Abstract
The survivin protein, a member of the inhibitors of apoptosis (IAP) family, has gained popularity as a therapeutic target for cancer due to its selective expression in tumor cells and its significant involvement in tumor cell viability. The aim of this study was to investigate the effect of the survivin-small interfering RNA (siRNA) plasmid on survivin expression in the human lung cancer cell line, A549, and to observe its effects on apoptosis and proliferation of A549 cells. A549 human lung cancer cells were transfected with survivin-targeting siRNA. The downregulation of survivin expression was determined by real-time polymerase chain reaction and western blotting. The proliferation of A549 cells was determined by MTT assay. The apoptotic rate and cell cycle distribution were analyzed by flow cytometry (FCM). Caspase-9 activity was also detected to study the apoptosis of lung cancer cells induced by siRNA against survivin. The sequence-specific siRNA efficiently and specifically downregulated the expression of survivin at both the mRNA and protein levels. Downregulation of survivin expression dramatically suppressed the proliferation of A549 cells and arrested the cells at the G (1)/G (0) phase. Caspase-9 activity was significantly increased in A549 cells transfected with siRNA against survivin. In this study, we found that survivin-specific siRNA can efficiently suppress the expression of survivin, increase apoptosis and inhibit A549 cell proliferation. Our findings further indicate the possibility that the antitumor effects of survivin-siRNA are mediated through the activation of caspase-9.
Keywords: RNA interference, survivin, lung cancer, proliferation, apoptosis
Introduction
Considering the rapid development of comprehensive treatment for lung cancer, it is regrettable that no breakthrough has yet been made concerning the long-term efficacy of treatment and prognosis (1,2). Owing to the development of molecular biology, gene therapy, following other traditional treatments such as surgery, radiotherapy and chemotherapy, has become an attractive therapeutic model. RNA interference, an evolution-conserving cell defense mechanism mediated by small interferencing RNA (siRNA), can induce post-transcriptional gene silencing. The advantages of this method include stability, specificity, efficacy and low cytotoxicity. Thus it is extensively used on functional genome and gene therapy (3,4). Survivin, a new member of the inhibitors of apoptosis (IAP) family, was expressed highly in most tumor tissue; however, it is not expressed in terminal undifferentiated mature tissue. Survivin is closely related to the generation and development of tumors (5,6). We successfully constructed a siRNA lentiviral vector targeting the survivin gene and then transfected it to A549 cells. We also explored the effect of survivin on the proliferation and apoptosis of human lung cancer cells and investigated the mechanisms underlying this process.
Materials and methods
Materials
The human lung cancer lines, A549 and 293T, were purchased from the Shanghai Cell Resource Center of the Chinese Academy of Sciences. The pGC-LV vector, pHelper 1.0 vector and pHelper 2.0 vector were purchased from Shanghai GeneChem Co., Ltd. The Qiagen Plasmid Midi Kit was purchased from Qiagen (Valencia, CA, USA). Trypsase was purchased from Shanghai Chemical Reagent Co., Ltd. E.coli DH5α, SYBR Master Mixture, T4 DNA ligase and TaqDNA polymerase were purchased from Takara, (Shiga, Japan). Age I restriction enzyme and EcoRI restriction enzyme were purchased from New England Biolabs Co. Liposome Lipfectamine 2000 was purchased from Invitrogen (Carlsbad, CA, USA). DMSO was purchased from Shanghai Biological Reagent Co., Ltd. DMEM culture medium was purchased from Gibco (Carlsbad, CA, USA). FBS was purchased from Shanghai Weike Biochemical Reagent Co., Ltd. Oligo(dT) was purchased from Sangon Biotech Co., Ltd (Shanghai, China). M-MLV reverse transcriptase and dNTP were purchased from Promega (Madison, WI, USA). A flow cytometer and its software were purchased from the USA. An inverted microscope was purchased from Germany. A flow cytometer and its software were purchased from Beckman-coulter (Fullerton, CA, USA). An inverted microscope was purchased from Leica (Bensheim, Germany).
