Abstract
Fatty acid-binding proteins are cytosolic fatty acid chaperones, and the adipocyte isoform, aP2, plays an important role in obesity and glucose metabolism. Recently, this protein has been detected in macrophages where it strongly contributes to the development of atherosclerosis. Here, we investigated the role of aP2 in macrophage biology and the molecular mechanisms underlying its actions. We demonstrate that aP2-deficient macrophages display defects in cholesterol accumulation and alterations in pro-inflammatory responsiveness. Deficiency of aP2 alters the lipid composition in macrophages and enhances peroxisome proliferator-activated receptor γ activity, leading to elevated CD36 expression and enhanced uptake of modified low density lipoprotein. The increased peroxisome proliferator-activated receptor γ activity in aP2-deficient macrophages is also accompanied by a significant stimulation of the liver X receptor α-ATP-binding cassette transporter A1-mediated cholesterol efflux pathway. In parallel, aP2-deficient macrophages display reduced IκB kinase and NF-κB activity, resulting in suppression of inflammatory function including reduced cyclooxygenase-2 and inducible nitric-oxide synthase expression and impaired production of inflammatory cytokines. Our results demonstrate that aP2 regulates two central molecular pathways to coordinate macrophage cholesterol trafficking and inflammatory activity.
Fatty acid-binding proteins (FABPs)1 constitute a family of highly homologous cytosolic proteins capable of binding a variety of hydrophobic compounds. FABPs display distinct patterns of tissue expression and have been the subject of extensive investigation at the biochemical level in terms of structure and ligand binding properties (1). However, fundamental questions remain unanswered regarding the primary functional role of these proteins and their mechanisms of action. Adipocyte FABP, also designated aP2 and FABP4, is an important contributor to the maintenance of systemic glucose metabolism (2–4). Because aP2 has been traditionally regarded as a protein expressed exclusively in differentiated adipocytes, the physiological consequences of its deficiency have been predominantly linked to changes in adipocytes. However, recent publications have reported the presence of aP2 in macrophages and have shown that aP2 expression can be induced by peroxisome proliferator-activated receptor γ (PPARγ) agonists, oxidized low density lipoprotein (oxLDL), and the differentiation of monocytes to macrophages and can be decreased by treatment with a cholesterol-lowering statin (5–9). aP2 deficiency was shown to provide remarkable protection against atherosclerosis in apolipoprotein E (apoE)-deficient models of atherosclerosis. Bone marrow transplantation studies demonstrated that this atheroprotective effect of aP2 is predominantly related to its actions in the macrophage (9, 10). Thus, it is clear that aP2 plays a vital role in the local macrophage responses in atherogenesis but the cellular mechanisms of action remain unknown.
aP2 binds a number of hydrophobic ligands known to influence macrophage function including fatty acids, various cyclooxygenase (COX) and lipoxygenase metabolites, and retinoic acid as well as several synthetic PPAR agonists (1). Therefore, we postulated that aP2 may regulate the accessibility of lipid ligands to their molecular targets in macrophages. A number of lipid-regulated pathways have been shown to play an important role in macrophage biology as it relates to cardiovascular and inflammatory disease. For example, the PPAR family of nuclear receptors, activated by multiple long chain fatty acids and eicosanoids, regulates the expression of numerous genes including those encoding proteins involved in cholesterol trafficking and the inflammatory response (11–15). In addition, the IKK-NF-κB pathway, which is critical to the control of macrophage pro-inflammatory activity, has been shown to be highly sensitive to inactivation by cyclopentenone prostaglandins (16, 17). Because the impact of aP2 deficiency on atherosclerosis equals or surpasses that of the manipulations targeting individual components of these pathways, we investigated the possibility that aP2 acts as a common upstream coordinator of these lipid-regulated pathways. In this report, we provide evidence that aP2 modulates signaling pathways that regulate both macrophage cholesterol trafficking and inflammatory activity.
EXPERIMENTAL PROCEDURES
Cell Culture
All of the cell culture was performed using low endotoxin regents in RPMI 1640 medium (Hyclone, Logan, UT) supplemented with 5% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, GA). Immortalized macrophage cell lines were established from aP2+/+ and aP2−/− mice (2) as previously described (9, 18, 19). Primary macrophages used in these studies included 4% thioglycollate-elicited (BD Biosciences) peritoneal macrophages or splenic macrophages prepared as previously described (20). Chinese hamster ovary (CHO) cells transfected with murine CD154 expression vector (CHO-CD154) and empty vector (CHO-Control) were a gift from Dr. Gail Bishop (Department of Microbiology, University of Iowa) (21).
Transfection of Macrophages and Reporter Assays
Cell lines were transfected by electroporation using the Eppendorf Multiporator (Eppendorf Scientific, Inc., Westbury, NY). Macrophages were co-transfected with 20 μg of endotoxin-free prepared firefly luciferase (Luc) plasmids, pPPRE-Luc, pGST-Luc (a gift from Dr. Russell Prough, Department of Biochemistry, University of Louisville), pNF-κB-Luc, or pMCS-Luc (Stratagene, La Jolla, CA) and 2 μg of Renilla luciferase plasmid (phRL-null-Luc) (Promega Corporation, Madison, WI). For NF-κB activity, macrophages were left untreated or treated for 16 h with indicated doses of LPS or 25, 50, and 75 μg of membrane proteins derived from CHO-control or CHO-CD154 isolated as previously described (22). For PPARγ activity, macrophages were stimulated for 24 h with vehicle and 25 μm ciglitazone (Biomol Research Laboratories Inc., Plymouth Meeting, PA) or pretreated for 1 h using the PPARγ-specific antagonist GW9662 (a gift from Dr. Peter Tontonoz). Statistical analysis of reporter assays was performed using the Student's t test.
Reconstitution of aP2 Expression into aP2-deficient Macrophages
The murine aP2 cDNA was ligated into plasmid pcDNA3.1Zeo (Invitrogen), resulting in plasmid pcDNA-aP2. Endotoxin-free pcDNA-aP2 (20 μg) was transfected by electroporation into the aP2−/− macrophage cell line (henceforth referred to as aP2−/−R) and selected by resistance to zeocin. Expression of aP2 by the aP2−/−R cell line was confirmed at the mRNA level (as shown in Fig. 2C, left panel). The levels of aP2 protein, as assessed by Western blot, were similar in the aP2−/−R line as compared with the aP2+/+ macrophage line (data not shown).
