Abstract
The biosynthesis of monolignols can potentially occur via two parallel pathways involving free acids or their coenzyme A (CoA) esters. Caffeic acid 3-O-methyltransferase (COMT) and caffeoyl CoA 3-O-methyltransferase (CCOMT) catalyze functionally identical reactions in these two pathways, resulting in the formation of mono- or dimethoxylated lignin precursors. The activities of the two enzymes increase from the first to the sixth internode in stems of alfalfa (Medicago sativa L.), preceding the deposition of lignin. Alfalfa CCOMT is highly similar at the amino acid sequence level to the CCOMT from parsley, although it contains a six-amino acid insertion near the N terminus. Transcripts encoding both COMT and CCOMT are primarily localized to vascular tissue in alfalfa stems. Alfalfa CCOMT expressed in Escherichia coli catalyzes O-methylation of caffeoyl and 5-hydroxyferuloyl CoA, with preference for caffeoyl CoA. It has low activity against the free acids. COMT expressed in E. coli is active against both caffeic and 5-hydroxyferulic acids, with preference for the latter compound. Surprisingly, very little extractable O-methyltransferase activity versus 5-hydroxyferuloyl CoA is present in alfalfa stem internodes, in which relative O-methyltransferase activity against 5-hy-droxyferulic acid increases with increasing maturity, correlating with increased lignin methoxyl content.
Lignin is a complex phenylpropanoid polymer that is located in the cell walls of conducting and supporting tissues such as vascular elements and phloem fibers, where it provides hydrophobicity and mechanical strength. It is also utilized by plants as an inducible physical barrier against pathogen infection (Vance et al., 1980). The chemical treatments needed to remove lignin during the paper- pulping process are expensive and environmentally unfriendly (Boudet and Grima-Pettenati, 1996). Lignin also negatively affects forage digestibility (Albrecht et al., 1987; Sewalt et al., 1997c), the extent of which depends on its monomeric composition (Buxton and Russell, 1988), tissue distribution (Akin, 1989), and phenolic functionality (Sewalt et al., 1997a). Thus, there is currently considerable interest in the prospects for altering lignin quantity or quality by genetic engineering (Boudet and Grima-Pettenati, 1996; Campbell and Sederoff, 1996; Sewalt et al., 1997b, 1997c).
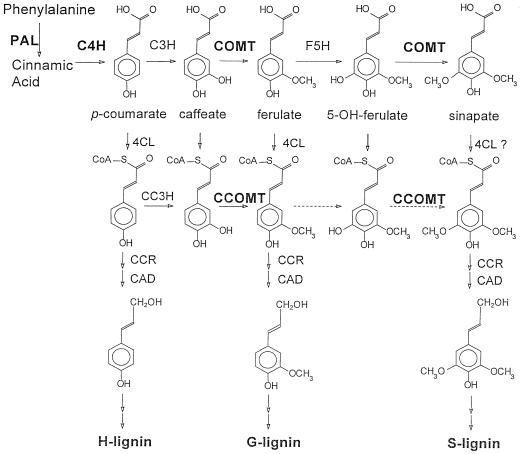
The building blocks of lignin are hydroxylated and methoxylated monomers derived from cinnamic acid. In dicotyledonous angiosperms, the major precursors are coniferyl and sinapyl alcohols, giving rise to the G (monohydroxy, monomethoxy) and S (monohydroxy, dimethoxy) components of the copolymer. A well-accepted pathway for the synthesis of these monomers involves methylation of caffeic acid to yield ferulic acid, followed by 5-hydroxylation of ferulate and a second methylation to yield sinapate (Fig. 1). In angiosperms, a bifunctional OMT appears to be involved in these conversions (Davin and Lewis, 1992), catalyzing the methylation of both caffeic and 5-hydroxyferulic acids, although it is generally referred to as COMT (EC 2.1.1.6). COMT has been purified from a wide range of plant species, including alfalfa (Medicago sativa L.) and poplar (Edwards and Dixon, 1991; Van Doorsselaere et al., 1993), and has been cloned from alfalfa, aspen, corn, tobacco, poplar, eucalyptus, and zinnia (Bugos et al., 1991; Gowri et al., 1991; Collazo et al., 1992; Dumas et al., 1992; Jaeck et al., 1992; Poeydomenge et al., 1994; Ye and Varner, 1995). COMT transcripts are present at highest levels in organs containing vascular tissue (Bugos et al., 1991; Gowri et al., 1991; Collazo et al., 1992). OMTs active against caffeic acid are induced at the onset of lignification in plants responding to infection by viruses (Jaeck et al., 1992) and fungi (Maule and Ride, 1976). Reduction of COMT activity by antisense expression in transgenic plants results in changes in lignin content and/or composition (Dwivedi et al., 1994; Ni et al., 1994; Atanassova et al., 1995; Van Doorsselaere et al., 1995; Sewalt et al., 1997c). Brown-mid-rib mutants of maize and sorghum, which have altered lignin composition and reduced lignin concentration, have been shown to be deficient in COMT (Grand et al., 1985; Vignols et al., 1995) or COMT and cinnamyl alcohol dehydrogenase (Pillonel et al., 1991). Thus, a considerable body of evidence implicates COMT as an important enzyme of lignin biosynthesis.
Figure 1.
Alternative pathways for the methylation of monolignols. The enzymes are: PAL, l-phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; C3H, coumarate 3-hydroxylase; COMT, bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase; F5H, ferulate 5-hydroxylase; CC3H, coumaroyl CoA 3-hydroxylase; CCOMT, bispecific caffeoyl/5-hydroxyferuolyl CoA O-methyltransferase; 4CL, coumarate (hydroxycinnamate) CoA ligase; CCR, cinnamoyl CoA reductase; and CAD, coniferyl alcohol dehydrogenase. Dashed arrows represent reactions that have yet to be clearly demonstrated in vitro. cDNA clones encoding enzymes marked in bold were obtained from alfalfa. In some species, 4CL has very little activity with sinapate as substrate.
