be a cancer stem cell marker for tongue squamous cell carcinoma.
Keywords: cancer stem cell, aldehyde dehydrogenase, tongue squamous cell carcinoma, suppression subtractive hybridization
Introduction
Oral cavity cancer is among the leading 10 most commonly diagnosed cancers. Squamous cell carcinoma of the tongue is one of the most common tumors in head and neck cancers (1). During the past several decades, great progress has been made in surgery, radiation therapy and chemotherapy, yet the 5-year survival rate of oral cancer has remained approximately 50% (2). In addition, these patients are at high risk for tumor recurrence and second primary tumors after regular treatment. Furthermore, the molecular mechanisms underlying the pathogenesis of oral cancer remain elusive.
Cancer stem cells (CSCs) were first discovered in 1997 in acute myeloid leukemia, and have recently been found in several tumor types, including lung, breast, brain, liver, pancreas and colon cancers (3,4). This unique subpopulation of cancer cells possesses the ability to self-renew, proliferate and differentiate through developmental signaling pathways in aberrant ways (4). CSCs have also been reported to be involved in tumor metastasis and recurrence. Accumulating data indicate that tongue squamous cell carcinoma harbors tumor-initiating cells or CSCs (5–7). Certain CSC-related genes, such as bone morphogenetic protein-4 (BMP-4), octamer-binding transcription factor 4 (Oct4), the homeobox protein Nanog, CD133 and B-cell-specific Moloney murine leukemia virus insertion site 1 (Bmi-1), have been identified and characterized in tongue squamous cell carcinoma (5,7–10).
Aldehyde dehydrogenase (ALDH) enzymes play a critical role in the metabolism of many molecules, and in the detoxification of external and internal substances, such as alcohol and toxins. Studies have shown that ALDH is a CSC marker in certain solid tumors, including colon, breast and lung cancers (11–13). In addition, the 19 ALDH family members may function differently, depending on the type of tissue and tumors (14). In this study, we observed that the Tca8113 tongue squamous cell carcinoma cell line harbored 1.3% of ALDH+ cells, which displayed CSC characteristics. The ALDH+ subpopulation possessed an elevated capacity to proliferate, differentiate and self-renew in comparison to their ALDH− counterparts. Our data suggest that ALDH activity is a CSC marker for tongue squamous cell carcinoma; therefore, targeting ALDH may be a potential therapeutic strategy.
Materials and methods
Cell line and reagents
The Tca8113 human tongue squamous cell carcinoma cell line was provided by the State Key Laboratory of Oral Diseases of Sichuan University, China. Tca8113 cells were cultured in RPMI-1640 (Life Technologies Inc.) and supplemented with 10% fetal calf serum (Gibco) at 37°C in a humidified atmosphere containing 5% CO2. Epidermal growth factors and basic fibroblast growth factors were purchased from PeproTech.
Aldefluor assay and flow cytometry
The Aldefluor kit (Stem Cell Technologies) was utilized to profile and isolate cells with high and low ALDH activity as previously described (15). Cells were incubated in Aldefluor assay buffer containing the ALDH substrate, BODIPY-aminoacetaldehyde (BAAA), at 37°C for 45 min. Cells that were able to catalyze BAAA to its fluorescent product, BODIPY-aminoacetate (BAA), were considered ALDH+ cells. The enzymatic activity of ALDH was blocked by a specific inhibitor, DEAB. Sorting gates for fluorescence-activated cell sorting (FACS) were drawn relative to baseline fluorescence, which was determined by DEAB-treated samples. After incubation, cells were resuspended in fresh assay buffer. ALDH+ and ALDH− cells were sorted by a BD Aria (BD Biosciences).
Proliferation assay
Sorted ALDH+, ALDH− cells and their parental cells were seeded in 96-well plates with a density of 1,000 cells/ml. Cell proliferation was detected after culturing for 1, 3, 5 and 7 days using the Cell Counting kit-8 (Dojindo) according to the manufacturer’s instructions. Data are shown as the means ± SD.
Differentiation assay
Unsorted and sorted single cells were cultured in regular medium. ALDH activity was measured when the cells became 85% confluent.
Tumorsphere formation assay
Sorted ALDH+ and ALDH− single cells were cultured in serum-free medium in 6-well plates at a density of 20,000 cells/ml. Tumorspheres were detected and images were captured using a Zeiss inverted microscope.
