Abstract
The assembly and disassembly of chromatin impacts all DNA dependent processes in eukaryotes. These processes are intricately regulated through stepwise mechanisms, requiring multiple proteins, post-translational modifications and remodeling enzymes, as well as specific proteins to chaperone the highly basic and aggregation-prone histone proteins. The histone chaperones are acidic proteins that perform the latter function by maintaining the stability of the histones when they are not associated with DNA and guiding the deposition and removal of histones from DNA. Understanding the thermodynamics of these processes provides deeper insights into the mechanisms of chromatin assembly and disassembly. Here we describe complementary thermodynamic and biochemical approaches for analysis of the interactions of a major chaperone of the H3/H4 dimer, Anti-silencing function 1 (Asf1) with histones H3/H4 and DNA. Fluorescence quenching approaches are useful for measuring the binding affinity of Asf1 for histones H3/H4 under equilibrium conditions. Electrophoretic mobility shift analyses are useful for examining Asf1 mediated tetrasome (H3/H4-DNA) assembly and disassembly processes. These approaches potentially can be used more generally for the study of other histone chaperone-histone interactions and provide a means to dissect the role of post-translational modifications and other factors that participate in chromatin dynamics.
Keywords: Anti-silencing function 1 (Asf1), histone H3/H4, tetrasome, binding affinity, fluorescence, electrophoretic mobility shift assay
1. Introduction
In eukaryotes, the nucleosome is the fundamental repeating unit of chromatin, in which the DNA wraps nearly two turns about a core histone octamer formed by one H3/H4 heterotetramer flanked by two H2A/H2B heterodimers (Jorcano and Ruiz-Carrillo 1979; Kornberg 1977; Luger et al. 1997). In a stepwise process, histone chaperones and ATP-dependent remodeling complexes guide both the assembly of nucleosomes from newly synthesized and recycled histones and the disassembly of chromatin for all DNA dependent processes (reviewed in (Clapier and Cairns 2009; Ransom et al. 2010)). The highly acidic histone chaperone proteins bind directly to the basic histone proteins and aid in directing histone deposition, exchange and eviction during nucleosome assembly and disassembly (reviewed in (Akey and Luger 2003; Das et al. 2010; Eitoku et al. 2008; Park and Luger 2008; Ransom et al. 2010)).
Anti-silencing Function 1 (Asf1) is a central histone chaperone for histones H3 and H4 once they have adopted the form of an H3/H4 dimer (Campos et al. 2010). Asf1 is a suppressor of gene silencing when overexpressed in the budding yeast Saccharomyces cerevisiae and its deletion in yeast results in DNA replication, repair, transcription, and histone modification defects (reviewed in (Mousson et al. 2007)). How Asf1 chaperones histones H3/H4 in vitro became much clearer with the determination of crystal structures of Asf1 bound to the H3/H4 dimer. These illustrated how Asf1 physically blocks formation of the H3/H4 heterotetramer by binding to the H3/H4 dimerization surface (English et al. 2006; English et al. 2005; Natsume et al. 2007). However, the strength of Asf1-histone interactions was unknown until the determination of the binding energetics of Asf1 with H3/H4 (Donham et al. 2011; Park et al. 2008) revealed a tight association that is nearly as tight as the interaction of H3/H4 with DNA (Andrews et al. 2010; Das et al. 2010).
Asf1 participates in nucleosome assembly and disassembly through pathways mediated by downstream histone chaperones, such as HIRA and CAF-1, but the mechanisms that regulate this flow of histones are still relatively poorly understood (Das et al. 2010). The earliest step in nucleosome assembly is regarded as the deposition of two H3/H4 dimers on the DNA resulting in the intermediate known as the tetrasome (reviewed in (Lavelle and Prunell 2007), (Zlatanova et al. 2009), and (Das and Tyler 2011)). As Asf1 carries H3/H4 in the cytosol and nucleus, the potential exists for Asf1 to directly deposit or evict H3/H4 from DNA. Therefore, understanding the intrinsic capabilities of histone chaperones to participate in nucleosome assembly and disassembly processes is important for a complete understanding of chromatin dynamics. This chapter focuses on the actions of Asf1 in both H3/H4 binding and tetrasome dynamics in vitro. Equilibrium fluorescence quenching methods that can be used to measure association constants and electrophoretic mobility assays for assessing tetrasome assembly and disassembly in near physiological conditions are detailed here.