Work with parsley and carrot cell-suspension cultures has drawn attention to an alternative pathway for methylation of hydroxycinnamic acid derivatives. An enzyme that converts caffeoyl CoA to feruloyl CoA (CCOMT, EC 2.1.1.104; Fig. 1) was shown to be induced by elicitor treatment in both systems (Kühnl et al., 1989; Pakusch et al., 1989). This enzyme was proposed to be involved in the synthesis of wall-esterified ferulic acid as a component of the plant's defense response (Kühnl et al., 1989; Pakusch et al., 1989). More recently, however, studies of vascular differentiation in isolated mesophyll protoplasts of zinnia have shown that CCOMT transcripts and activity are highly induced during appearance of tracheary elements, whereas COMT activity did not correlate with lignification of these elements in this system (Ye et al., 1994). COMT transcripts appeared to be localized to phloem and xylem fibers rather than to tracheary elements in zinnia stems (Ye and Varner, 1995). Strong down-regulation of bispecific COMT by antisense expression in tobacco and poplar leads to the production of lignin with a decreased S:G ratio that also contains 5-hydroxy-G residues (Atanassova et al., 1995; Van Doorsselaere et al., 1995). This suggests that COMT and CCOMT may be functionally redundant with respect to methylation of the caffeate moiety but that CCOMT does not effectively methylate the 5-hydroxyferulate moiety in vivo. Clearly, more studies of the biochemical properties of CCOMT and its expression relative to that of COMT are needed.
Little is known about the comparative developmental expression of both COMT and CCOMT in the same species. Although it has been shown that the two enzymes are differentially expressed in zinnia stems (Ye and Varner, 1995), this study did not address how the enzymes change during stem development in relation to changes in lignin content and composition. After characterizing and cloning alfalfa COMT (Edwards and Dixon, 1991; Gowri et al., 1991), we have now isolated and functionally characterized a cDNA clone encoding the alfalfa CCOMT. We present here a comparative study of the developmental expression and catalytic properties of alfalfa COMT and CCOMT. These studies provide a basis for the targeted genetic manipulation of lignin biosynthesis in a key forage crop by altering expression of multiple OMTs.
MATERIALS AND METHODS
Sampling of Alfalfa (Medicago sativa L.) Tissue
Internodes from greenhouse-grown alfalfa plants (cv Apollo) were ground under liquid nitrogen and part of the powdered tissue was stored at −70°C for enzyme assays. The remaining portion was freeze dried for lignin analyses.
Chemicals
5-Hydroxyferulic acid was synthesized via 5-hydroxyvanillin by the methods of Banerjee et al. (1962) and Pearl and Beyer (1951). Its structure was confirmed by 1H- and 13C-NMR analysis. CoA esters of caffeic and 5-hydroxyferulic acids were prepared according to the method of Stöckigt and Zenk (1975), and identified and quantified spectrophotometrically as described by Lüderitz et al. (1982).
Enzyme Extraction and Assay
Powdered plant tissue was extracted for 20 min at 4°C in extraction buffer (100 mm Tris-HCl, 0.2 mm MgCl2, 2.0 mm DTT, and 10% [v/v] glycerol). The extraction buffers for COMT and CCOMT differed in pH (7.2 for COMT, 7.5 for CCOMT). After the sample was centrifuged (12,000g, 4°C, 10 min), extracts were desalted on a PD-10 column (Pharmacia). Soluble-protein concentration in the enzyme extracts was determined using the Bradford dye-binding reagent (Bio-Rad) with BSA as the standard. Enzyme activities were assayed as described previously for COMT (Gowri et al., 1991) and CCOMT (Ni et al., 1996).
When reaction products were monitored by HPLC, nonlabeled S-adenosyl l-Met was used as a methyl donor. The reaction was terminated by addition of 10 μL of 1 n HCl. After the insoluble material was removed by centrifugation at 10,000g for 5 min, 20 μL of supernatant was subjected directly to HPLC as follows: column, ODS2 (Waters, 5 μm, 250 × 4.6 mm); gradient elution, solvent A (1% H3PO4) and solvent B (CH3CN) (gradient: 0–5 min, 5% solvent B; 5–10 min, 5–10% solvent B; 10–25 min, 10–17% solvent B; 25–35 min, 17–29% solvent B; 35–36 min, 29–100% solvent B); flow rate, 1.0 mL/min; detection, diode array.
Determination of Lignin Concentration
Powdered tissue was freeze dried, ground to pass a 1-mm sieve, and extracted with boiling neutral detergent (Van Soest et al., 1991) using filter bags in a batch fiber analyzer (ANKOM, Fairport, NY). The residual neutral detergent fiber, a pectin-free cell wall preparation, was oven dried (55°C) and used for quantitation of Klason lignin according to the work of Kaar et al. (1991), modified for microanalytical scale. Briefly, 100 mg of neutral detergent fiber was suspended in 1 mL of 72% H2SO4 in 50-mL reaction tubes kept in a water bath at 30°C for 1 h. The initial hydrolysis was followed by dilution to 4% H2SO4 and autoclaving at 121°C for 1 h. The hydrolysis mixture was passed through a previously tared glass-fiber filter (Whatman grade 934 AH, particle retention 1.5 mm) in a tared 30-mL gooch crucible of medium porosity (pore size, 4–5.5 μm). The residue (Klason lignin) and filter were oven dried overnight (105°C) and weighed. Acid-soluble lignin was estimated in the filtrate by UV absorption at 205 nm (Technical Association of the Pulp and Paper Industry, 1989).