Suppression subtractive hybridization (SSH)
Total RNA from sorted ALDH+ and ALDH− cells was extracted with TRIzol (Invitrogen) according to the manufacturer’s instructions. Double-stranded cDNA was generated using PCR-Select cDNA Subtraction kit (Clontech). cDNAs were digested with RsaI (New England Biolabs). The tester cDNA (ALDH+ cells) was subdivided into two portions and each was ligated to a different double-stranded adaptor (1 and 2). Enrichment of differentially expressed transcripts was achieved in two successive rounds of PCR amplifications that employed different adapters (1 and 2), and sequential subtractive hybridization of the tester to an excess of driver cDNA (ALDH− cells). Purified cDNA was ligated into the pGEM-T easy vector system (Promega) and DH5α-competent cells were transformed. Selected clones were grown in LB medium with 1 μg/ml ampicillin. For sequencing, the plasmids were extracted and sequenced using BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) with M13 primers on an ABI-PRISM 3100 (Applied Biosystems) automated gene analyzer.
Statistical methods
Two-way ANOVA was performed to determine the significant differences in cell growth between sorted and unsorted cells in vitro.
Results
Identification of ALDH+ subpopulation in Tca8113 tongue squamous cell carcinoma cells
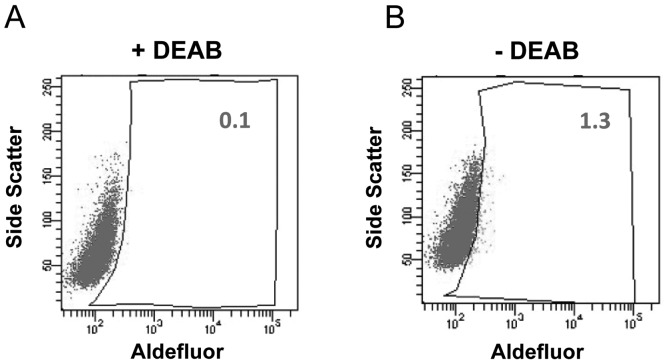
The enzymatic activity of ALDH has been demonstrated to be a CSC marker in several cancer cell lines and patient tumor samples (4). To examine whether human tongue squamous cell carcinoma contains a subpopulation with high ALDH activity, Aldefluor assays were performed to identify ALDH+ cells in the Tca8113 cell line. In the presence of the ALDH-specific inhibitor DEAB, ALDH+ cells were barely observed. Notably, ~1.3% of Tca8113 cells displayed very bright fluorescence in the absence of DEAB (Fig. 1). This result indicates that the Tca8113 human tongue squamous cell carcinoma cell line contains a subset of cells with high ALDH enzymatic activity. Our data are consistent with a previous report that the Tca8113 cell line harbors ~1% of CD133+ cells, which is another well-studied CSC marker (7).
Figure 1.
ALDH+ cells detected in the Tca8113 tongue squamous cell carcinoma cell line. ALDH+ Tca8113 cells were identified using a flow cytometry-based Aldefluor assay. (A) Baseline fluorescence was established in the presence of ALDH inhibitor DEAB. (B) ALDH+ cells detected in Tca8113 cells without DEAB.
Enhanced proliferation of ALDH+ tongue squamous cell carcinoma cells
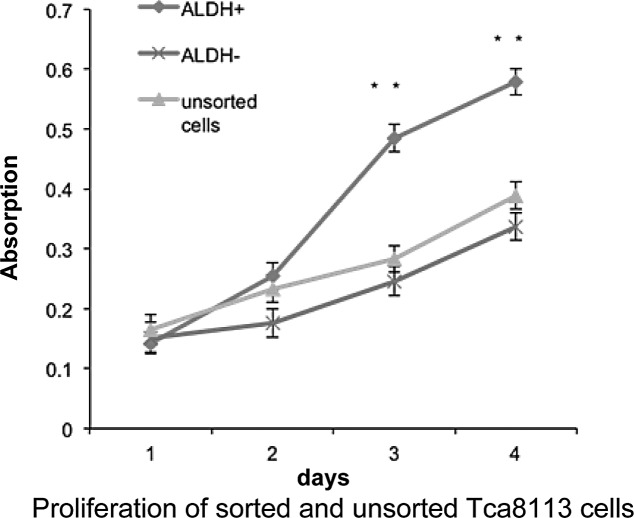
CSCs possess an increased capacity for proliferation, differentiation and tumorigenicity. We hypothesized that ALDH+ cells in the Tca8113 cell line are CSCs. To test this hypothesis, we compared the proliferation rate of sorted ALDH+, ALDH− cells and parental Tca8113 cells using Cell Counting Kit-8. After a 5-day culture, ALDH+ cells proliferated significantly faster than their ALDH− counterparts, whereas the unsorted cells exhibited a similar proliferation rate as that of the ALDH− cells (Fig. 2). This result indicates that ALDH+ cells in the Tca8113 cell line have enhanced capacity to proliferate relative to ALDH− cells in vitro.