Methods
Cell culture
Human lung cancer A549 cells were cultured in DMEM culture medium containing 10% FBS. The temperature of the thermostat was 37˚C and CO2 concentration was 5%. The cells were digested and passaged by 0.25% lipase every 2–3 days. The cells at log phase were selected for the next step.
Design and screening of siRNA against survivin
We designed the target sequence according to survivin mRNA sequence in GenBank and the principles of siRNA design (7,8). Three pairs of siRNA targeting survivin and 1 pair of siRNA with the negative control were designed (Table I). The synthesis of siRNA was carried out by Shanghai GeneChem Co., Ltd. The siRNA was then transfected into 293T cells, according to the guidelines of Lipfectamine 2000 (Invitrogen). The transfecting results were observed under a fluorescence microscope after 24 h and the cell-collection and protein-abstraction were carried out 36 h later. The most efficient siRNA was chosen by western blot analysis.
Table I.
siRNA sequence specific to survivin.
Construction and transfection of survivin-siRNA lentiviral vector
Western blot analysis proved that the first pair of siRNA was the most efficient. A double-stranded DNA fragment, with cohesive termini of Age I restriction enzyme and EcoRI restriction enzyme, and an internal hairpin sequence of 5′-GGCTGGCTTCATCCACTGCTTCAAGAGAGCAGTGG ATGAAGCCAGCC-3′, was synthesized in vitro. The fragment was ligated into pGC-LV and then transfected into E.coli DH5α. Following amplification and screening, the construction was confirmed by sequencing. The plasmid was extracted and survivin-siRNA lentiviral vector was recombined, transfecting the A549 cells into a knockdown group (KD). The A549 cells transfected with the negative control and no sequence were labelled negative control (NC) and control group (CON), respectively.
Isolation of total RNA and RT-qPCR
Total RNA was extracted by TRIzol and then reverse-transcribed into cDNA, for which real-time quantitative PCR (RT-qPCR) was then performed. The survivin and actin primers (as the internal control) were synthesized by Shanghai GeneChem Co., Ltd. The sequences are shown in Table II. The reaction conditions of PCR were: pre-denaturation was at 95˚C for 15 sec; denaturation was at 95˚C for 5 sec; annealing was at 60˚C for 30 sec; 45 cycles were completed. The mixture was denatured for 1 min at the end of PCR and then cooled to 55˚C, at which the double strands of DNA could combine sufficiently. From 55 to 95˚C the light absorption value was recorded for 4 sec at every 0.5˚C. From this step the melting curve was depicted. The quantitative analysis was performed with the ratio of the target gene to actin. The 2−Δ ΔCt method was used for statistical analysis.
Table II.
Primer sequences of survivin and actin.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Product size (bp) |
|---|---|---|---|
| Survivin | ACCGCATCTCTACATTCAAG | CAAGTCTGGCTCGTTCTC | 113 |
| Actin | GTGGACATCCGCAAAGAC | AAAGGGTGTAACGCAACTA | 302 |
Detection of protein expression by western blotting
Total protein of A549 cells was isolated 72 h after transfection. Protein quantification was performed by BCA. The protein sample was normalized at the same time. The sample load was 30 μg total protein per lane. Protein from 10% SDS-PAGE gel was transferred to a PVDF membrane following electrophoresis. The protein was blocked with 5% non-fat dry milk at 4˚C. The primary antibodies, survivin (1:1000) and GAPDH (1:1000), were then added, and the mixture was subsequently incubated overnight at 4˚C on a rocking platform. After washing the membrane, HRP-conjugated secondary antibody (1:5000) was added to it and it was then incubated for 2 h. Protein bands were detected (the colored membranes) with the enhanced chemiluminescence (ECL) system and exposed to X-ray film. The membranes with no color (gray) were scanned using the image analytical system.
Cell proliferation by MTT assay
At the log phase of each group, A549 cells were inoculated into 96-well plates at 100 μl per well. The inoculating density was 1×104/well. The plates were incubated at 37˚C, 5% CO2 and saturated humidity. MTT assay was performed on days 1 to 5 following incubation. A value at a wavelength of 570 nm was detected by a microplate spectrophotometer. The mean value of 5 wells was the final OD value. The cell proliferating curve was sketched with the time as the horizontal axis and OD value as the vertical axis. The suppression rate of A549 cell proliferation = (1 − OD value of KD)/OD value of CON ×100%.