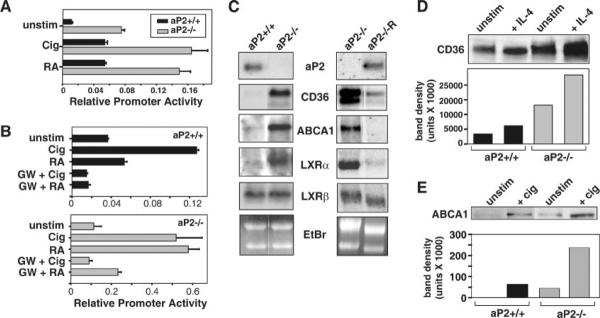
Fig. 2. PPARγ activity is enhanced by the absence of aP2.
A, PPARγ driven reporter activity is increased by aP2 deficiency. Macrophage cell lines were co-transfected with pPPRE-Luc and phRL-null-Luc reporter constructs and stimulated for 24 h with 25 μm ciglitazone (Cig) or 25 μm 9-cis-retinoic acid (RA). Relative promoter activities are reported as a ratio of firefly to Renilla luciferase activities ± S.D. of triplicate determinations. B, the PPARγ-specific antagonist, GW9662 (GW), represses agonist driven reporter activity. Macrophage cell lines were transfected as in A and preincubated for 1 h with 100 nm GW9662 before treatment with Cig and 9-cis-RA. Relative promoter activities are reported as a ratio of firefly to Renilla luciferase activities ± S.D. of triplicate determinations. C, aP2 deficiency increases mRNA expression of PPARγ-regulated genes. The expression of CD36, LXRα, LXRβ, and ABCA1 was detected by Northern blot analysis using RNA isolated from macrophage cell lines, untreated aP2+/+ and aP2−/− (left), and aP2−/− and aP2−/− R (right) with the exception of LXRα on the right (treated for 24 h with 10 ng/ml IL-4). D, aP2 deficiency increases CD36 protein expression. Western blot was used to determine CD36 protein expression in splenic macrophages (unstim) treated with 20 ng/ml IL-4 for 48 h. E, expression of ABCA1 protein is elevated by aP2 deficiency. Western blot was used to determine ABCA1 protein expression in aP2+/+ and aP2−/− macrophage cell lines (unstim) incubated for 24 h with 25 μm Cig.
Analysis of Intracellular Lipids
For quantification of individual lipid species (cholesterol, cholesterol esters, triglycerides, and free fatty acids), lipids were analyzed as previously described (9). An analysis of free fatty acids in untreated macrophage cell lines was also performed by Lipomics Technologies (West Sacramento, CA) (23).
Cholesterol Trafficking
Oxidation of LDL and 125I-LDL (Biomedical Technologies, Inc., Stoughton, MA) was performed using 5 μm CuSO4 for 18–24 h (24). Degradation of modified LDL was assessed as a measure of particle uptake at 37 °C for 24 h using 10 μg/ml 125I-oxLDL in the presence or absence of a 20-fold excess of unlabeled oxLDL competitor as described previously (25). Specific degradation was calculated by subtracting the radioactivity measured in the presence of a 20-fold excess of unlabeled oxLDL from the total radioactivity and normalized to cellular protein content. For analysis of cholesterol efflux, radiolabeled modified LDL particles were generated in serum-free medium containing 1 μCi/ml 1,2-3H(N)-cholesterol (PerkinElmer Life Sciences) incubated with 50 μg/ml acetylated LDL (Biomedical Technologies, Inc.) in RPMI 1640 medium supplemented with 0.2% low endotoxin fatty acid-free bovine serum albumin (Sigma) for 30 min at 37 °C. aP2+/+ and aP2−/− macrophages were loaded with radiolabeled lipoproteins for 24 h and then equilibrated overnight in RPMI 1640 medium supplemented with 0.1% low endotoxin fatty acid-free bovine serum albumin with no lipoproteins (25). Efflux to apolipoprotein A1 (apoA1) (0–15 μg/ml) proceeded for 3 h. Radioactivity of clarified culture supernatants and cellular lysate was measured separately by liquid scintillation. Percent efflux was calculated as counts per minute in [medium/(medium + lysate)], normalized to cellular protein content, and expressed as a percentage relative to aP2+/+ cultured with no apoA1 (100%). Statistical analysis was performed using the Student's t test.
Analysis of mRNA and Protein Levels
aP2+/+ and aP2−/− macrophage cell lines were untreated or treated with LPS (Escherichia coli serotype O111:B4, Sigma) for the indicated times before harvesting RNA using TRIzol (Invitrogen). The RiboQuant® multi-probe RNase protection assay system using probe set mCK2b (BD Biosciences) was used for analysis of cytokine RNA. Standard Northern analysis was performed on untreated macrophage cell lines or with 10 ng/ml IL-4 for 24 h for LXRα image of reconstituted macrophages only.
Macrophage proteins were harvested in lysis buffer (25 mm Tris-HCl, pH 7.4, 1% Triton X-100, 1% deoxycholate, 0.35 m NaCl, 10 mm EDTA, and Calbiochem protease inhibitor mixture III (Calbiochem)). Total protein content of the samples was assessed by micro-BCA assay (Pierce Biotechnology, Rockford, IL), and equal amounts of protein per sample were subjected to SDS-PAGE. Western blots were performed using mouse anti-COX-2 and mouse anti-iNOS (BD Transduction Laboratories, Lexington, KY), rabbit anti-ABCA1 (NB 400–105, Novus Biologicals, Littleton, CO), guinea pig anti-CD36 (a gift from Dr. Chistopher K. Glass, Department of Cellular and Molecular Medicine, University of California), mouse anti-β-actin (Sigma), rabbit anti-aP2 (a gift from Bristol-Myers Squibb Co., New York, NY), anti-p65 (Santa Cruz Biotechnologies, Santa Cruz, CA), and horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA).