Figure 2.
ALDH+ cells proliferate faster than ALDH− Tca8113 cells in vitro. Cell growth curves were generated from sorted ALDH+, ALDH− cells and unsorted Tca8113 cells (n=3, **P<0.01).
ALDH+ tongue squamous cell carcinoma cells form tumorspheres
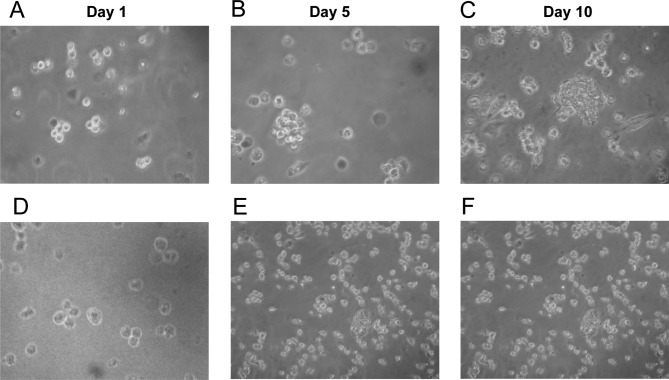
Similar to normal stem cells, CSCs are able to self-renew through asymmetric cell divisions. Tumorsphere formation assays are widely used to evaluate self-renewal and differentiation at the single-cell level in vitro. Separated ALDH+ and ALDH− single cells were maintained in serum-free medium, in which non-CSCs cannot survive. On day 5, ALDH+ cells started to aggregate with each other. Clear tumorspheres were observed on day 10, while most single cells were already dead (Fig. 3B and C). By contrast, tumorspheres were barely found in the plates seeded with ALDH− cells (Fig. 3E and F).
Figure 3.
ALDH+ Tca8113 cells form tumorspheres in vitro. Isolated single ALDH+ and ALDH− Tca8113 cells were cultured in serum-free medium. (A and D) Cells were monitored the next day after being seeded in the plates. Tumorspheres were observed after a 5- or 10-day incubation of the ALDH+ cells (B and C), while tumorspheres were not noted in the cultured ALDH− cells (E and F).
Differentiation of ALDH+ tongue squamous cell carcinoma cells
One critical characteristic of CSCs is the ability to differentiate and form all types of tumor cells. To evaluate the differentiation capacity of ALDH+ cells, the percentage of ALDH+ cells was continuously measured during the culture of isolated ALDH+ cells in regular medium. We observed that the percentage of ALDH+ cells gradually reduced to the level of parental Tca8113 cells, whereas the amount of ALDH− counterparts increased (Fig. 4). Taken together, these data demonstrate that ALDH+ cells self-renew, differentiate and generate ALDH− subpopulations in vitro.
Figure 4.
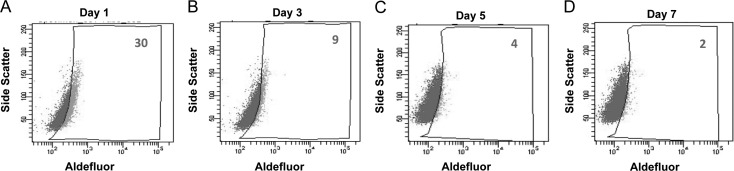
ALDH+ Tca8113 cells generate ALDH− Tca8113 cells. (A-D) ALDH+ Tca8113 cells were maintained in normal medium and ALDH activity was examined using Aldefluor assay on days 1, 3, 5 and 7.