Determination of Lignin Methoxyl Content
Lignin methoxyl groups were determined by reaction of Klason lignin with 3 mL of hydriodic acid (57%) in 22-mL vials equipped with Teflon-lined valves and heated for 25 to 30 min at 130°C in a dry bath (Baker, 1996). After the sample was cooled on ice an appropriate amount of internal standard (ethyl iodide) and 3 mL of pentane were added (through the valve). The mixture was vortexed and placed on ice for 2 min to minimize volatilization, and 1 mL of the pentane phase was placed in glass vials for GC analysis of methyl iodide (released from lignin methoxyl groups) and ethyl iodide (internal standard). The GC column and conditions were as described by Baker (1996) with the following adaptations: the column temperature was 40°C for 2 min, then increased to 80°C (10°C/min), and then held at 80°C for 1 min; the run time was 7 min. Accuracy of the quantitation was confirmed by comparison of the GC method with standard T 209 su-72 (Technical Association of the Pulp and Paper Industry, 1972), using a mixed hardwood lignin sample.
RNA-Blot Analysis
RNA was prepared according to the method of Chomczynski and Sacchi (1987). Samples of total RNA (10 μg) were fractionated on a formaldehyde/denaturing gel according to standard procedures (Sambrook et al., 1989) and blotted to a Hybond N nylon membrane (Amersham) according to the manufacturer's instructions. Blots were probed with 32P-labeled alfalfa COMT and CCOMT cDNAs and washed at high stringency (final wash 0.2× SSC and 0.1% SDS at 68°C).
Tissue Print Analysis
Plants with at least eight internodes were sampled. Counting from the top (the youngest internode), tissue prints and corresponding stem sections were prepared for the second, third, fifth, and seventh internodes.
Nylon membranes (GeneScreen, DuPont) were used without pretreatment. The membrane was placed on top of three layers of Whatman 1MM paper. The face of the stem freshly cut with a double-edge razor blade was printed onto a membrane for 15 to 30 s. After printing, the stem was sectioned to a 100-mm thickness and then stained for 1 min with 1% aqueous safranin O for observation of stem anatomy. The printed membrane was UV illuminated using a UV Stratalinker (Stratagene), air dried overnight, washed in 0.2× SSC and 1% SDS for 2 h at 65°C, and prehybridized for 2 h at 68°C in DIG Easy Hyb (Boehringer Mannheim).
Plasmids containing alfalfa COMT and CCOMT cDNAs in pBluescript SK(−) were linearized by digestion with either HindIII or SmaI and used as templates for the in vitro synthesis of digoxigenin-labeled sense and antisense RNA probes. The probes were synthesized according to the manufacturer's protocol (Boehringer Mannheim). Hybridization of the probes to the membrane was carried out at 68°C for 16 h. The membrane was washed two times for 10 min each in 2× SSC and 0.1% SDS at room temperature and then two times for 20 min each in 0.2× SCC and 0.1% SDS at 68°C. Immunological color detection of digoxigenin-labeled probes was performed using anti-digoxigenin-alkaline phosphatase and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate p-toluidine salt (Boehringer Mannheim) according to the manufacturer's instructions.
Expression of COMT and CCOMT in E. coli
COMT and CCOMT cDNAs were expressed in E. coli from the pBluescript SK(−) (Stratagene) vector, as described previously for alfalfa COMT (Gowri et al., 1991). The polylinker region at the 5′ end of the pCCOMT1 open reading frame was modified by the addition of 4 bp to place the CCOMT-coding sequence in-frame with the lacZ initiation codon, with no intervening stop codons. This was done by filling in BamHI recessed ends followed by blunt- end re-ligation.
All procedures were performed at 4°C unless otherwise stated. E. coli DH5α transformed with pBluescript SK(−), pCOMT1, or pCCOMT1 was grown at 37°C in Luria-Bertani medium (Bio101, Inc., Vista, CA) with 50 μg/mL ampicillin until the culture reached stationary phase. The cells were harvested by centrifugation for 15 min at 3,800g and resuspended in a buffer containing 100 mm Tris-HCl, pH 7.5, 2.0 mm DTT, 0.20 mm MgCl2, and 10% glycerol. Insoluble materials were removed by centrifugation at 10,000g for 5 min. The resultant supernatant was desalted on a PD-10 column (Pharmacia) and then concentrated using Centricon-10 (Amicon, Beverly, MA). OMT activities of the protein solution were examined as described above with the following modifications: the reaction was performed for 20 min at 30°C, and the reaction mixtures (50 μL) contained 100 mm Tris-HCl, pH 7.5, 2.0 mm DTT, 0.20 mm MgCl2, 10% glycerol, 0.1 mm each phenolic substrate, 0.06 mm [14C]S-adenosyl l-Met (13 mCi/mmol), and 20 μg of protein.
RESULTS AND DISCUSSION
Activity of COMT and CCOMT in Relation to Lignification
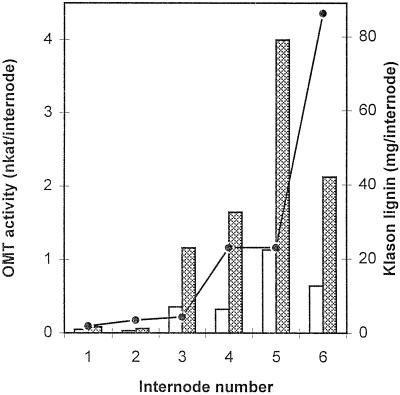
Individual alfalfa internodes of progressive maturity were assayed for COMT, CCOMT, and Klason lignin. A typical profile is shown in Figure 2, in which total amounts of enzyme activity per internode are compared with total lignin accumulation (in milligrams) per internode. A large increase in activity of both COMT and CCOMT occurred before complete elongation (in the third visible internode) and was maintained during active lignification. Apparently, the first large increment in lignin accumulation lags behind the initial increase in activity of methylating enzymes by about 36 to 48 h (the approximate time difference in development of consecutive internodes). CCOMT activity appeared to be higher than that of COMT in this experiment, although activities of the two enzymes were more similar to each other in subsequent experiments (see below). This apparent discrepancy might be due to the fact that the concentration of caffeic acid in the COMT assay was 0.5 mm in the experiment shown in Figure 2 but 0.1 mm (the concentration of caffeoyl CoA in both experiments) in the experiment shown in Figure 7.
Figure 2.