Analysis of IKK and NF-κB Activity
Macrophage lines were stimulated with 1 μg/ml LPS for the indicated times before harvest in lysis buffer (1% Triton X-100, 1 mm EGTA, 5 mm EDTA, 20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 20 mm β-glycerophosphate, 10 mm p-nitrophenyl phosphate, 1.5 mm Na3VO4, and 1 mm phenylmethylsulfonyl fluoride) and assay of IKK activity was performed. For a comparison of kinase activity in aP2+/+, aP2−/−, and aP2−/−R, macrophages were treated with LPS for 10 min. IKKα/β was immunoprecipitated using anti-IKKα/β (Santa Cruz Biotechnologies). One-third of the immunoprecipitate was analyzed for total IKKα/β content, and the rest was incubated for 30 min at 30 °C in kinase buffer (20 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 20 μm ATP, 2 mm dithiothreitol, 5 μCi/sample [γ-32P]ATP, 1 μg/sample IκBα-GST) to phosphorylate 1–54 amino acids of IκBα-GST substrate (a gift from Ebrahim Zandi, Department of Molecular Microbiology and Immunology, University of California). Proteins were separated, transferred, and visualized by standard methods.
DNA binding activity of NF-κB was determined using the Gelshift Plus Rel Family electrophoretic mobility shift assay (EMSA) kit (Geneka Biotechnology, Inc., Montreal, Canada). aP2+/+ and aP2−/− macrophages lines were stimulated with 1 μg/ml LPS for 30 min. Nuclear proteins were extracted using a modified Dignam method (26). Nuclear protein extract (20 μg) from each sample and a Raji positive control nuclear extract were incubated with a 100-fold excess of unlabeled (cold competition) or [γ-32P]ATP-labeled double-stranded DNA containing NF-κBp65 consensus-binding sequence for 20 min. Protein-DNA complexes were separated by gel electrophoresis and visualized by autoradiography.
Assay of Inflammatory Mediators
aP2+/+ and aP2−/− macrophages were treated with LPS, CHO-CD154-transfected cells, or CHO-control cells (18 h for IL-6 and MCP-1 and 6 h for tumor necrosis factor α) before culture supernatants were analyzed by ELISA (OptEIA™, BD Biosciences). Supernatants from macrophage cell lines were evaluated for MCP-1 secretion by ELISA after 0 (unstimulated) or 25 nm phorbol 12-myristate 13-acetate (PMA) treatment for 6 h. For analysis of PGE2 secretion, cells were stimulated with LPS for 24 h, after which supernatants were assayed for PGE2 content via competitive ELISA (Neogen, Lexington, KY). For analysis of nitric oxide production, macrophages were treated with 500 ng/ml LPS and/or 10 units/ml IFNγ for 48 h. Supernatants were assayed for nitrite content by the addition of Greiss reagent as described previously (27). ELISAs and nitrite assays were analyzed using an E-max Precision microplate reader (Molecular Devices, Sunnyvale, CA). Statistical analysis was performed using the Student's t test.
RESULTS
aP2 Modulates the Intracellular Lipid Profile of Macrophages
Atherosclerotic lesions in both mice and humans express high levels of aP2 (6, 9, 10). In the absence of aP2, macrophages exhibit reduced capacity for foam cell formation (9). To investigate this phenotype further, we examined the macrophage intracellular lipid milieu in the presence and absence of aP2. Total cholesterol (C) and cholesterol ester (CE) content was decreased substantially in aP2-deficient macrophages as compared with wild type at base line (C, 37.3 versus 45.5; CE, 19.5 versus 28.3 μg/mg, respectively) as well as after acLDL treatment for 72 h (C, 37.2 versus 50.0; CE, 38.5 versus 56.5 μg/mg, respectively). Interestingly, total cellular triglyceride (TG) and non-esterified free fatty acid (FFA) levels were increased in aP2−/− macrophages compared with aP2+/+ controls at base line (TG, 26.5 versus 19.8; FFA, 26.8 versus 16.9 μg/mg, respectively) and after acLDL loading (TG, 37.0 versus 23.3; FFA, 33.8 versus 24.7 μg/mg, respectively). The increase in FFA levels in macrophages is consistent with earlier reports on aP2−/− adipocytes (28). Importantly, in a separate experiment, reconstitution of aP2−/− macrophages with aP2 reduced the fatty acid content in macrophages toward wild-type levels (aP2+/+, 574; aP2−/−, 1013, and aP2−/−R, 820 nm FFA/109 cells).
aP2 Deficiency Enhances Cholesterol Trafficking in Macrophages
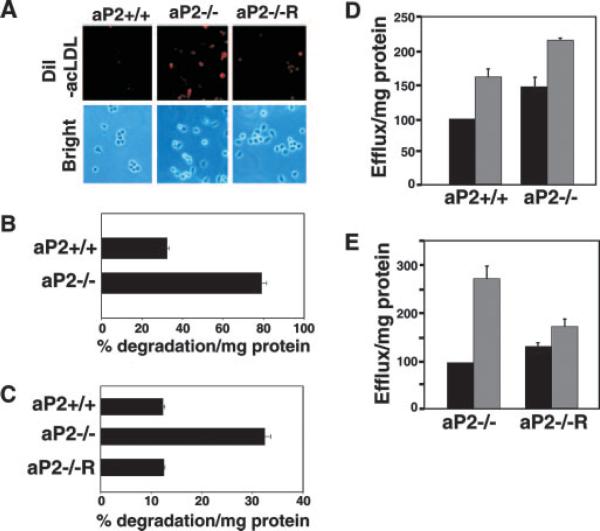
To determine the mechanism for aP2-mediated cholesterol accumulation in macrophages, we first examined cholesterol influx by visualizing and quantifying modified LDL (DiI-acLDL) uptake (25). aP2+/+, aP2−/−, and aP2−/− macrophages reconstituted with aP2 were loaded for 4 h with fluorescent-labeled acLDL particles. Surprisingly, aP2−/− macrophages internalized more modified LDL particles. The enhanced modified LDL uptake by aP2−/− macrophages was reversed by restoration of aP2 expression, substantiating a role of aP2 in particle uptake (Fig. 1A). As shown in Fig. 1B, thioglycollate-elicited primary aP2−/− macrophages internalized significantly more modified LDL as compared with aP2+/+ cells (p < 0.00001). Similarly, in the macrophage cell lines, aP2 deficiency resulted in a substantial increase in internalization of particles over that of wild type (data not shown). aP2 reconstitution of aP2−/− macrophages completely reversed this alteration in cholesterol uptake compared with aP2−/− cells, demonstrating the specificity of aP2 in this pathway (Fig. 1C).