Cell cycle and apoptosis by flow cytometry (FCM)
A549 cells (1×106) of each group were digested and centrifuged for 5 min. Supernatants were discarded. Cells were washed with ice-cold PBS, fixated with 70% ethanol, centrifuged and collected. The sedimentation was washed with PBS. PI dye (1000 μl of 2 mg/ml solution) was added and mixed. The mixture was dyed for 30 min at 4˚C lucifugally. Red fluorescence at a wavelength of 488 nm was tested by FACScalibur and recorded. The data were analyzed with cell cycle fitting software.
Statistical analysis
Data were processed by SPSS15.0 statistical software. Quantitative data was expressed as the means ± standard deviation. One-way ANOVA was performed between different groups. Dunnett’ t-test (when homogeneity of variances existed) or Dunnett T3 (when heterogeneity of variances existed) were used to calculate the values between the different groups. P<0.05 indicated a statistically significant difference.
Results
The screening outcome of survivin-siRNA lentiviral vector
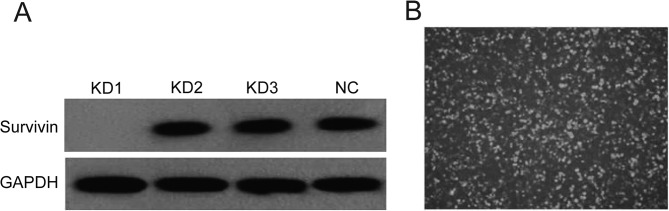
As the results of western blot analysis showed, the first pair of siRNA was the most efficient to downregulate the expression (Fig. 1A). Therefore we selected the first pair for the recombination of the survivin-siRNA lentiviral vector. After the survivin-siRNA lentiviral vector was transfected into A549 cells, the cells had green fluorescence (Fig. 1B). Screening and amplification with G418 was then carried out.
Figure 1.
(A) KD1 shows the clearly inhibition of the expression of survivin as detected by western blot analysis; (B) Evaluation of transfection efficiencies showed the high transfection efficiency 24 h after survivin-siRNA transfection.
The effect of survivin-siRNA on survival of lung cancer cells
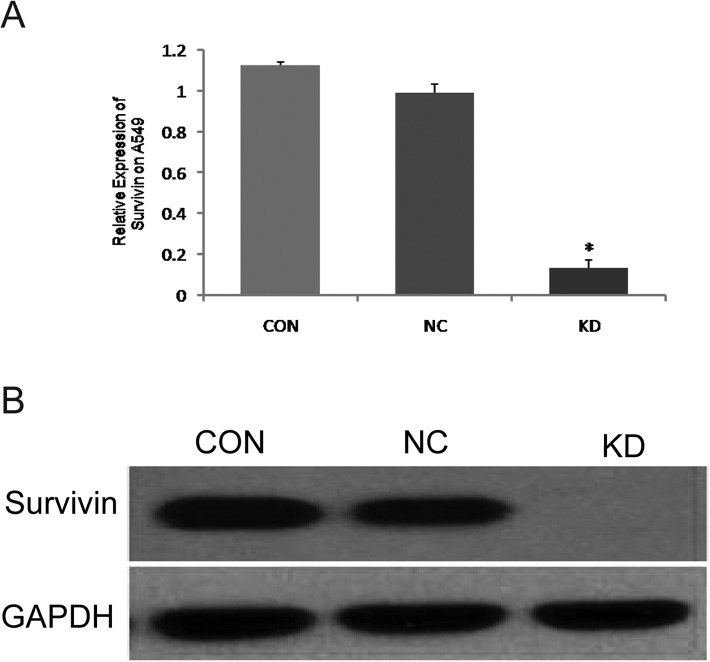
As the results of RT-qPCR showed, the expression of survivin mRNA in the KD group was significantly lower than that in the NC and CON groups (P<0.05) (Fig. 2A), which was consistent with the results of western blot analysis. Both results signified that the expression of survivin protein was significantly lower in the KD than in the NC and CON groups (P<0.05) (Fig. 2B).