2. Preparation of Asf1
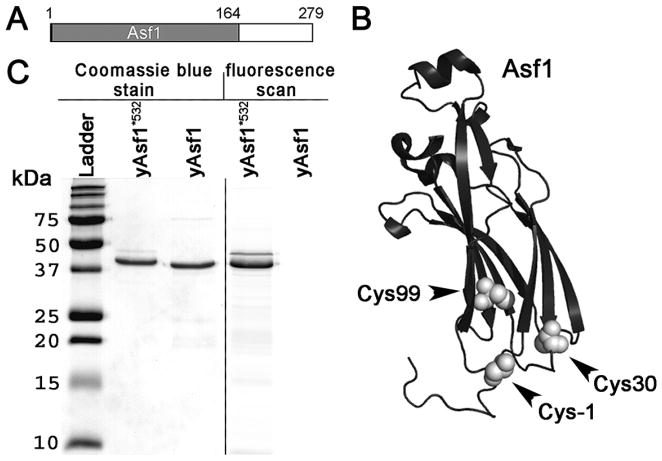
Yeast Asf1 (yAsf1) is a modular protein composed of 279 amino acids residues, with an N-terminal globular core (aa 1–155) and an intrinsically disordered C-terminal tail (155–279) (Fig. 2.1A). Therefore, in order to reliably obtain full-length Asf1, the protein constructs included a C-terminal His6-tag for a final metal affinity purification step, as described previously (Donham et al. 2011). The full-length protein is somewhat sticky and prone to aggregation, most likely due to the highly acidic intrinsically disordered C-terminal tail. The addition of glycerol and detergent greatly aids in solubility and prevention sticking to surfaces. The native cysteines in Asf1 (Cys30 and Cys99) are not near the histone-binding interface and in addition were very inefficiently labeled due to their partial burial in the hydrophobic core of the protein (as verified by mass spectrometry, data not shown) (Fig. 2.1B). Therefore, a cysteine substitution was introduced at the position one residue preceding the first amino acid in the protein at position -1. The position of the labeled cysteine is shown in Fig. 2.1B.
Figure 2.1. Asf1 protein.
(A) Schematic diagram showing the globular core of Asf1 in grey and the C-terminal intrinsically disordered tail in white. (B) Schematic diagram showing Asf1 in black with the positions of the cysteines in the globular core of Asf1 and the cysteine introduced at −1 position shown as white spheres. (C) 15% SDS-PAGE analysis of proteins. Coomassie Blue stain (lanes: molecular weight ladder, yAsf1*532, and yAsf1) and fluorescent image showing Alexa Fluor 532 labeling of Asf1 (Lane yAsf1*532, and yAsf1).
2.1. Expression and purification of full-length yeast Asf1
yAsf1, and truncated versions, can be expressed and purified as described as described previously (Donham et al. 2011).
2.2. Fluorescent labeling of Asf1
The yAsf1 protein can be efficiently labeled using maleimide chemistry (Fig. 2.1C) with Alexa Fluor 532 (Invitrogen) yielding yAsf1*532. The protocol is adapted from Invitrogen’s “Thiol reactive probes” information.
-
1
Dialyze Asf1 into Fluorophore Labeling buffer (FLB: 25 mM HEPES pH 7.5, 0.5 M NaCl, 0.5 mM TCEP, 10% glycerol, 0.05% BRIJ-35) overnight at 4°C. After dialysis, concentrate protein to 20–50 μM using Vivaspin 10,000 MWCO centrifugal concentrator.
-
2
Prepare a 10 mM stock solution of the reactive dye in DMSO immediately prior to use. Add a 10–15 molar excess of dye:protein slowly while stirring the reaction.
-
3
Allow the reaction to proceed for 2 hours at room temperature, rotating constantly in a foil-covered tube. Upon completion of the labeling reaction, add 20 mM βME to quench the reaction.
-
4
To separate labeled yAsf1 from excess dye, apply the reaction (volume no greater than 500μl) to an analytical Superdex S200 column equilibrated in Gel Filtration buffer (GFB: 10 mM Tris-HCl pH 7.5, 0.5 M NaCl, 0.5 mM TCEP, 10% glycerol, 0.05% BRIJ-35) at 4°C. Run column at a flow rate of 0.5 ml/min. Full length labeled Asf1 will elute at approximately 17 ml, with smaller molecular weight species and free dye eluting at approximately 25 ml.
-
5
Analyze fractions by 12.5% SDS-PAGE. Scan the gel using an AlphaImager or Typhoon to confirm the presence of labeled full-length yAsf1*532. After fluorescence imaging, the gel is stained gel with Coomassie blue to confirm the presence of pure yAsf1 free of unlabeled protein contaminants (Fig. 2.1C).