Fig. 1. aP2 deficiency accelerates cholesterol trafficking.
A, internalization of acLDL is enhanced by aP2 deficiency. aP2+/+, aP2−/−, and aP2−/−R macrophages (Brightfield) were treated with 10 μg/ml DiI-acLDL (red) for 4 h. B, aP2 deficiency enhances flux of cholesterol into macrophages. oxLDL internalization by thioglycollate-elicited macrophages was assessed by treatment with 125I-oxLDL in the presence or absence of excess unlabeled oxLDL for 24 h. Specific degradation is the percent uptake normalized to milligram of cellular protein ± S.E. C, stable expression of aP2 into aP2−/− macrophages (aP2−/−R) restores oxLDL uptake to aP2+/+ levels. Macrophage cell lines were treated the same as in B. D, cholesterol efflux from macrophages is increased by aP2 deficiency at base line and with induction. Thiogly-collate-elicited aP2+/+ and aP2−/− macrophages were loaded for 24 h with [3H]AcLDL, and cholesterol efflux to 0 (black bar) or 10 μg/ml (gray bar) apoA1 proceeded for 3 h. Percent efflux was normalized to cellular protein content and expressed as a percentage relative to aP2+/+ cultured with no apoA1 (100%) ± S.E. E, stable restoration of aP2 into aP2−/− macrophages reduces induced cholesterol efflux. Macrophage cell lines were treated as in D with the exception that 0 (black bar) or 15 μg/ml (gray bar) apoA1 was used.
The increased internalization of modified lipoprotein particles by aP2−/− cells appears paradoxical considering the reduced cholesterol and cholesterol ester content of the aP2−/− macrophages and protection of aP2−/− mice against atherosclerosis (9, 10). However, macrophages are equipped with a cholesterol efflux pathway that provides a mechanism to limit foam cell formation. Thus, we considered the possibility that the reduced cholesterol content of aP2−/− macrophages may be due to the acceleration of cholesterol efflux (25, 29). Fig. 1D demonstrates that cholesterol efflux from primary thioglycollate-elicited macrophages isolated from aP2−/− mice was increased at base line and upon the addition of apoA1 as compared with macrophages isolated from aP2+/+ mice. The cholesterol efflux rate in the aP2−/− macrophage cell line was also substantially higher than in the aP2+/+ cells (data not shown). The elevated apoA1-mediated efflux capacity in aP2−/− macrophages was abrogated with reconstitution of aP2 (Fig. 1E). Overall, these data demonstrate that aP2-deficient macrophages are protected from excessive cholesterol accumulation, primarily through activation of the cholesterol efflux pathway. The restoration of aP2 expression into aP2−/− macrophages reversed the hyperactivated cholesterol influx and efflux activity, demonstrating a specific link between aP2 expression and cholesterol trafficking in macrophages.
The simultaneous increase in modified cholesterol uptake and efflux has also been observed in macrophages exposed to agonists of PPARγ, a key nuclear receptor that initiates the transcription of genes involved in cholesterol trafficking (29–32). Given the ability of aP2 to bind known PPARγ ligands, we postulated that aP2 may act as a negative regulator of PPARγ activity in macrophages. We tested this hypothesis by examining the influence of aP2 on PPARγ transcriptional activity using a PPAR response element (PPRE)-driven reporter. Macrophages were treated with a PPARγ-specific synthetic agonist, ciglitazone, or with 9-cis-retinoic acid, an agonist for RXRα, the binding partner of PPARγ. Both background and agonist-induced PPRE-driven transcriptional activity was significantly higher in the aP2−/− macrophages as compared with wild type (Fig. 2A) with p values of 0.013 and 0.0003 for ciglitazone and retinoic acid, respectively. Agonist-induced reporter activity was inhibited by the irreversible PPARγ-specific antagonist, GW9662 (33), in both aP2+/+ and aP2−/− macrophages, demonstrating that PPARγ was responsible for the increased reporter activity observed (Fig. 2B). No differences in PPRE reporter activity were seen between aP2+/+ and aP2−/− macrophages transfected with the negative control reporter plasmid, pGST-Luc, or when treated with vehicle alone (data not shown). Treatment with the PPARα agonist WY14643 did not induce reporter activity above background levels in either aP2+/+ or aP2−/− macrophage (data not shown).
Activation of PPARγ leads to an increase in scavenger receptor CD36 expression, which mediates internalization of modified LDL particles (11, 13, 25). CD36 mRNA expression was dramatically increased in the absence of aP2, consistent with increased uptake of modified LDL particles and PPARγ transcriptional activity in aP2−/− macrophages (Fig. 2C, left panel). Upon stable reconstitution of aP2 in aP2−/− macrophages, the expression of CD36 was reduced to the levels observed in wild-type macrophages (Fig. 2C, right panel). This reduction in CD36 mRNA in the aP2−/−R line is accompanied by a significant decrease in cholesterol influx (Fig. 1C, p < 0.00003), demonstrating that aP2, specifically, plays a regulatory role in CD36 expression and cholesterol particle influx in macrophages. Elevated CD36 mRNA expression in the aP2−/− macrophages correlated with elevated protein levels as assessed by Western blot analyses (Fig. 2D). In addition, we demonstrated that levels of CD36 protein were enhanced by treatment with IL-4, which induces production of lipid ligands for PPARγ (34), and that IL-4 enhancement of CD36 was ~5-fold greater in aP2−/− macrophages than in wild-type cells (Fig. 2D).
Consistent with the elevated PPARγ activity observed in aP2−/− macrophages, the expression of the PPARγ-regulated transcription factor LXRα was also higher in aP2−/− macrophages than in wild-type cells, as was expression of the LXRα-regulated gene, ABCA1. The expression level of LXRβ, which is independent of the PPARγ pathway (15), is indistinguishable between genotypes (Fig. 2C, left panel). Restoration of aP2 in aP2−/− macrophages reduced the expression of both the LXRα and ABCA1 genes to the levels present in wild-type macrophages (Fig. 2C, right panel). Furthermore, both unstimulated and ciglitazone-treated aP2−/− macrophages displayed higher levels of ABCA1 protein than did aP2+/+ macrophages (Fig. 2E). The elevated activity of the PPAR-LXRα-ABCA1 pathway provides a potential mechanism underlying the atheroprotective phenotype of decreased cholesterol and cholesterol ester accumulation in aP2-deficient macrophages.