Figure 2.
siRNA-survivin transfectants showed a remarkably decreased expression of survivin in A549 cells.
The effect of survivin-siRNA on lung cancer cell proliferation
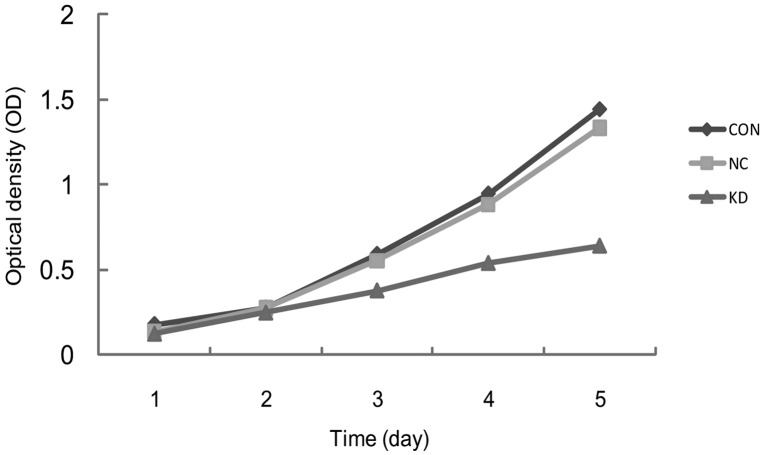
The cells’ proliferative curves were sketched based on the OD values from days 1 to 5 of each group. The differences of the initial OD values among the three groups were not significant, (CON, 0.18±0.02; NC, 0.17±0.01; KD, 0.16±0.01; P>0.05). However, the difference in OD values on day 5 between the CON and NC group was not significant (CON, 1.44±0.01; NC, 1.13±0.44; P>0.05) and the proliferative activity of the KD group significantly decreased (KD, 0.80±0.03; P<0.05). The OD values of the KD group on days 3, 4 and 5 were 36.0, 43.1 and 44.6%, respectively (Fig. 3).
Figure 3.
The proliferative activity of siRNA for the inhibition of survivin expression in A459 cells on days 1–5 assessed by the MTS assay.
The effect of survivin-siRNA on cell cycle of lung cancer cells and apoptosis
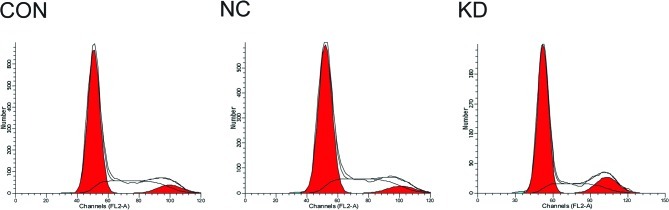
FCM showed that the cell cycle of A549 cells in the KD group changed significantly: the proportion in the G1 phase increased and the proportion in the S phase decreased. The differences were significant (P<0.05). G2/M phase did not change significantly, though it also showed a reductive tendency (Table III and Fig. 4). The apoptical rate of A549 cells in the KD group was 10.68±0.28%, which was significantly higher than that in the NC (6.68±0.44%) and CON (4.97±0.33%) groups (P<0.05).
Table III.
Effect of survivin-siRNA on cell cycle of A549 cells (%, mean ± SD, n=3).
| Group | G0/G1 | S | G2/M |
|---|---|---|---|
| CON | 61.30±0.59a | 30.43±0.68a | 8.28±0.79b |
| NC | 59.19±1.46a | 33.98±1.91a | 6.83±1.63b |
| KD | 66.62±0.59 | 27.34±0.92 | 6.03±1.12 |
P<0.05,
P>0.05.
Figure 4.
Analysis of the cell cycle showed that following the downregulation of survivin expression, the proportion of cells in the G0/G1 phase increased whereas the proportion of cells in the S phase decreased.