-
6Pool fractions containing pure full-length, labeled protein and concentrate to approximately 10 μM. Check concentration and degree of labeling by measuring the absorbance of yAsf1*532 at 280 nm and the absorbance of the dye at its excitation maximum (Amax). The concentration of the protein is determined using the Beer-Lambert law after correcting for the contribution of the dye to the absorbance at A280.
The calculated Aprotein is then divided by the extinction coefficient of yAsf1 at 280 nm, and multiplied by the molecular weight to provide the protein concentration in mg/ml.
The degree of labeling (DOL) is calculated as follows:where MW = the molecular weight of the protein, εdye = the extinction coefficient of the dye at its absorbance maximum, and the protein concentration is in mg/ml. Labeling efficiencies of 0.8–1.0 are regularly observed using this method. The protein can also be analyzed using mass spectrometry (MALDI-TOF; not shown). Yields of pure labeled yAsf1 are usually 50–60% of the input.
-
8
Make 10–20 μl aliquots of protein into Protein Lobind tubes (Eppendorf). Flash freeze and store at −80°C.
3. Preparation of histone components
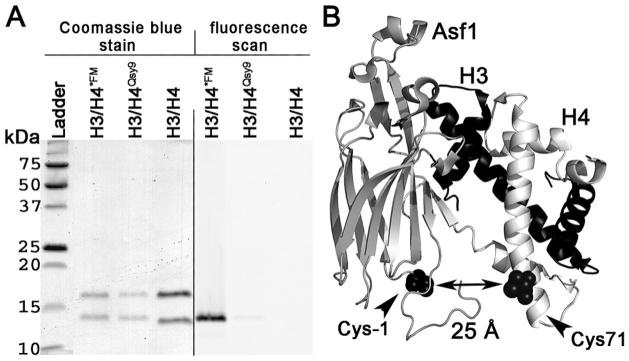
Xenopus laevis histones H3 and H4 with amino acid residue substitutions H3 C110A and H4 T71C, were expressed and purified as previously described (Dyer et al. 2004; Luger et al. 1999) with minor modifications (Fig. 3.1A). The H3C110A mutant (xH3C110A) was used to mimic the yeast histone dimerization surface and to avoid any unwanted cysteine/dye reaction (Park et al. 2004). Histone H4 with a mutation of T71C was used to label H4 without interfering with the H3/H4 dimerization interface or Asf1-H3/H4 binding (Fig. 3.1B) (Park et al. 2004). Histone H4T71C mutant (xH4T71C) was fluorescently labeled (English et al. 2006; Park et al. 2004) with the Qsy9 fluorescence quencher or fluorescein (FM) (Invitrogen) and then assembled into H3/H4 tetramers (Dyer et al. 2004). The distance between the cysteines to which the fluorophores and/or quenchers are attached in Asf1 and H4 is predicted to be approximately 25 Å, which is closer than the Förster radius for the FM-Alexa-532 fluorophore pair.
Figure 3.1. Histone Proteins.
(A) 15% SDS-PAGE analysis of histone proteins and fluorophore labeling. Coomassie Blue stain (left hand side) and fluorescent image (right hand side) showing H3/H4*FM, H3/H4Qsy9, and H3/H4 samples. (B) Schematic diagram of Asf1-H3/H4 complex with the positions used for fluorophore labeling indicated. Asf1, shown in grey, is labeled at cysteine -1 with Alexa Fluor 532 shown in a black space filling representation. Histone H3 is shown in black. Histone H4, shown in white, is labeled at Cysteine 71 with Qsy9 or FM shown in a black space filling representation.
3.1. Expression, purification, and labeling of histone proteins
The expression and inclusion body preparation of Xenopus laevis histones H3 and H4 with amino acid residue substitutions H3 C110A and H4 T71C, were as previously reported (Dyer et al. 2004) with modifications (Donham et al. 2011). The modifications to the procedure are noted below.
Individual denatured histone proteins in 8M Urea are passed through Q Sepharose Fast Flow resin (GE Healthcare) to remove DNA and acidic contaminants.
Collect the flow through from Q Sepharose Fast Flow resin, and pass this solution through SP Sepharose Fast Flow resin (GE Healthcare) to bind the histone proteins. Elute histone proteins by a stepwise increase in the concentration of NaCl from 0.2 to 1 M NaCl in 8M Urea, increasing by 100 mM NaCl at each step.