IKK and NF-κB Activity Is Impaired in aP2-deficient Macrophages
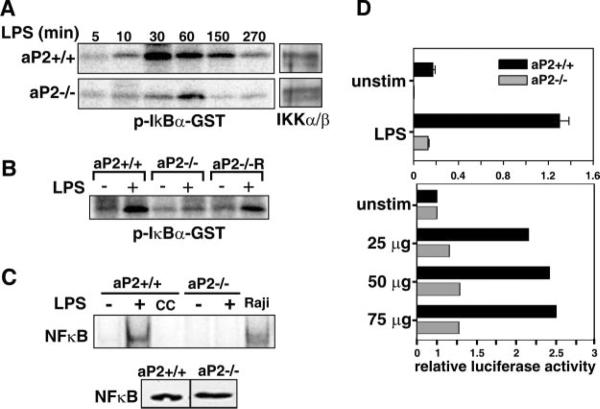
Recent studies have shown that signaling through the IKK-NF-κB pathway is highly sensitive to the intracellular lipid status of the cell. For example, cyclopentenone prostaglandins can inhibit the NF-κB pathway through direct inactivation of IKK and NF-κB (16, 17). To test the potential impact of aP2 deficiency on this pathway, we examined the activity of IKK and NFκB in aP2+/+ and aP2−/− macrophages. As shown in Fig. 3A, in vitro kinase assays demonstrate that LPS-induced activation of IKK in aP2−/− macrophages was greatly diminished and delayed as compared with that of wild-type macrophages. These changes occurred without alterations in the levels of IKKα or IKKβ, suggesting that the reduced IKK activity in aP2−/− macrophages is not due to decreased levels of IKK protein itself (Fig. 3A, right). Thus, the absence of aP2 exerts a negative influence on IKK activity. As shown in Fig. 3B, IKK activity in response to LPS was regained in the aP2−/−R line.
Fig. 3. aP2 deficiency inhibits the NF-κB signaling pathway in macrophages.
A, kinase activity of IKK is impaired by aP2 deficiency. The activity of IKK was determined by in vitro kinase phosphorylation of an IκBα-GST substrate after macrophage lines were stimulated by 1 μg/ml LPS for the indicated times. Western blot analysis of immunoprecipitated IKKα/β is shown on the right. B, reconstitution of aP2 restores kinase activity of IKK. aP2+/+, aP2−/−, and aP2−/−R macrophage cell lines were stimulated by 1 μg/ml LPS for 10 min, and kinase activity of IKK was assayed as in A. Data shown are representative of three independent kinase assays. C, NF-κB binding to DNA is abrogated by aP2 deficiency. EMSA was used to determine NF-κB DNA binding activity of nuclear protein extracts from aP2+/+ and aP2−/− macrophages (−) stimulated with 1 μg/ml LPS (+) for 30 min using radiolabeled or non-labeled (cold competition, CC) NF-κB consensus-binding sequences (top panel). Nuclear extracts from the Burkitt's lymphoma cell line, Raji, served as a positive control. Western blot analysis of NF-κB in untreated macrophage lines (bottom panel). D, NF-κB driven reporter activity is impaired by aP2 deficiency. Macrophage lines were co-transfected with pNF-κB-Luc and phRL-null-Luc reporter constructs and activated with 1 μg/ml LPS (top panel) or with 0 (unstim), 25, 50, or 75 μg of membrane extracts from CHO-control cells or CHO-CD154 (bottom panel) for 16 h. Relative promoter activities are reported as the ratio of firefly to Renilla luciferase activity ± S.D. of triplicate samples (top) or fold induction by CHO-CD154 relative to CHO control (bottom).
To address the consequences of these alterations in IKK activity further, NF-κB DNA binding and transcriptional activity were evaluated by EMSA and reporter assays. Fig. 3C, top panel, demonstrates that LPS stimulation of aP2+/+, but not aP2−/− macrophages, results in increased NF-κBp65 DNA binding. Western blot analysis demonstrated no difference in NF-κB protein expression levels between genotypes (Fig. 3C, bottom panel). Thus, the decreased expression of NF-κB-regulated genes was not due to depressed expression of NF-κB. Likewise, aP2−/− macrophages did not demonstrate NF-κB transactivation when stimulated with LPS, as compared with high activity levels observed in aP2+/+ macrophages (Fig. 3D, top panel, p = 0.0026).
We also examined NF-κB activation in response to stimulation of macrophages through the CD40-CD154 receptor-ligand interaction, which has been shown to be closely linked with the formation of atherosclerotic lesions (35). Stimulation of macrophages through CD40 was achieved with the use of plasma membranes isolated from CD154-transfected CHO cells (21). CD40-stimulated aP2−/− macrophages showed no appreciable NF-κB-driven reporter activity, whereas aP2+/+ macrophages responded with a 2-fold increase in NF-κB-driven reporter activity above control levels (Fig. 3D, bottom panel). No differences were seen in firefly luciferase activity in macrophages transfected with the negative control vector pMCS-Luc under control or LPS-stimulated and CD154-stimulated conditions (data not shown).
aP2 Deficiency Diminishes Macrophage Inflammatory Activity
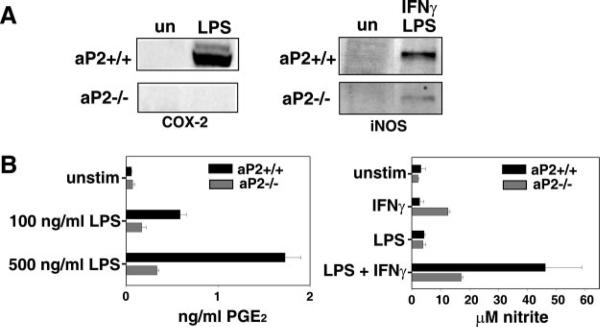
We next investigated the downstream inflammatory responses of macrophages including COX-2 and iNOS expression as well as LPS and CD154 induction of inflammatory cytokine production. Treatment with PPARγ ligands has been shown to repress expression of COX-2 and iNOS in macrophages (36, 37). Because aP2 regulates both IKK and PPARγ activity, we evaluated the influence of aP2 deficiency on the synthesis of COX-2 and iNOS. Macrophages were stimulated with either LPS or a combination of LPS and IFNγ for 12 h. As shown in Fig. 4A, aP2−/− macrophages were deficient in activation-induced COX-2 and iNOS expression. Flow cytometric analysis revealed that wild-type and aP2−/− macrophages did not differ in the level of CD40 receptor expression or in the level of CD14, a membrane protein required for LPS binding and subsequent signaling through Toll-like receptor 4, suggesting that the reduced responsiveness to LPS and CD154 stimulation is not probably due to reduced receptor expression by aP2−/− macrophages (data not shown). The diminished COX-2 expression by aP2−/− macrophages was accompanied by a functional defect resulting in a significant reduction in PGE2 secretion (Fig. 4B, left panel, p < 0.001). In addition, a dramatic defect in nitrite production is evident in aP2−/− macrophages as compared with wild-type cells, representative of decreased iNOS expression (Fig. 4B, right panel, p = 0.017).