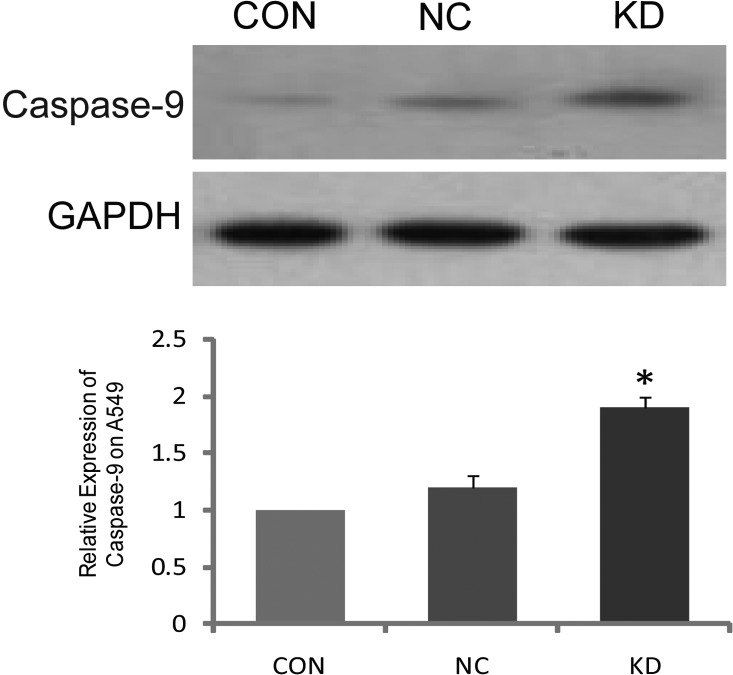
The activity of caspase-9 after survivin interference
The expression of caspase-9 protein in the KD group was significantly higher than that in the CON and NC groups after survivin-siRNA transfection (P<0.05). However, the differences between the CON and NC group were not significant (P>0.05) (Fig. 5).
Figure 5.
Expression of caspase-9 in A549 cells shown by western blotting and analyzing the gradation. (A) Survivin inhibition increased caspase-9 expression in A549 carcinoma cells. (B) The gradation ratio of caspase-9.
Discussion
Currently, lung cancer has the highest rates of morbidity and mortality of all malignant tumors worldwide. Since the number of lung cancer patients in China ranked first in the world, it is an urgent and important task to prevent and treat lung cancer, which should be the key point in the tumor-control plan of China. However, the controversial pathogenesis of lung cancer hinders the methods of prevention and treatment (9,10). As modern tumor molecular biology shows, the pathogenesis and development of lung cancer is a complex biological process, in which multiple genes are involved, multiple factors react and multiple phases exist. The pathogenesis and development of lung cancer could be attributed to the activation of an oncogene or the inactivation of a tumor inhibitor gene, which can induce abnormal proliferation, differentiation and apoptosis. Therefore, targeted therapy could be a new method for the prevention and treatment of lung cancer (11,12).
The IAP protein family (IAP) plays a key role in inhibiting apoptosis and is, therefore, more important than the BCL family. Survivin, a new member of the IAP family, has the least molecular weight, but the strongest capacity. Survivin can not only regulate the apoptosis, but also control cell cycle. These dual functions are an exclusive characteristic of the IAP family. Therefore, survivin plays a significant role in maintaining the quick proliferation, growth and the malignant functions of cells (13). Survivin is specifically expressed in embryo tissue and in developing fetal tissue, but is not expressed in adult terminal differentiated tissue. It is also widely expressed in certain cancer tissue, such as non-small cell lung cancer (NSCLC), gastric and intestinal cancer. Singha et al (14) concluded that survivin is one of the most significant independent prognostic factors of NSCLC. The unique distribution of survivin in the tissue means that normal tissue is almost not affected by the inhibition of survivin protein expression. Therefore survivin could be an ideal target gene for antitumor antisense therapy. To date, possible mechanisms for inhibiting apoptosis by survivin have been investigated (15,16). These include: i) the direct or indirect inhibition of the activities of caspases (caspase-3, caspase-7, caspase-6, caspase-8, caspase-9 and caspase-10); ii) competitive interaction with cyclin-dependent kinase (CDK) cdk4/P21 complex, and release of P21; then P21 is conjugated with caspase-3; conjugated with the second mitochondrial-derived activator/direct IAP binding protein with low PI (Smac/DIABLO) and isolated from other IAPs. Studies on survivin in cell mitosis have shown that it has a mutual function with the inner centromere protein (INCENP) and Aurora B. These genes have been shown to inhibit the expression of survivin, destroy microtubule organization and induce the failure of polyploidy formation and cell mitosis. Yonesaka et al (17) transfected the lung cancer cell line PC9 (including mutant p53) with survivin-siRNA, and found that cell proliferation was retarded. Chao et al (18) found that the expression of survivin mRNA was downregulated, the polykaryocyte formed and apoptosis occured 48 h after transfecting the A549 cells with survivin-siRNA.