Analyze the eluted fractions using 15% SDS-PAGE. Extensively dialyze fractions containing the pure protein into water and then lyophilize to dryness.
Labeling of histone H4T71C mutant (xH4T71C) (English et al. 2006; Park et al. 2004) with the Qsy9 fluorescence quencher or fluorescein (FM) (Invitrogen) is performed essentially as described in (Park et al. 2004).
3.2. Preparation of H3/H4 tetramers
H3/H4 tetramers are prepared and purified as detailed in (Luger et al. 1997). Fluorescently labeled tetramers are prepared in the same fashion. Histone H4 is labeled at cysteine 71 prior to being combined with histone H3, for the refolding. After the H3/H4 tetramers are prepared, they are purified over an S200 size exclusion column, and the fractions eluting from the column are analyzed by 15% SDS-PAGE. In the case where FM labeled H3/H4*FM tetramers are formed, the gel is first fluorescently imaged to confirm FM labeling of H4. The gel is then stained with Coomassie blue to confirm the presence of equal amounts of H3 and H4. Analysis of labeled H3/H4*FM tetramers in Fig. 3.1 shows equal amounts of H3 and H4 with clear fluorophore labeling of H4. Yields of H3/H4 tetramers from the size exclusion were improved from 10–20% of input to around 50% by adding 10% glycerol to the gel filtration buffer. The glycerol most likely increases solubility of the tetramers as well as preventing aggregation and adherence to the size exclusion resin.
4. Fluorescence spectroscopy analysis of Asf1 binding interactions with histones
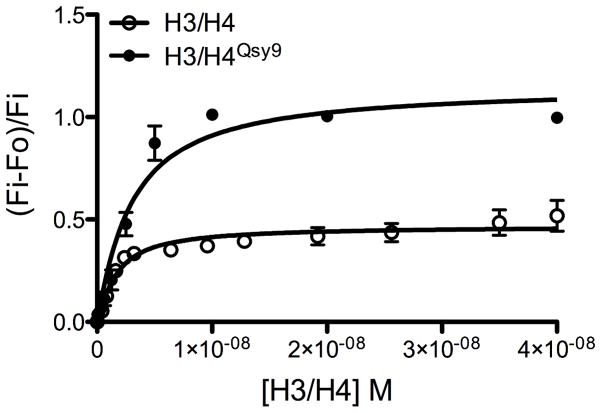
Tetramers prepared from H3 and unlabeled H4or Qsy9-labeled H4 (H4Qsy9) were prepared as tetramers yielding either H3/H4Qsy9 or H3/H4for fluorescence quenching of Asf1*532 for determination of the binding affinity (KD). The use of lo-bind tubes, and buffers containing Brij-35 and 1% glycerol greatly reduce histone aggregation and sticking to surfaces. We consistently observed fluorescence quenching of the fluorophore labeled Asf1 with the unlabeled H3/H4 as well as the Qsy9-quencher labeled H3/H4Qsy9 (Fig. 4.1). This suggests that the histones themselves, perhaps via the histone tails, may partially quench the yAsf*532 signal through collisional quenching, and that complete quenching can be observed with the specific quencher present at T71 of H4. Fluorescence quenching experiments with each of these experimental configurations gave similar KD values for yAsf1-H3/H4 binding (Table 4.1).
Figure 4.1. KD determination using a fluorescence quenching approach.
Fluorescence quenching of the yAsf1*532 fluorescence signal by H3/H4Qsy9 and unlabeled H3/H4 was observed and quantitated to determine the KD of the yAsf1-H3/H4 interaction. H3/H4 or H3/H4*Qsy9 was titrated into 1.0 nM yAsf1*532. The dilution corrected and background subtracted fluorescence data were fitted with a ligand-depleted binding model (Equation 1; GraphPad Prism) because the concentration of yAsf1*532 was within 10-fold of the KD value.
Table 4.1.
Fluorescence quenching of yAsf1*532 with unlabeled and quencher-labeled H3/H4
| Protein pair | KD (M) | Degree of quench |
|---|---|---|
| yAsf1*532-H3/H4 | 1.2 × 10−9 ± .09 | 0.518 |
| yAsf1*532-H3/H4Qsy9 | 1.6 × 10−9 ± .09 | 0.997 |
Before any experiments are to be conducted the quartz cuvette must be thoroughly cleaned. Concentrated cuvette cleaner (Starna Cells Inc.) is added to the cuvette and diluted with water to fill the cuvette and incubated for 5 minutes. The cuvette is washed using a cuvette washer, first extensively with Milli-Q water, followed by HPLC grade Ethanol, and lastly with HPLC grade Methanol. Any particles remaining on the outside of the cuvette and wiped away with a lint free cloth soaked in 100% MeOH. This washing procedure is performed prior to each experiment.