Fig. 4. aP2−/− macrophages show reduced inflammatory activity.
A, deficiency of aP2 diminishes production of proteins from NF-κB-regulated genes. aP2+/+ and aP2−/− macrophages were left unstimulated (un) or incubated for 12 h with 100 ng/ml LPS (left panel) or 100 ng/ml LPS + 10 units/ml murine IFNγ (right panel) before Western blot analysis. B, production of COX-2 and iNOS enzymatic products is reduced by aP2 deficiency. aP2+/+ and aP2−/− macrophage cell lines were stimulated with LPS at the concentrations shown for 24 h, and supernatants were assayed for PGE2 content by ELISA (left panel). Data shown are mean ± S.D. of triplicate determinations. aP2+/+ and aP2−/− macrophage cell lines were stimulated with LPS (500 ng/ml) and/or IFNγ (1 units/ml) for 48 h before measuring nitrite content of supernatants. Data shown are mean ± S.D. of triplicate determinations (right panel).
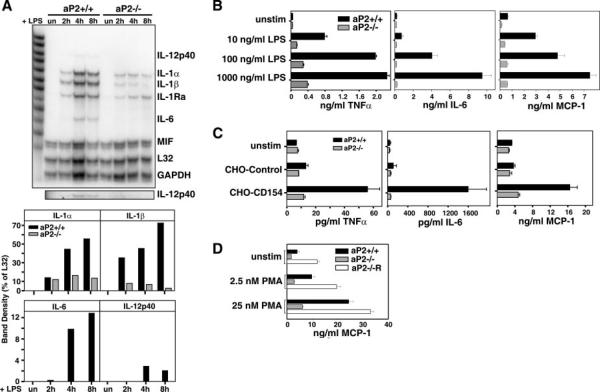
To further address the impact of aP2 on inflammatory activity, macrophage inflammatory cytokine production in response to LPS and CD154 was evaluated. First, aP2+/+ and aP2−/− macrophages were stimulated with LPS for 2, 4, and 8 h and pro-inflammatory cytokine mRNA was analyzed by ribonuclease protection assay (Fig. 5A). Overall, aP2−/− macrophages expressed lower amounts of IL-1α, IL-1β, IL-6, and IL-12p40 mRNA compared with aP2+/+ macrophages. No differences were seen in macrophage migration inhibitory factor mRNA. In addition, cytokine protein secretion was analyzed by ELISA. Wild-type and aP2−/− macrophage cell lines were stimulated with LPS, CHO-CD154 transfectants (Fig. 5, B and C), or CHO control cells as negative controls. We found that tumor necrosis factor α, IL-6, and MCP-1 secretion by aP2−/− macrophages was greatly reduced as compared with aP2+/+ cells in response to both stimuli (p values <0.0025). Finally, impaired PMA-induced MCP-1 secretion in aP2−/− macrophages could be restored with reconstitution of aP2 expression in aP2−/− macrophages, demonstrating the specificity of this response to the presence of aP2 (Fig. 5D).
Fig. 5. LPS and CD154-induced expression of inflammatory cytokines are impaired in aP2−/− macrophages.
A, production of inflammatory cytokine RNA is reduced by aP2 deficiency. aP2+/+ and aP2−/− macrophages were stimulated by 1 μg/ml LPS for 0, 2, 4, and 8 h, and cytokine mRNA expression was analyzed by RNase protection assay (top panel). IL-12p40 is shown in a darker exposure in lower panel. Band density of specific RNA is expressed as a percentage of L32 density (bottom panel). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MIF, migration inhibitory factor. B, LPS induction of inflammatory cytokines and chemokine is reduced by aP2 deficiency. LPS stimulation of cytokines and chemokine production from aP2+/+ and aP2−/− macrophages stimulated for 18 h for IL-6 and MCP-1 and 6 h for tumor necrosis factor α were analyzed by ELISA. C, macrophages were stimulated with CD154 and assayed as in B. Data shown are mean ± S.D. of triplicate determinations. D, aP2 reconstitution into aP2−/− macrophages restores MCP-1 production. Supernatants from aP2+/+ and aP2−/− macrophage cell lines were evaluated for MCP-1 secretion by ELISA after 0 (unstim), 2.5, or 25 nm PMA treatment for 6 h. Data are presented as mean ± S.E. of three determinations.
DISCUSSION
Although it has been established that aP2 plays a critical role in atherosclerosis (9, 10), the mechanisms underlying its action are largely unknown. Here, we provide evidence that aP2 expression impacts two key macrophage functions involved in atherogenesis, cholesterol trafficking and inflammatory activity (Fig. 6). We demonstrate that, in the absence of aP2, macrophage PPARγ activity is enhanced, whereas pro-inflammatory signaling pathways are suppressed. Part of the role of PPARγ in atherogenesis involves CD36-mediated stimulation of lipoprotein entry, which triggers the PPARγ-driven LXRα-ABCA1 cholesterol efflux pathway (15, 25, 30, 38–41). Our data suggest that aP2 acts upstream of the PPARγ-LXRα-ABCA1 pathway and contributes to the control of cholesterol trafficking in macrophages with the lack of aP2 resulting in accelerated uptake of modified LDL particles coupled with enhanced cholesterol efflux.