Activities of OMTs in developing alfalfa stem internodes. Total activities of COMT (white bars) and CCOMT (shaded bars) are shown in individual alfalfa internodes in relation to the total amount of lignin (line) per internode.
Figure 7.
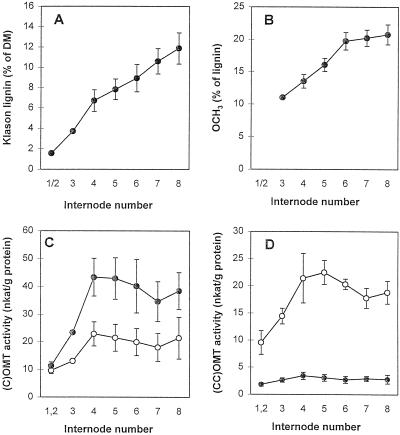
OMT substrate preference in alfalfa stem internodes in relation to lignin composition. A, Lignin content in individual internodes (counting from the top). DM, Dry matter. B, Lignin methoxyl content. C, OMT activity against free acid substrates. ○, Activity against caffeic acid; •, activity against 5-OH ferulic acid. D, OMT activity against CoA esters. ○, Activity against caffeoyl CoA; •, activity against 5-hydroxyferuloyl CoA. Error bars represent sd above and below the mean (n = 3 individual plants).
Molecular Cloning of Alfalfa CCOMT
PCR amplification of parsley genomic DNA was used to obtain a partial-length CCOMT sequence for screening an alfalfa cDNA library prepared from RNA isolated from elicited cell cultures (Dalkin et al., 1990). The primers used for PCR amplification corresponded to nucleotides 470 to 490 and 896 to 916 of the parsley CCOMT sequence (Schmitt et al., 1991) and resulted in the amplification of a 900-bp fragment, as opposed to the predicted 446-bp fragment, suggesting the presence of an intron. After the identity was confirmed by sequence analysis of 3′ and 5′ ends, the partial clone was used to screen the alfalfa cDNA library. A single, full-length alfalfa CCOMT cDNA was isolated, which was 83% identical and 93% similar to the parsley CCOMT at the amino acid level (Fig. 3). The nucleotide sequence of the alfalfa CCOMT cDNA can be found in the GenBank database, accession no. U20736. The alfalfa CCOMT had a six-amino acid insertion in the N-terminal region compared with the parsley enzyme. This insertion is not present in CCOMT from zinnia (Ye et al., 1994) or Stellaria longipes (Zhang et al., 1995). Its presence in alfalfa CCOMT was confirmed by sequencing two additional independent clones from the cDNA library.
Figure 3.
Comparison of the deduced amino acid sequences of alfalfa (Ms) and parsley (Pc) CCOMT. Six additional amino acids were found in the N-terminal region of the alfalfa sequence. Asterisks indicate nonconservative differences.
Southern-blot analysis indicated three hybridizing fragments recognized by the CCOMT probe in alfalfa genomic DNA digested with DraI, three fragments in EcoRI digests, and six fragments in EcoRV digests (data not shown). None of these enzymes cuts within the CCOMT open reading frame. In view of the tetraploid nature of alfalfa, CCOMT is probably encoded by at least two genes in the alfalfa genome.
Developmental Expression of COMT and CCOMT Transcripts in Alfalfa
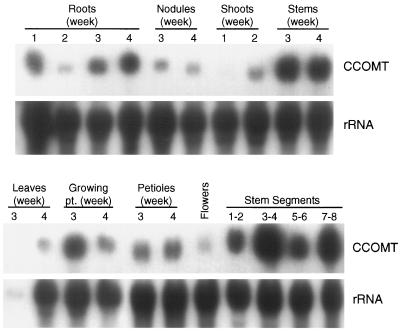
The expression patterns of CCOMT transcripts in tissues of developing alfalfa plants were first determined using RNA-blot analysis (Fig. 4). CCOMT transcripts were most strongly expressed in stems, roots, and petioles, with low expression in nodules, flowers, and leaves. Maximal levels in stems were observed at 3 to 4 weeks of development, and the highest level of transcripts in stem tissue appeared to be in the third and fourth internodes. Measurement of CCOMT activities in the same internode samples as analyzed in Figure 4 gave values (pkat/mg protein) of 6.9 (internodes 1 and 2), 20.7 (internodes 3 and 4), 19.6 (internodes 5 and 6), and 17.4 (internodes 7 and 8), showing a good correlation between enzyme activity and transcript level except in internodes 5 and 6. Probing the same blot with a COMT probe revealed a nearly identical pattern of developmental expression, with highest levels of expression in stems and roots after 3 to 4 weeks of development (data not shown). These observations confirm the results of previous alfalfa COMT transcript expression studies of Gowri et al. (1991).
Figure 4.
Tissue distribution of CCOMT transcripts in developing alfalfa seedlings. Total RNA from various alfalfa organs harvested from plants grown for the number of weeks shown was subjected to northern-blot analysis, using the full-length alfalfa CCOMT sequence as probe. The numbers for stem segments indicate internode number from the top of the plant. The blot was reprobed with an rRNA sequence to check for loading and transfer efficiency. pt., Point.