Thaw all protein stocks to be used. Centrifuge at 13,200 K rpm for 5 minutes to remove any precipitated proteins.
Measure protein concentration using a spectrophotometer (ND-1000 Spectrophotometer, NanoDrop Technologies). Three separate measurements are taken and averaged for each protein that will be used. Calculate the concentration of labeled and unlabeled proteins as described in section 2.2.
Make a 30 nM dilute stock of yAsf1*532 by diluting the concentrated protein stock with 150 mM Fluorescence buffer (FB: 150 mM KCl, 2 mM MgCl2, 10 mM Tris-HCl pH 7.5, 1% glycerol, 0.05% Brij-35, 0.5 mM TCEP). Also make a 100 nM dilute stock of H3/H4 (or H3/H4Qsy9) by diluting the concentrated H3/H4 tetramer stock with 150 mM fluorescence buffer.
Using a Horiba Fluorolog-3 spectrometer conduct a blank measurement by scanning 3 ml of FB alone in the cuvette with the specific parameters for the experiment. Double measurements should be taken with an integration time of 0.5 seconds at 20°C, using a 0.5 cm path-length cuvette. In the case of yAsf1*532: excitation wavelength is 528 nm, with a slit width of 7 mm; emission is recorded at 547 nm with a slit width of 7 mm.
Remove 100 μl of buffer from the cuvette, discard and add 100 μl of 30 nM Asf1 stock. Mix gently but thoroughly.
Incubate 10 minutes and then collect a fluorescence scan of yAsf1*532 alone.
Add the calculated amount of 100 nM H3/H4 (or H3/H4Qsy9) stock for first titration point.
Incubate 10 minutes and collect a fluorescence scan. When working with new proteins, initial experiments should be done to confirm how long after the addition of histones it will take for the sample to reach equilibrium and for the fluorescence signal to remain stable.
Repeat steps 7 and 8 until well after the fluorescent quench has saturated.
-
Each raw data point is then multiplied by the dilution factor.
Each dilution corrected data point then has the buffer scan subtracted from it. The data points at 548 nm are then normalized by the following method: (fluorescence observed-fluorescence initial)/fluorescence initial. The normalized data points are then plotted on a graph as a function of the concentration of H3/H4 at each point.
-
The data were fitted with the ligand-depleted binding model (Equation 1) in cases where the concentration of yAsf1* was within 10-fold of the KD value.
Eq. 1 with the variable i indicating the varying concentrations of H3/H4 that were titrated into the yAsf1*.
5. Electrophoretic mobility shift analysis of Asf1 binding interactions with histones
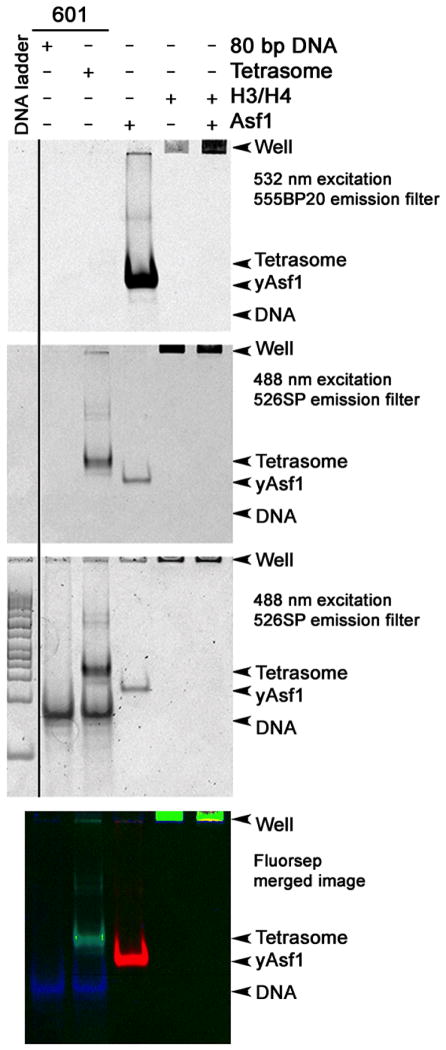
Under a variety of conditions in vitro, H3/H4 can interact productively with DNA to form tetrasomes in the absence of histone chaperones (Alilat et al. 1999; Jackson 1995). In order to learn about the intrinsic properties of Asf1 in tetrasome and disome assembly and disassembly in vitro, we have tested the assembly properties of yAsf1 in a minimal tetrasome system (Donham et al. 2011). Electrophoretic tetrasome assembly and disassembly assays were developed that take advantage of the observation that Asf1 electrophoreses well in non-denaturing-PAGE and the species that it forms and various tetrasome species can be easily visualized, with fluorophore labeled proteins and fluorescent DNA stains.