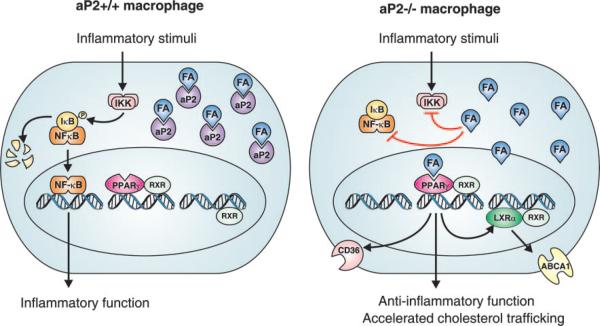
Fig. 6. aP2 coordination of cholesterol trafficking and inflammatory pathways in macrophages.
A hypothetical model of the role of aP2 in macrophages is depicted. In the presence of aP2 (left panel), the availability of fatty acids (FA) or other bioactive lipid mediators is restricted, hence fatty acids are unable to interact with targets such as PPARγ and components of the IKK-NF-κB pathway. As such, aP2 may act to control signaling through these pathways. In the absence of aP2 (right panel), increased availability of FA inhibits the NF-κB pathway, resulting in the blockade of inflammatory responses, and enhances PPARγ activity, resulting in accelerated cholesterol trafficking. Although the identity of the key aP2 ligands and the specific mechanisms of action remain to be determined, this model suggests that aP2 may coordinately control macrophage signaling pathways that contribute to the pathogenesis of metabolic syndrome and atherosclerosis.
In parallel, signaling via the IKK-NF-κB pathway is impaired and, as a consequence, the products of NF-κB-regulated genes are reduced in aP2−/− macrophages. The reduced inflammatory potential of aP2−/− macrophages may also be related to the enhanced PPARγ and LXRα activity or expression in aP2−/− macrophages, because these nuclear receptors have been shown to antagonize transcription of genes encoding some inflammatory proteins (12, 37, 42, 43). However, certain fatty acids, including lipoic acid and the cyclopentenone family of prostaglandins (PGA1, PGA2, PGJ2, and 15d-PGJ2) that may signal through PPARγ, can also inhibit IKK activation and the ability of NF-κB to bind to its response element independent of PPARγ (16, 17, 44–47). The extent to which these lipid mediators may play a role in the anti-inflammatory phenotype of aP2 deficiency is under investigation.
Our observations indicate that aP2 impacts multiple signaling pathways and suggest that this may occur via the control of availability of lipids that regulate these pathways. Thus, when aP2 expression is up-regulated in macrophages, lipid ligands are probably sequestered from their targets including nuclear receptors (e.g. PPARγ). In the absence of aP2, lipid ligands are free to interact with nuclear receptors, as well as other targets, including IKK (Fig. 6). Although our studies have identified PPARγ and IKK as two pathways regulated by aP2, preliminary data (not shown) suggest that the expression of additional regulators involved in cholesterol and fatty acid metabolism, including sterol regulatory element-binding protein-1c, is up-regulated in the absence of aP2, probably due to increased PPAR-LXR activity in aP2−/− macrophages (48). Furthermore, we have also considered the possibility that aP2 may be involved in protein-protein interactions independent of its role in binding lipids to generate its biological functions. Alternatively, aP2 may be involved in the specific delivery of ligands, bioactive lipids, or other metabolic substrates to locations such as the mitochondria, endoplasmic reticulum, or nucleus. These possible scenarios are currently under investigation.
Importantly, the data implicating aP2 in the control of lipid homeostasis tied to both cholesterol trafficking and inflammatory function in macrophages suggest that its function may impact a wide variety of disease states, particularly those associated with metabolic syndrome. In recent years, it has become clear that integrated regulation of inflammatory and metabolic pathways plays a central mechanistic role in many of the pathologies associated with this syndrome. Interestingly, another FABP, mal1, is also present in macrophages but it is not yet clear whether this isoform plays a similar role in atherosclerotic lesion formation or may be involved in other inflammatory or metabolic diseases. It will be important to elucidate the coordinated function of these FABPs in macrophage and adipocyte biology toward their utility as an attractive therapeutic target in a broad range of pathologies including obesity, insulin resistance, type 2 diabetes, atherosclerosis, and other inflammatory conditions.
Acknowledgments
We acknowledge the expert technical assistance of Aaron denDekker and Lihua Zhang.
Footnotes
This work was supported by National Institutes of Health Grant R01 AI48850 (to J. S. and G. S. H.), National Multiple Sclerosis Society Grant RG 3374 (to J. S. and G. S. H.), American Heart Association Predoctoral Fellowship (to K. C. B.), National Institutes of Health, NRSA F32 HL075970-01 (to L. M.), and in part by the Commonwealth of Kentucky Research Challenge Trust Fund (to J. S.).
The abbreviations used are: FABP, fatty acid-binding protein; apoA1, apolipoprotein A1; LDL, low density lipoprotein; acLDL, acetylated LDL; CHO, Chinese hamster ovary cell; C, cholesterol; CE, cholesterol ester; PGE2, prostaglandin; COX, cyclooxygenase; EMSA, electrophoretic mobility shift assay; IL, interleukin; iNOS, inducible nitric-oxide synthase; Luc, luciferase; FFA, non-esterified free fatty acid; oxLDL, oxidized low density lipoprotein; PPARγ, peroxisome proliferator-activated receptor gamma; PPRE, PPAR response element; PMA, phorbol 12-myristate 13-acetate; TG, triglycerides; IFNγ, interferon γ; IKK, IκB; GST, glutathione S-transferase; LPS, lipopolysaccharide; ELISA, enzyme-linked immunosorbent assay; LXR, liver X receptor; ABCA1, ATP-binding cassette transporter A1; MCP, monocyte chemoattractant protein.