ALDH+ tongue squamous cell carcinoma cells express CSC-related genes
Previous studies have reported that certain signaling pathways, such as Notch, Wnt and Hedgehog pathways, play critical roles in regulating CSCs for self-renewal, differentiation and proliferation (4,12). Therefore, we compared gene expression profiles between ALDH+ and ALDH− fractions using the suppression subtraction hybridization assay. Based on sequencing and bioinformatics analyses, 62 genes were found to be differentially expressed in the two fractions isolated from Tca8113 cells, among which 9 genes are involved in CSC biology (Table I). For example, the Notch2 signaling pathway is known to be essential for the growth of tumor neurospheres and xenografts (16). In summary, there were 28 upregulated and 34 downregulated genes in the ALDH+ subpopulation relative to the ALDH− fraction. Our results provide insight into understanding the molecular mechanisms of CSCs in tongue squamous cell carcinoma.
Table I.
Differentially expressed cancer stem cell-related genes.
| Gene name | Accession no. | Identified | E-value | Gene ID | Gene symbol | Chromosome location |
|---|---|---|---|---|---|---|
| Homo sapiens inositol 1,4,5-triphosphate receptor, type 1 (ITPR1), RefSeqGene on chromosome 3 | NG_016144.1 | 253/254 (99%) | 1.00E-127 | 269954693 | ITPR1 | 3p26-p25 |
| Homo sapiens notch 2 (NOTCH2), RefSeqGene on chromosome 1 | NG_008163.1 | 512/514 (99%) | 0 | 4853 | NOTCH2 | 1p13-p11 |
| Homo sapiens nuclear receptor corepressor 1 (NCOR1), transcript variant 3, mRNA | NM_001190440.1 | 213/214 (99%) | 1.00E-106 | 9611 | NCOR1 | 17p11.2 |
| Homo sapiens retinoblastoma 1 (RB1), mRNA | NM_000321.2 | 269/270 (99%) | 1.00E-137 | 5925 | RB1 | 13q14.2 |
| Homo sapiens CREB binding protein (CREBBP), RefSeqGene on chromosome | NG_009873.1 | 197/198 (99%) | 1.00E-96 | 1387 | CREBBP | 16p13.3 |
| Homo sapiens SMAD family member 1 (SMAD1), transcript variant 2, mRNA | NM_001003688.1 | 262/263 (99%) | 1.00E-133 | 4086 | SMAD1 | 7p15 |
| Homo sapiens Niemann-Pick disease, type C1 (NPC1), RefSeqGene on chromosome 18 | NG_012795.1 | 174/175 (99%) | 3.00E-83 | 4864 | NPC1 | 18q11-q12 |
| Homo sapiens epidermal growth factor receptor (EGFR), RefSeqGene on chromosome 7 | NG_007726.1 | 207/208 (99%) | 3.00E-102 | 1956 | EGFR | 7p12 |
| Homo sapiens septin 9 (SEPT9), transcript variant 4, mRNA | NM_001113495.1 | 379/380 (99%) | 0 | 10801 SEPT9 | SEPT9 | 17q25 |
Discussion
Cancer is a complex disease that results from dozens of genetic and epigenetic mutations. Although it is known that cancer cells are heterogeneous, the mechanism by which cancer cells become heterogeneous during the mulitstep process of tumorigenesis is largely unknown. The potential role of CSCs in this process has been the source of great interest in cancer biology and translational cancer research since the first identification of CSCs in leukemia by John Dick in 1997 (3), and in solid tumors by Michael Clarke in 2003 (17). The CSC model states that only a small subpopulation of cancer cells functions as cancer-initiating cells with unlimited proliferation potential, whereas the majority of cancer cells are differentiated cells with limited proliferation capacity. Thereby, the CSC hypothesis provides a rational explanation for the recurrence of tumors after chemotherapy or radiation therapy, and drug resistance. Targeting CSCs may be the key for complete inhibition or killing of cancer cells. Thus, identification and characterization of CSCs in different types of cancers can help our understanding of cancer biology and anticancer drug development.