Electrophoretic analysis using the fluorophore labeled proteins permits the observation of a role for yAsf1 in aiding the assembly of H3/H4-DNA complexes, and provides a means to study whether yAsf1 can disrupt pre-formed tetrasomes. For example, direct binding of yAsf1*532 to H3/H4*FM shifts the electrophoretic mobility of yAsf1*532 so that it no longer enters the gel and remains bound in the well to H3/H4*FM, which does not enter the gel (Fig. 5.1). The free 80 bp DNA, free Asf1 and tetrasomes have distinct electrophoretic mobilities, but the false colored image permits a clear determination of the content of each band in the experiment in cases where there may be overlap.
Figure 5.1. Non-denaturing gel electrophoretic analysis of histones, Asf1, and tetrasomes.
7% non-denaturing PAGE illustrates the migration of DNA, proteins and complexes in electrophoretic assays. The images were obtained using a Typhoon 9400 Variable mode imager (GE Healthcare) in two modes. First the fluorescence of the labeled proteins was imaged using two excitation and emission filter sets to detect the signal of the histone H4 FM label (488 nm excitation and 526SP emission filter) and the Alexa Fluor 532 signal of yAsf1 (532 nm excitation and 555BP20 emission filter). The gel was then stained with SYBR Green I (Invitrogen) nucleic acid stain and imaged again (488 nm excitation and 526SP emission filter). Typically, 80 ng of DNA was loaded and the PMT was set to 300 V. Finally, the sequential scans were merged with FluorSep, and band intensity was quantitated with ImageQuant. Migration of yAsf1*532 (top), Tetrasomes (FM) (middle), and SYBR Green I stained DNA (bottom) are shown with a DNA ladder. While the tetrasomes migrate at approximately the same position as 200 bp linear DNA in this system, H3/H4*FM and the Asf1-H3/H4 complex do not enter the gel.
Moderate amounts of pure 80 bp DNA fragments are needed for salt dialysis tetrasome reconstitutions, with an amount of approximately 15 μg used for each 100 μl reconstitution. Tetrasome formation by direct addition of histone to DNA uses much less DNA with around 0.2μg per reaction using 0.4 μM 80 bp DNA. Larger fragments, such as 146 bp or 208 bp, can be used to study the movement of the histone tetramer on DNA and for studying nucleosomal and histone chaperone/remodeler interactions. In brief, the DNA is produced using PCR with subsequent purification and assembly into tetrasomes using two different approaches. The traditional salt dialysis method allows for formation of relatively homogeneous tetrasomes (Fig. 5.1), whereas, the direct assembly or reconstitution methods allow for analysis of the effects of yAsf1 on H3/H4 deposition onto DNA (Fig. 5.2).
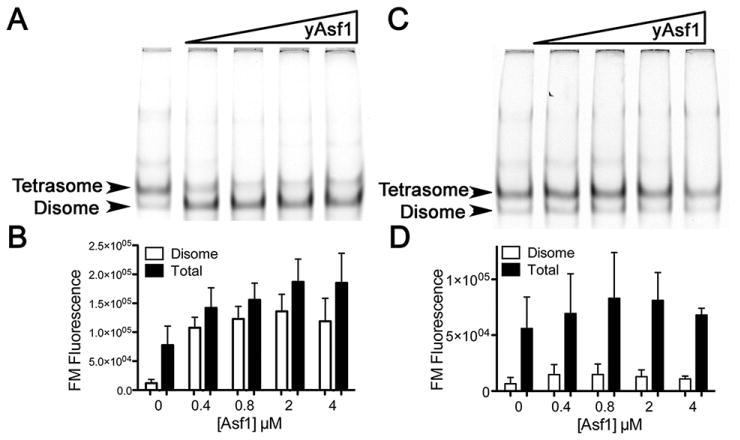
Figure 5.2. Activities of Asf1 in the assembly and disassembly of H3/H4-DNA complexes.