REFERENCES
- 1.Makowski L, Hotamisligil GS. J. Nutr. 2004;134:2464S–2468S. doi: 10.1093/jn/134.9.2464S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 3.Scheja L, Makowski L, Uysal KT, Wiesbrock SM, Shimshek DR, Meyers DS, Morgan M, Parker RA, Hotamisligil GS. Diabetes. 1999;48:1987–1994. doi: 10.2337/diabetes.48.10.1987. [DOI] [PubMed] [Google Scholar]
- 4.Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS. Endocrinology. 2000;141:3388–3396. doi: 10.1210/endo.141.9.7637. [DOI] [PubMed] [Google Scholar]
- 5.Pelton PD, Zhou L, Demarest KT, Burris TP. Biochem. Biophys. Res. Commun. 1999;261:456–458. doi: 10.1006/bbrc.1999.1071. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y, Luo N, Lopes-Virella MF, Garvey WT. Atherosclerosis. 2002;165:259–269. doi: 10.1016/s0021-9150(02)00305-2. [DOI] [PubMed] [Google Scholar]
- 7.Fu Y, Luo N, Lopes-Virella MF. J. Lipid Res. 2000;41:2017–2023. [PubMed] [Google Scholar]
- 8.Llaverias G, Noe V, Penuelas S, Vazquez-Carrera M, Sanchez RM, Laguna JC, Ciudad CJ, Alegret M. Biochem. Biophys. Res. Commun. 2004;318:265–274. doi: 10.1016/j.bbrc.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF. Nat. Med. 2001;7:699–705. doi: 10.1038/89076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, Hotamisligil GS. Arterioscler. Thromb. Vasc. Biol. 2002;22:1686–1691. doi: 10.1161/01.atv.0000033090.81345.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 12.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 13.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 14.Jiang C, Ting AT, Seed B. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 15.Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, Liao D, Nagy L, Edwards PA, Curtiss LK, Evans RM, Tontonoz P. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 16.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro MG. Nature. 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]
- 17.Straus DS, Pascual G, Li M, Welch JS, Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G, Glass CK. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4844–4849. doi: 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. Nature. 1986;318:667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- 19.Clemons-Miller AR, Cox GW, Suttles J, Stout RD. Immunobiology. 2000;202:477–492. doi: 10.1016/s0171-2985(00)80105-9. [DOI] [PubMed] [Google Scholar]
- 20.Stout RD, Suttles J. Cell. Immunol. 1992;139:363–374. doi: 10.1016/0008-8749(92)90078-4. [DOI] [PubMed] [Google Scholar]
- 21.Baccam M, Bishop GA. Eur. J. Immunol. 1999;29:3855–3866. doi: 10.1002/(SICI)1521-4141(199912)29:12<3855::AID-IMMU3855>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.McLeish KR, Klein JB, Lederer ED, Head KZ, Ward RA. Kidney Int. 1996;50:407–416. doi: 10.1038/ki.1996.330. [DOI] [PubMed] [Google Scholar]
- 23.Watkins SM, Reifsnyder PR, Pan HJ, German JB, Leiter EH. J. Lipid Res. 2002;43:1809–1817. doi: 10.1194/jlr.m200169-jlr200. [DOI] [PubMed] [Google Scholar]
- 24.Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HF, Freeman MW. J. Biol. Chem. 2002;277:49982–49988. doi: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- 25.Moore KJ, Rosen ED, Fitzgerald ML, Randow F, Andersson LP, Altshuler D, Milstone DS, Mortensen RM, Spiegelman BM, Freeman MW. Nat. Med. 2001;7:41–47. doi: 10.1038/83328. [DOI] [PubMed] [Google Scholar]
- 26.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding AH, Nathan CF, Stuehr DJ. J. Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 28.Coe NR, Simpson MA, Bernlohr DA. J. Lipid Res. 1999;40:967–972. [PubMed] [Google Scholar]
- 29.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama TE, Sakai S, Lambert G, Nicol CJ, Matsusue K, Pimprale S, Lee YH, Ricote M, Glass CK, Brewer HB, Jr., Gonzalez FJ. Mol. Cell. Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM. Nat. Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 32.Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, Teissier E, Minnich A, Jaye M, Duverger N, Brewer HB, Fruchart JC, Clavey V, Staels B. Nat. Med. 2001;7:53–58. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- 33.Leesnitzer LM, Parks DJ, Bledsoe RK, Cobb JE, Collins JL, Consler TG, Davis RG, Hull-Ryde EA, Lenhard JM, Patel L, Plunket KD, Shenk JL, Stimmel JB, Therapontos C, Wilson TM, Blanchard SG. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 34.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, Witztum JL, Funk CD, Conrad D, Glass CK. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 35.Lutgens E, Daemen MJ. Trends Cardiovasc. Med. 2002;12:27–32. doi: 10.1016/s1050-1738(01)00142-6. [DOI] [PubMed] [Google Scholar]
- 36.Inoue H, Tanabe T, Umesono K. J. Biol. Chem. 2000;275:28028–28032. doi: 10.1074/jbc.M001387200. [DOI] [PubMed] [Google Scholar]
- 37.Li M, Pascual G, Glass CK. Mol. Cell. Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Ishibashi S, Perrey S, Osuga J, Gotoda T, Kitamine T, Tamura Y, Okazaki H, Yahagi N, Iizuka Y, Shionoiri F, Ohashi K, Harada K, Shimano H, Nagai R, Yamada N. Arterioscler. Thromb. Vasc. Biol. 2001;21:372–377. doi: 10.1161/01.atv.21.3.372. [DOI] [PubMed] [Google Scholar]
- 39.Collins AR, Meehan WP, Kintscher U, Jackson S, Wakino S, Noh G, Palinski W, Hsueh WA, Law RE. Arterioscler. Thromb. Vasc. Biol. 2001;21:365–371. doi: 10.1161/01.atv.21.3.365. [DOI] [PubMed] [Google Scholar]
- 40.Joseph SB, McKilligin E, Pei L, Watson MA, Collins AR, Laffitte BA, Chen M, Noh G, Goodman J, Hagger GN, Tran J, Tippin TK, Wang X, Lusis AJ, Hsueh WA, Law RE, Collins JL, Willson TM, Tontonoz P. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7604–7609. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. J. Clin. Investig. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Nat. Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 44.Zhang WJ, Frei B. FASEB J. 2001;15:2423–2432. doi: 10.1096/fj.01-0260com. [DOI] [PubMed] [Google Scholar]
- 45.Bernardo A, Levi G, Minghetti L. Eur. J. Neurosci. 2000;12:2215–2223. doi: 10.1046/j.1460-9568.2000.00110.x. [DOI] [PubMed] [Google Scholar]
- 46.Kawahito Y, Kondo M, Tsubouchi Y, Hashiramoto A, Bishop-Bailey D, Inoue K, Kohno M, Yamada R, Hla T, Sano H. J. Clin. Investig. 2000;106:189–197. doi: 10.1172/JCI9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrova TV, Akama KT, Van Eldik LJ. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4668–4673. doi: 10.1073/pnas.96.8.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]