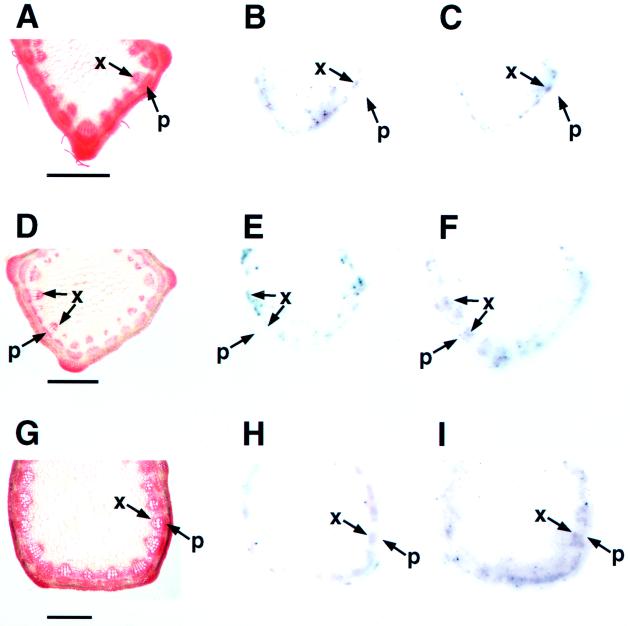
In zinnia, COMT and CCOMT transcripts are differentially localized to different cell types during vascular development (Ye and Varner, 1995). To examine the cellular distribution of COMT and CCOMT transcripts in alfalfa stems, tissue prints were made of alfalfa stem internodes at different developmental stages. The location of the hybridization signal was determined by superimposition of the signals on the tissue prints with sections stained to show the cellular anatomy. Transcripts encoding both COMT and CCOMT were localized to the vascular tissue where lignin is deposited (Fig. 5). However, the temporal and spatial expression of the OMT transcripts was not identical. In the second internode, both COMT and CCOMT transcripts were localized to xylem tissue, most probably in the region in which xylem was differentiating. In contrast, COMT transcripts in the third and fifth internodes were mainly localized to xylem, whereas CCOMT transcripts were detected in both xylem and phloem. Signals for both COMT and CCOMT transcripts were faint in tissue prints of the seventh internode but were still clearly localized to xylem (data not shown). Control hybridizations with COMT and CCOMT sense RNA probes gave no signal on prints of any of the internodes.
Figure 5.
Tissue print localization of COMT and CCOMT transcripts in alfalfa stem internodes. Pictures indicate anatomy and COMT and CCOMT transcript distribution in the second (A–C), third (D–F), and fifth (G–I) internodes from the top of the stem. A, D, and G show sections stained with safranin O. B, E, and H are corresponding tissue prints hybridized with a COMT antisense probe, and C, F, and I are prints hybridized with a CCOMT antisense probe. p, Phloem; and x, xylem. Bars = 1.0 mm.
The above localization pattern is different from that reported in zinnia, where COMT transcripts were predominantly localized to phloem fibers and CCOMT transcripts were mainly present in the xylem of the younger internodes (Ye and Varner, 1995). However, in addition to demonstrating that both COMT and CCOMT can be expressed in the same cell types in alfalfa, our data are also consistent with the hypothesis of Ye and Varner (1995) that the two enzymes may be involved in the formation of different types of lignin in different cell types.
Substrate Specificities of Alfalfa COMT and CCOMT
Expression studies in E. coli were performed for two reasons: first, to provide functional evidence for the identity of the presumed CCOMT cDNA, especially in view of the sequence insertion at the N terminus, and second, to obtain information concerning the relative substrate specificities of COMT and CCOMT. In particular, the activities of the two enzymes against 5-hydroxyferuloyl CoA had not been tested until now. Alfalfa COMT (Gowri et al., 1991) and CCOMT cDNAs were engineered into the pBluescript E. coli expression vector. The target protein accumulation in the bacterial cell lysate was not induced significantly by addition of isopropyl-β-thiogalactopyranoside (up to 1 mm) as previously demonstrated (Gowri et al., 1991). Therefore, stationary-phase culture without isopropyl-β-thiogalactopyranoside treatment was used as an enzyme source.
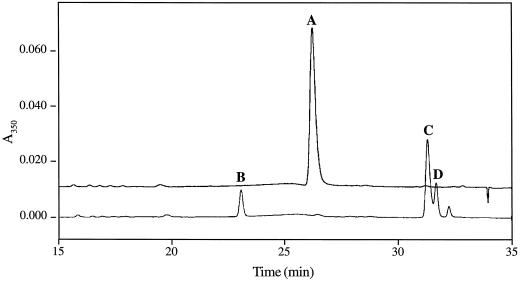
The protein extracts from E. coli harboring pCCOMT1 methylated CoA esters efficiently (Table I). The activity was absent in controls using the cell lysate from E. coli transformed with the vector pBluescript SK(−). The CCOMT-mediated reaction was also monitored by HPLC (Fig. 6). A small but significant conversion of the CoA esters to free acids was observed, which might be due to esterases present in the protein extracts. However, the UV/visible spectrum of the compound corresponding to product peak C in Figure 6 confirmed that it was indeed a CoA ester (sinapoyl CoA, data not shown). These results confirm that the pCCOMT1 cDNA clone encodes a protein with CCOMT activity.
Table I.
Relative activities of E. coli expressed alfalfa COMT and CCOMT against hydroxycinnamic acids and their corresponding CoA esters
| Plasmid | Substrate
|
|||
|---|---|---|---|---|
| Caffeic acid | 5-Hydroxyferulic acid | Caffeoyl CoA | 5-Hydroxyferuloyl CoA | |
| pBluescript SK(−) | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 |
| pCOMT1 | 8.8 ± 0.6 (1)a | 19.5 ± 0.4 (2.2) | 3.8 ± 0.0 (0.4) | 10.1 ± 0.9 (1.1) |
| pCCOMT1 | 0.4 ± 0.0 (1) | 5.4 ± 0.4 (15.5) | 46.0 ± 1.0 (131) | 26.5 ± 1.7 (75) |
Activities are expressed as specific activities (pkat/mg protein), means ± sd (n = 2).
Numbers in parentheses indicate activities relative to that for caffeic acid.
Figure 6.
O-Methylation of hydroxycinnamoyl CoA esters by alfalfa CCOMT expressed in E. coli. HPLC chromatogram of the OMT reaction mixture using 5-hydroxyferuloyl CoA as substrate. For conditions, see Methods. Each peak was identified by direct comparison with authentic standards (B and D) and/or its UV/visible spectrum. A, 5-Hydroxyferuloyl CoA; B, 5-hydroxyferulic acid; C, sinapoyl CoA; and D, sinapic acid. No free acids (B and D) or sinapoyl CoA (C) were detected when the assay mixture was incubated without protein solution (upper trace).