(A) Effect of prior addition of Asf1 on tetrasome and disome formation. H3/H4*FM histones at 0.8 μM were incubated in the absence and presence of increasing concentrations of unlabeled yAsf1 (0, 0.4, 0.8, 2.0, or 4.0 μM) for 30 minutes at 20 °C prior to addition of 0.4 μM concentration 80 bp DNA fragments of 5SDNA. After further incubation for 60 minutes at 20 °C the products were analyzed by non-denaturing PAGE. The gel was scanned to obtain the H3/H4*FM fluorescence before the gel was stained with SYBR Green I nucleic acid stain and then rescanned. The position of each species is indicated with an arrow. (B) The data from at least three independent experiments were quantitated and are presented in graphical form (bottom panel). (C) Effect of the addition of Asf1 on preformed tetrasomes and disomes. H3/H4*FM histones at 0.8 μM were incubated with 0.4 μM concentration 80 bp DNA fragments of 5SDNA prior to the addition of buffer or increasing concentrations of unlabeled yAsf1 (0, 0.4 0.8. 2, and 4 μM) for 90 minutes at 20 °C. The products were analyzed by non-denaturing PAGE and scanned for FM fluorescence, and the data from three independent experiments are presented in graphical form (D).
5.1. Preparation of DNA fragments
Assemble PCR reaction. The template DNA is the 601 plasmid for the 80 and 146 bp 601 sequences (Lowary and Widom 1998). For the 80 bp 601 sequence the primers used are as follows: 601primerI (GTC GTA GAC AGC TCT AGC A) and 601primerII (TAG GGAGTA ATC CCC TTG). For the 146 bp 601 sequence: 601146 F (CTG GAG AAT CCC GGT GC) and 601146R (CAG GAT GTA TAT ATC TGA CAC GTG C). Large reaction volumes are made, 10 ml at a time. The reactions are aliquoted into 96 well PCR plates (Thermo-Fast Low-Profile, ABgene PCR plates, ThermoScientific), 100 μl/well.
PCR program: Initial Denaturation: 94 °C for 5 minutes, followed by 35 cycles of: 94 °C for 1 minute, 56 °C 1 minute, 72 °C 3 minutes. Final step: 72 °C for 5 minutes.
The PCR product is then ethanol precipitated, by adding 3M sodium acetate pH 5.2 at 1/10 the volume of the PCR product. The product and sodium acetate are mixed and 2.5–3 volumes of ice-cold ethanol is added. The EtOH, sodium acetate, and PCR product is mixed well and aliquoted into COREX tubes.
The COREX tubes are then put at −80°C for 1 hour.
After 1 hour, the tubes are centrifuged at 12,000 rpm for 30 minutes.
The supernatant is immediately decanted and tubes are dried inverted for 5–10 minutes.
The pellets are resuspended in a total of 10 ml TE pH 8 (10 mM Tris, 1 mM EDTA).
The resuspended PCR products are then purified by ion-exchange chromatography using a DEAE column (Tyopearl MD-G DEAE SPW-1; Tosoh Bioscience LLC). Elute with a gradient of 0–1 M NaCl in TE pH 8.
Analyze peak fractions on 6% non-denaturing PAGE. The Fermentas 50 bp ladder was used as a molecular weight marker. Make sure to dilute each sample, as high amounts of NaCl will interfere with sample mobilities in the native gel.
Pool pure fractions of proper size, and ethanol precipitate as described earlier.
Resuspend the DNA pellet in 2M FB at a final concentration of 500 ng/μl. Aliquot into 50 μl aliquots. Store at −20° C.
5.2a. Preparation of H3/H4 tetrasomes using salt dialysis
To form tetrasomes, combine purified 80 bp or 146 bp DNA and H3/H4 in Protein Lobind tubes (Eppendorf) in a ratio of 1:1 with a final DNA concentration of 0.15 mg/ml.
Transfer the mixture into a 3500 MWCO Slide-A-Lyzer Dialysis Cassette (Thermo Scientific) and dialyze against 300 ml of 10 mM Tris-HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, and 1 mM TCEP in a 4 L graduated cylinder. Add 3700 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 1mM TCEP to the 300 ml at a rate of 1 ml/min to give a final salt concentration of 150 mM. The solution should be stirring at all times and kept at 4°C. If using fluorescently labeled tetramers, all containers must be covered in foil and protected from exposure to light.
After dialysis, remove the tetrasome sample from the Slide-A-Lyzer Dialysis Cassette. Aliquot the sample in Protein Lobind tube (Eppendorf) and centrifuge at 13,200 rpm for 5 minutes to remove any precipitate. Transfer supernatant to fresh tube.