In the present study, RT-qPCR and western blot analysis proved that the expressions of survivin transcription and translation were significantly inhibited after transfecting A549 cells with the survivin-siRNA lentiviral vector. These results also confirmed the successful construction of survivin-siRNA lentiviral vector. Furthermore, the results also showed that the expressions of survivin mRNA and protein were downregulated in the KD group, and the proliferation and growth of lung cancer cells were clearly inhibited. FCM showed the significant apoptosis of lung cancer cells. Most cells were blocked in the G1 phase, leading to the decrease of cells in the S phase. The results of FCM proved that survivin played an important role in the shift from the G1 to the S phase; it also demonstrated that after silencing the expression of survivin, the inhibition of cell proliferation was caused not only by the increase of apoptosis, but also by the decrease in the mitotic activity of the cells. These results are consistent with those from the study by Suzuki et al (19), who proved that the overexpression of survivin can accelerate the shift from the G1 to the S phase and resist G1 inhibition. The classical passages of cell apoptosis included the mitochondrial and death receptor pathways. At present, the anti-apoptotic mechanisms of survivin have not been fully elucidated. Some studies have shown that survivin can directly suppress the activities of caspase-3 and caspase-7, while others have shown survivin to mainly suppress the caspase-9-dependent apoptotic pathway. Thr34 phosphorylated survivin can directly bind with caspase-9 and inactivate it or indirectly block apoptosis by binding with SMAC (20). Banks et al and Shi (21,22), observed that the activity of caspase-3 did not change after suppressing the expression of survivin. They found that the keratinocytes isolated from K14-survivin mice highly expressed survivin, and that the keratinocytes could resist apoptosis mediated by UVB, an activator of the caspase-9-dependent apoptotic pathway. Apoptosis induced by a mutant type of survivin (T34A) could be blocked by the antagonist of caspase-9. All the results proved that survivin should suppress caspase-9-dependent apoptosis, not caspase-3-dependent. O’Connor et al (23) showed the direct interaction between survivin and caspase-9. Our research revealed that in the KD group, the activity of caspase-9 clearly increased while the expressions of survivin mRNA and protein decreased. Therefore, it may be through the activation of caspase-9 that the silencing expression of survivin could decrease the resistance of lung cancer cells to apoptosis and increase the susceptibility of the cells to apoptotic factors.
In brief, our study shows that the RNA interference technique can effectively silence the expression of survivin, inhibit lung cancer cell growth and induce apoptosis in lung cancer cells. It may also be related to the activation of caspase-9. The details of this mechanism require further clarification. Nevertheless, survivin may be promising for antitumor targeted therapy (24,25).
Acknowledgements
This study was supported by grants from the Natural Science Foundation of Fujian Province (No. 2008J0284) and Science and Technology Project of the Fujian Provincial Department of Education (No. JA09121).