Cell-free extracts of the CCOMT-expressing bacteria also methylated free hydroxycinnamic acids. The activity with caffeic acid was less than 1% of that with caffeoyl CoA, whereas activity with 5-hydroxyferulic acid was about 20% of that with the corresponding CoA ester. The ratio of methylation of caffeoyl CoA to 5-hydroxyferuloyl CoA in the extracts of pCCOMT1-transformed E. coli was 1.7, whereas that of caffeic acid to 5-hydroxyferulic acid in pCOMT1-transformed bacteria was 0.45 (Table I). Thus, CCOMT preferentially methylates at the caffeoyl level, and COMT preferentially methylates at the 5-hydroxyferuloyl level.
The cell-free extracts of E. coli transformed with pCOMT1 showed significant activity with the CoA esters (Table I), in good agreement with previous results (Meng and Campbell, 1996). However, no CoA ester products could be detected by HPLC (data not shown). Therefore, the substrates that are methylated by COMT in assays in which products are not individually separated are most likely the free acids formed by hydrolysis of the CoA esters in the protein extracts.
Few studies have determined the relative substrate preferences of COMT and CCOMT against both caffeic and 5-hydroxyferulic acids and their CoA esters. Aspen lignin OMT purified after expression in E. coli has relative activity ratios of 100:220:36 against caffeic acid, 5-hydroxyferulic acid, and caffeoyl CoA, respectively (Meng and Campbell, 1996), similar to the results reported here for alfalfa COMT. In contrast, a novel multifunctional OMT has recently been cloned from loblolly pine (Li et al., 1997). This enzyme, when expressed in yeast, catalyzes the O-methylation of caffeic acid, 5-hydroxyferulic acid, caffeoyl CoA, and 5-hydroxyferuloyl CoA in relative activity ratios of 100:76:86:68.
Developmental Patterns of OMT Substrate Preference in Alfalfa Stem Tissue in Relation to Lignin Composition
Protein extracts from individual internodes of young alfalfa stems were assayed for OMT activity using caffeic acid, 5-hydroxyferulic acid, caffeoyl CoA, and 5-hydroxyferuloyl CoA as substrates under the appropriate assay conditions for COMT or CCOMT. Activities with caffeic acid and caffeoyl CoA were similar in this experiment (in contrast to the results shown in Fig. 2) and increased in parallel, preceding the changes in lignin deposition as shown in Figure 2. However, the highest activity occurred with 5-hydroxyferulic acid, and the ratio of activity with 5-hydroxyferulic acid to that with caffeic acid increased with increasing developmental age (Fig. 7). This correlated with a steady increase in overall lignin methoxyl group content in successive internodes, suggesting an increased lignin S:G ratio, in agreement with the results of recent detailed analyses of lignin deposition during alfalfa stem differentiation (Vallet et al., 1996). Surprisingly, however, in view of the activity of both COMT and CCOMT with 5-hydroxyferuloyl CoA when expressed in E. coli, only very low activity with this compound was observed in alfalfa stem extracts, and this did not change significantly with developmental age. This contrasts with the situation in loblolly pine stem extracts, in which activity with 5-hydroxyferuloyl CoA is higher than with either caffeic or 5-hydroxyferulic acids (Li et al., 1997). In zinnia cells differentiating tracheary elements, OMT activity with 5-hydroxyferuloyl CoA is approximately one-half that with caffeoyl CoA, but both activities increase in parallel during vascular differentiation (Ye et al., 1994).
It was recently demonstrated that another enzyme of lignin biosynthesis, 4-coumarate:CoA ligase, has a different substrate specificity for the (hydroxy) cinnamate moiety when expressed in E. coli than it does when assayed in stem extracts (Lee and Douglas, 1996), and it was suggested that additional cellular factors may be involved in controlling the substrate specificity of this enzyme. It is therefore now important to re-evaluate the substrate specificity of COMT and CCOMT following exhaustive purification from alfalfa stem extracts.
In developing wheat seedlings, OMT activity against caffeic acid peaks early in development, prior to lignin deposition, and then declines, whereas OMT activity against 5-hydroxyferulic acid parallels the later process of lignification (Lam et al., 1996). In monocots, early esterification of cell wall arabinoxylans with ferulate residues may require a different OMT from that involved in lignification. Although the ratio of OMT activity with 5-hydroxyferulic acid compared with caffeic acid increases during development in alfalfa stems, the coordinated increase in both activities is still consistent with the involvement of a single class of bifunctional OMT for methylation of caffeic and 5-hydroxyferulic acids.
Implications for the Genetic Engineering of Lignin
The data indicating that CCOMT activity is at least as high as COMT activity throughout development in alfalfa stems and that both COMT and CCOMT are preferentially expressed in stem vascular tissue point to the probable importance of both OMTs in lignification. These observations also suggest the possible operation of a metabolic grid for the formation of monolignols. For example, 5-hy-droxyferulate could in theory be converted to sinapoyl CoA through the COMT reaction or, following CoA esterification, via the CCOMT reaction. Such a metabolic grid would provide a route for by-passing a metabolic block imposed via antisense down-regulation of one of the OMTs in transgenic plants.
Experiments in which COMT has been down-regulated by antisense technology have sometimes, but not always, resulted in a decrease in the lignin S:G ratio (Dwivedi et al., 1994; Ni et al., 1994; Atanassova et al., 1995; Van Doorsselaere et al., 1995; Sewalt et al., 1997c). The differences in the results of different groups might be explained on the basis of different relative activities of COMT and CCOMT in the species under study or in tissues of the same species examined at different developmental stages. Our observation of the different substrate (caffeate moiety versus 5-hydroxyferulate moiety) preferences for COMT compared with CCOMT predicted a decrease in the S:G ratio on down-regulation of COMT in transgenic alfalfa, in view of the preferential activity of COMT in the production of the S moiety. The relatively poor utilization of the CoA ester pathway for the formation of S lignin that would be predicted from our results is consistent with the observation that a mutation in ferulate 5-hydroxylase in Arabidopsis leads to formation of lignin lacking S units (Chapple et al., 1992).