Identification of CSC markers is the first step in CSC research. Many molecules have been used to identify a subpopulation of cancer cell lines or tumor samples which possess CSC properties. For example, in breast cancer both CD44 high/CD24 low and ALDH are CSC markers. CD133 is also a promising CSC marker for brain tumors and colon cancers (4). The ALDH superfamily of NAD(P)+-dependent multifunctional enzymes catalyze the oxidation of various aldehydes to their corresponding carboxylic acids. ALDH isozymes are widely distributed in tissues and organs. ALDH was first demonstrated to be highly enriched in hematopoietic stem cells. Stem cells isolated using Aldefluor assays have been utilized in regenerative medicine, such as bone marrow transplantation (14). Recently, ALDH activity has been reported to be a CSC marker in certain solid tumors, including breast, lung, liver, pancreas, prostate and colon cancers. In this study, Aldefluor assays were applied to enrich a subset of cells with CSC characteristics from the Tca8113 tongue squamous cell carcinoma cell line. To determine the functional differences between sorted ALDH+ and ALDH− fractions, we compared their proliferation and found that ALDH+ Tca8113 cells proliferated faster than ALDH− cells in vitro.
Self-renewal and differentiation are essential properties of CSCs. Our in vitro differentiation assays showed that ALDH+ cells were able to differentiate into ALDH− cells, and to maintain undifferentiated ALDH+ parts. By contrast, ALDH− cells did not generate ALDH+ cells under the same conditions. Non-adherent sphere formation assays have been accepted to evaluate putative CSC activity. This assay predicts that a CSC can be serially passaged for many cycles and generates a tumorsphere resembling the primary sphere. Our sphere formation and differentiation results demonstrated that ALDH+ cells could self-renew and differentiate to ALDH− cells. We also observed that a small portion of sorted ALDH− cells survived in serum-free medium and formed spheres, which may be caused by the contamination during FACS sorting procedure. Together, these data confirmed that ALDH activity is valuable in identifying tongue squamous cell carcinoma stem cells. Kang et al (7) recently reported that CD133 may be a potential tumor-initiating marker for the Tca183 cell line. They carried out similar experiments and showed that CD133+ cells possessed a higher ability of proliferation, differentiation and sphere formation than CD133− cells. It will be interesting to test whether these two markers identify the same subpopulation of Tca183 cells.
In this study, we not only showed that ALDH activity can serve as a marker to identify CSCs in tongue squamous cell carcinoma, but we also compared the gene expression patterns between isolated fractions using the suppression subtractive hybridization assay. It was interesting to find that CD44 was one of the 28 overexpressed genes in the ALDH+ fraction, indicating that ALDH+ cells may overlap with the CD44+ subpopulation. Notch2 is another upregulated gene in ALDH+ cells. It has been demonstrated that knockdown of Notch2 sensitizes glioma stem cells to radiation and impairs xenograft tumor formation (16). Further analysis of these differentially expressed genes in CSCs is required to identify new therapeutic targets of tongue squamous cell carcinoma.
Acknowledgements
This study was supported by grants from the ChongQing Science and Technology Commission Project (no. 2008-2-232). The authors thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645–662. doi: 10.1007/s10555-007-9082-y. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiao B, Johnson NW, Chen X, et al. Disclosure of a stem cell phenotype in an oral squamous cell carcinoma cell line induced by BMP-4 via an epithelial-mesenchymal transition. Oncol Rep. 2011;26:455–461. doi: 10.3892/or.2011.1299. [DOI] [PubMed] [Google Scholar]
- 7.Kang FW, Wang K, Wu M, et al. Biological characteristics of CD133+ subpopulation in tongue squamous cell carcinoma Tca8113 cell line. Hua Xi Kou Qiang Yi Xue Za Zhi. 2010;28:560–564. (In Chinese). [PubMed] [Google Scholar]
- 8.Du L, Yang Y, Xiao X, et al. Sox2 nuclear expression is closely associated with poor prognosis in patients with histologically node-negative oral tongue squamous cell carcinoma. Oral Oncol. 2011;47:709–713. doi: 10.1016/j.oraloncology.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Hayry V, Makinen LK, Atula T, et al. Bmi-1 expression predicts prognosis in squamous cell carcinoma of the tongue. Br J Cancer. 2010;102:892–897. doi: 10.1038/sj.bjc.6605544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack B, Gires O. CD44s and CD44v6 expression in head and neck epithelia. PLoS One. 2008;3:e3360. doi: 10.1371/journal.pone.0003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70:9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storms RW, Trujillo AP, Springer JB, et al. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA. 1999;96:9118–9123. doi: 10.1073/pnas.96.16.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcato P, Dean CA, Giacomantonio CA, et al. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 15.Pearce DJ, Taussig D, Simpson C, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752–760. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 16.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]