References
- 1.Seve P, Dumontet C. Chemoresistance in non-small cell lung cancer. Curr Med Chem Anticancer Agents. 2005;5:73–88. doi: 10.2174/1568011053352604. [DOI] [PubMed] [Google Scholar]
- 2.Lee MW, Kim DS, Min NY, Kim HT. Akt1 inhibition by RNA interference sensitizes human non-small cell lung cancer cells to cisplatin. Int J Cancer. 2008;122:2380–2384. doi: 10.1002/ijc.23371. [DOI] [PubMed] [Google Scholar]
- 3.Dong AQ, Kong MJ, Ma ZY, Qian JF, Xu XH. Down-regulation of IGF-IR using small, interfering, hairpin RNA (siRNA) inhibits growth of human lung cancer cell line A549 in vitro and in nude mice. Cell Biol Int. 2007;31:500–507. doi: 10.1016/j.cellbi.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Ashihara E. RNA interference for cancer therapies. Gan To Kagaku Ryoho. 2010;37:2033–2041. [PubMed] [Google Scholar]
- 5.Nakashima N, Huang CL, Liu D, Ueno M, Yokomise H. Intratumoral Wnt1 expression affects survivin gene expression in non-small cell lung cancer. Int J Oncol. 2010;37:687–694. doi: 10.3892/ijo_00000718. [DOI] [PubMed] [Google Scholar]
- 6.Porebska I, Sobanska E, Kosacka M, Jankowska R. Apoptotic regulators: P53 and survivin expression in non-small cell lung cancer. Cancer Genomics Proteomics. 2010;7:331–335. [PubMed] [Google Scholar]
- 7.Zheng W, Ma X, Wei D, Wang T, Ma Y, Yang S. Molecular cloning and bioinformatics analysis of a novel spliced variant of survivin from human breast cancer cells. DNA Seq. 2005;16:321–328. doi: 10.1080/10425170500226490. [DOI] [PubMed] [Google Scholar]
- 8.Horn ME, Waterhouse PM. Rapid match-searching for gene silencing assessment. Bioinformatics. 2010;26:1932–1937. doi: 10.1093/bioinformatics/btq318. [DOI] [PubMed] [Google Scholar]
- 9.Ichite N, Chougule M, Patel AR, Jackson T, Safe S, Singh M. Inhalation delivery of a novel diindolylmethane derivative for the treatment of lung cancer. Mol Cancer Ther. 2010;9:3003–3014. doi: 10.1158/1535-7163.MCT-09-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biran A, Brownstein M, Haklai R, Kloog Y. Downregulation of survivin and aurora A by histone deacetylase and RAS inhibitors: a new drug combination for cancer therapy. Int J Cancer. 2011;128:691–701. doi: 10.1002/ijc.25367. [DOI] [PubMed] [Google Scholar]
- 11.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 12.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 13.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 14.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki A, Ito T, Kawano H, et al. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 16.Chiou SK, Jones MK, Tarnawski AS. Survivin - an anti-apoptosis protein: its biological roles and implications for cancer and beyond. Med Sci Monit. 2003;9:PI25–29. [PubMed] [Google Scholar]
- 17.Yonesaka K, Tamura K, Kurata T, et al. Small interfering RNA targeting survivin sensitizes lung cancer cell with mutant p53 to adriamycin. Int J Cancer. 2006;118:812–820. doi: 10.1002/ijc.21350. [DOI] [PubMed] [Google Scholar]
- 18.Chao JI, Liu HF. The blockage of survivin and securin expression increases the cytochalasin B-induced cell death and growth inhibition in human cancer cells. Mol Pharmacol. 2006;69:154–164. doi: 10.1124/mol.105.015503. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki A, Hayashida M, Ito T, et al. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/cyclin E complex activation. Oncogene. 2000;19:3225–3234. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 20.Verhagen AM, Vaux DL. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis. 2002;7:163–166. doi: 10.1023/a:1014318615955. [DOI] [PubMed] [Google Scholar]
- 21.Banks DP, Plescia J, Altieri DC, et al. Survivin does not inhibit caspase-3 activity. Blood. 2000;96:4002–4003. [PubMed] [Google Scholar]
- 22.Shi Y. Survivin structure: crystal unclear. Nat Struct Biol. 2000;7:620–623. doi: 10.1038/77904. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor DS, Grossman D, Plescia J, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang JS, Kim KM, Kang KH, et al. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60:31–39. doi: 10.1016/j.lungcan.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]