Experiments are now in progress that utilize antisense strategies to down-regulate COMT and CCOMT singly and in combination. These studies should confirm whether each enzyme can compensate for reduced expression of the other, and whether they make identical or different contributions to the final pattern of lignin methoxylation.
ACKNOWLEDGMENTS
We thank Drs. San-Jung Lee and Tom Mabry for synthesis of 5-hydroxyferulic acid, David Huhman for assistance with GC analyses, Ralph Kowatsch for technical assistance, Cuc Ly and Darla Boydstone for help with graphics, and Drs. Dusty Post-Beittenmiller and William Schneider for critical reading of the manuscript.
Abbreviations:
- CCOMT
caffeoyl CoA 3-O-methyltransferase
- COMT
caffeic acid 3-O-methyltransferase
- G
guaiacyl
- OMT
O-methyltransferase
- S
syringyl
Footnotes
This work was supported by the Samuel Roberts Noble Foundation.
The nucleotide sequence data reported will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession no. U20736 (alfalfa S-adenosyl-1-Met:trans-caffeoyl CoA 3-O-methyltransferase).
LITERATURE CITED
- Akin DE. Histological and physical factors affecting digestibility of forages. Agron J. 1989;81:17–25. [Google Scholar]
- Albrecht KA, Wedin WF, Buxton DR. Cell-wall composition and digestibility of alfalfa stems and leaves. Crop Sci. 1987;27:735–741. [Google Scholar]
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier M-T, Monties B, Fritig B, Legrand M. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995;8:465–477. [Google Scholar]
- Baker SM. Rapid methoxyl analysis of lignins using gas chromatography. Holzforschung. 1996;50:573–574. [Google Scholar]
- Banerjee SK, Manolopoulo M, Pepper JM. The synthesis of lignin model substances: 5-hydroxyvanillin and 5-hydroxyacetoguaiacone. Can J Chem. 1962;40:2175–2177. [Google Scholar]
- Boudet AM, Grima-Pettenati J. Lignin genetic engineering. Mol Breeding. 1996;2:25–39. [Google Scholar]
- Bugos RC, Chiang VLC, Campbell WH. cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyl-transferase of aspen. Plant Mol Biol. 1991;17:1203–1215. doi: 10.1007/BF00028736. [DOI] [PubMed] [Google Scholar]
- Buxton DR, Russell JR. Lignin constituents and cell-wall digestibility of grass and legume stems. Crop Sci. 1988;28:553–558. [Google Scholar]
- Campbell MM, Sederoff RR. Variation in lignin content and composition. Mechanisms of control and implications for the genetic improvement of plants. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CCS, Vogt T, Ellis BE, Somerville CR. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collazo P, Montoliu L, Puigdoménech P, Rigau J. Structure and expression of the lignin O-methyl-transferase gene from Zea mays L. Plant Mol Biol. 1992;20:857–867. doi: 10.1007/BF00027157. [DOI] [PubMed] [Google Scholar]
- Dalkin K, Jorrin J, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). VII. Induction of defense-related mRNAs in elicitor-treated cell suspension cultures. Physiol Mol Plant Pathol. 1990;37:293–307. [Google Scholar]
- Davin LB, Lewis NG. Phenylpropanoid metabolism: biosynthesis of monolignols, lignans and neolignans, lignins and suberins. Rec Adv Phytochem. 1992;26:325–375. [Google Scholar]
- Dumas B, Van Doorsselaere J, Gielen J, Legrand M, Fritig B, Van Montagu M, Inzé D. Nucleotide sequence of a complementary DNA encoding O-methyltransferase from poplar. Plant Physiol. 1992;98:796–797. doi: 10.1104/pp.98.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi UN, Campbell WH, Yu J, Datla RSS, Bugos RC, Chiang VL, Podila GK. Modification of lignin biosynthesis in transgenic Nicotiana through expression of an antisense O-methyltransferase gene from Populus. Plant Mol Biol. 1994;26:61–71. doi: 10.1007/BF00039520. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon RA. Purification and characterization of S-adenosyl-1-methionine: caffeic acid 3-O-methyl-transferase from suspension cultures of alfalfa (Medicago sativa L.) Arch Biochem Biophys. 1991;287:372–379. doi: 10.1016/0003-9861(91)90492-2. [DOI] [PubMed] [Google Scholar]
- Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). X. Molecular cloning and expression of S-adenosyl-1-methionine: caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol. 1991;97:7–14. doi: 10.1104/pp.97.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand CP, Parmentier P, Boudet A, Boudet AM. Comparison of lignins and of enzymes involved in lignification in normal and brown midrib (bm3) mutant corn seedlings. Physiol Veg. 1985;23:905–911. [Google Scholar]
- Jaeck E, Dumas B, Geoffroy P, Favet N, Inzé D, Van Montagu M, Fritig B, Legrand M. Regulation of enzymes involved in lignin biosynthesis: induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol Plant-Microbe Interact. 1992;5:294–300. doi: 10.1094/mpmi-5-294. [DOI] [PubMed] [Google Scholar]
- Kaar WE, Cool LG, Merriman MM, Brink DL. The complete analysis of wood polysaccharides using HPLC. J Wood Chem Technol. 1991;11:447–463. [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E. Elicitor induced S-adenosyl-1-methionine: caffeoyl-CoA 3-O-methyltransferase from carrot cell suspension cultures. Plant Sci. 1989;60:21–25. [Google Scholar]
- Lam TBT, Iiyama K, Stone BA. Caffeic acid: O-me-thyltransferases and the biosynthesis of ferulic acid in primary cell walls of wheat seedlings. Phytochemistry. 1996;41:1507–1510. [Google Scholar]
- Lee D, Douglas CJ. Two divergent members of a tobacco 4-coumarate: coenzyme A ligase (4CL) gene family. cDNA structure, gene inheritance and expression, and properties of recombinant proteins. Plant Physiol. 1996;112:193–205. doi: 10.1104/pp.112.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Popko JL, Zhang X-H, Osakabe K, Tsai C-J, Joshi CP, Chiang VL. A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc Natl Acad Sci USA. 1997;94:5461–5466. doi: 10.1073/pnas.94.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüderitz T, Shatz G, Grisebach H. Enzymic synthesis of lignin precursors. Purification and properties of 4-coumarate: CoA ligase from cambial sap of spruce (Picea abies L.) Eur J Biochem. 1982;123:583–586. [PubMed] [Google Scholar]
- Maule AJ, Ride JP. Ammonia-lyase and O-methyl transferase activities related to lignification in wheat leaves infected with Botrytis. Phytochemistry. 1976;15:1661–1664. [Google Scholar]
- Meng H, Campbell WH. Characterization and site-directed mutagenesis of aspen lignin-specific O-methyltransferase expressed in Escherichia coli. Arch Biochem Biophys. 1996;330:329–341. doi: 10.1006/abbi.1996.0260. [DOI] [PubMed] [Google Scholar]
- Ni W, Paiva NL, Dixon RA. Reduced lignin in transgenic plants containing an engineered caffeic acid O-methyltransferase antisense gene. Transgenic Res. 1994;3:120–126. [Google Scholar]
- Ni W, Sewalt VJH, Korth KL, Blount JW, Ballance GM, Dixon RA. Stress responses in alfalfa (Medicago sativa L.). XXI. Activation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase genes does not contribute to changes in metabolite accumulation in elicitor-treated cell suspension cultures. Plant Physiol. 1996;112:717–726. doi: 10.1104/pp.112.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakusch AE, Kneusel RE, Matern U. S-Adenosyl-1-methionine: trans-caffeoyl-coenzyme A 3-O-methyltransferase from elicitor-treated parsley cell suspension cultures. Arch Biochem Biophys. 1989;271:488–494. doi: 10.1016/0003-9861(89)90299-3. [DOI] [PubMed] [Google Scholar]
- Pearl IA, Beyer DL. Reactions of vanillin and its derived compounds. XI. cinnamic acids derived from vanillin and its related compounds. J Org Chem. 1951;16:216–221. [Google Scholar]
- Pillonel C, Mulder MM, Boon JJ, Forster B, Binder A. Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor L. Moench. Planta. 1991;185:538–544. doi: 10.1007/BF00202964. [DOI] [PubMed] [Google Scholar]
- Poeydomenge O, Boudet AM, Grima-Pettenati J. A cDNA encoding S-adenosyl-1-methionine: caffeic acid 3-O-methyltransferase from Eucalyptus. Plant Physiol. 1994;105:749–750. doi: 10.1104/pp.105.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitt D, Pakusch A-E, Matern U. Molecular cloning, induction, and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem. 1991;266:17416–17423. [PubMed] [Google Scholar]
- Sewalt VJH, Beauchemin KA, Glasser WG. Lignin impact on fiber degradation. 3. Reversal of inhibition of enzymatic hydrolysis by chemical modification of lignin and by additives. J Agric Food Chem. 1997a;45:1823–1828. [Google Scholar]
- Sewalt VJH, Ni W, Blount JW, Jung HG, Howles PA, Masoud S, Lamb C, Dixon RA. Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of phenylalanine ammonia-lyase or cinnamate 4-hy-droxylase. Plant Physiol. 1997b;115:41–50. doi: 10.1104/pp.115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt VJH, Ni W, Jung H, Dixon RA. Lignin impact on fiber degradation: increased enzymatic digestibility of genetically engineered tobacco (Nicotiana tabacum) stems reduced in lignin content. J Agric Food Chem. 1997c;45:1977–1983. [Google Scholar]
- Srebotnik E, Messner K. A simple method that uses differential staining and light microscopy to assess the selectivity of wood delignification by white rot fungi. Appl Environ Microbiol. 1994;60:1383–1386. doi: 10.1128/aem.60.4.1383-1386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckigt J, Zenk MH. Chemical synthesis and properties of hydroxycinnamoyl-coenzyme A derivatives. Z Naturforsch. 1975;30c:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- Technical Association of the Pulp and Paper Industry (1972) Methoxyl content of pulp and wood. TAPPI standard method 209 su-72. Technical Association of the Pulp and Paper Industry, Atlanta, GA
- Technical Association of the Pulp and Paper Industry (1989) Acid-soluble lignin in wood and pulp. TAPPI useful method 250. Technical Association of the Pulp and Paper Industry, Atlanta, GA
- Vallet C, Chabbert B, Czaninski Y, Monties B. Histochemistry of lignin deposition during sclerenchyma differentiation in alfalfa stems. Ann Bot. 1996;78:625–632. [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT. Lignification as a mechanism of disease resistance. Annu Rev Phytopathol. 1980;18:259–288. [Google Scholar]
- Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B and others. A novel lignin in poplar trees with a reducedcaffeic acid/5-hydroxyferulic acid O-methyltransferase activity. Plant J. 1995;8:855–864. [Google Scholar]
- Van Doorsselaere J, Dumas B, Baucher M, Fritig B, Legrand M, Van Montagu M, Inzé D. One-step purification and characterization of a lignin-specific O-methyltransferase from poplar. Gene. 1993;133:213–217. doi: 10.1016/0378-1119(93)90640-o. [DOI] [PubMed] [Google Scholar]
- Van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdoménech P. The brown midrib3 (bm3) mutation in maize occurs in thegene encoding caffeic acid O-methyltransferase. Plant Cell. 1995;7:407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Kneusel RE, Matern U, Varner JE. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Differential expression of two O-methyltransferases in lignin biosynthesis in Zinnia elegans. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Dickson EE, Chinnappa CC (1995) Nucleotide sequence of a cDNA clone encoding caffeoyl-coenzyme A 3-O-methyltransferase of Stellaria longipes (Caryophyllaceae). Plant Physiol 108: 429–430 [DOI] [PMC free article] [PubMed]