Analyze concentration of tetrasomes and DNA by measuring absorbance at 260 nm.
Store at 4° C for no more than 2 weeks.
5.2b. Assembly of tetrasomes by reconstitution
Tetrasomes were alternatively formed by direct addition of H3/H4 tetramers to 80 bp DNA. Direct addition as opposed to salt dialysis allows one to observe the influence of other proteins, in this case yAsf1, on tetrasome formation. This direct addition of the histones to the DNA produces two bands when the reactions are electrophoresed on non-denaturing PAGE (Fig. 5.2A). The top band has the same electrophoretic mobility as tetrasomes formed by salt dialysis and the bottom band has a slightly greater mobility. By quantitation of the fluorescence of both bands before and after SYBR green I DNA staining, the histone:DNA ratio was found to be twice as large for the top band compared to the bottom band (Fig. 5.2A and shown in (Donham et al. 2011)). Thus, the top band is a true tetrasome ((H3/H4)2-DNA), whereas the bottom band is a disome (one H3/H4 dimer on 80 bp of DNA).
0.8 μM H3/H4 dimer is directly added to 0.4 μM 80 bp DNA in 150 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.5 mM TCEP and incubated for 30–60 minutes.
Electrophoresis is carried out as described in section 5.3 below.
5.3. Analysis of effects of Asf1 on tetrasome assembly
Thaw all protein stocks to be used. Centrifuge at 13,200 rpm for 5 minutes to remove any precipitate.
Measure UV-vis scan of proteins using nanodrop. Calculate concentration of labeled and unlabeled proteins as stated in section 2.2.
To prepare the reaction mixture. First add H2O, 10x EMSA Buffer (100 mM Tris-HCl, pH 7.5, 1.5 M NaCl, 5 mM TCEP) to Protein Lobind tube (Eppendorf) tube. For assembly reactions yAsf1 is added to the tube, followed by H3/H4. After the required incubation period, DNA is added to the assembly reaction.
Incubate the samples for 1 hour at desired temperature.
Add 10% glycerol to each sample to a final concentration of 5% glycerol. Mix well, and immediately load no more than 8 μl on a 7% polyacrylamide gel (59:1 acrylamide:bis-acrylamide) containing 0.2 x TBE that has been pre-run for 60 minutes in 0.2 x TBE at 4°C at 70V.
Perform gel electrophoresis at 4°C and 70 V for 2–3 hours. Bands are visualized by recording the signal of the histone H4 fluorescein (FM) label, the Alexa Fluor 532 signal of yAsf1*, and SYBR Green I (Invitrogen) nucleic acid stain fluorescence with a Typhoon 9400 Variable mode imager (GE Healthcare) (Figs. 5.1, 5.2A and C).
The bands are quantitated using ImageQuant. A grid is made with the number of lanes desired to quantitate plus one blank lane for background. The grid allows one to make equal volume boxes surrounding each band. A volume report is generated from the grid that gives the fluorescence of each box. Background fluorescence (the fluorescence from the blank box) is subtracted from each band. These corrected numbers can then be used directly or normalized (Fig. 5.2B and D).
5.4. Analysis of effects of Asf1 on tetrasome disassembly
For disassembly reactions, the same procedures are used as above in section 5.3, except that in step 2, the DNA is added first followed by H3/H4. Then, after the required incubation period, Asf1 is added to the disassembly reaction at the required concentrations or ratios. In Fig. 5.2C and D, it is clear that addition of unlabeled Asf1does not dissociate preformed tetrasomes. In addition, yAsf1 was not observed to bind to tetrasomes.
Summary and Implications
The methods here allowed for an analysis of the intrinsic histone chaperone activities of yAsf1 in vitro. The finding that yAsf1 binds histones tightly, but was not sufficient by itself to dissociate the tetrasome, is consistent with a role as a protector of free histones when they are either in excess over DNA or are not in the vicinity of DNA. The observation that yAsf1 can guide the assembly of H3/H4 with DNA into histone-DNA complexes by preventing non-specific aggregation of the histones with DNA (Donham et al. 2011) was analogous to recent findings with Nap1 (Andrews et al. 2010). However, the finding that Asf1 was even able to assist in the direct deposition of H3/H4 dimers onto DNA was novel and unexpected. Although the action of Asf1 directly in assembly of disomes was not necessarily anticipated, the relatively high affinity of the complex suggests that Asf1 has the potential to bias the species of histones that are available for direct deposition on to DNA, if they are not assembled through normal chromatin assembly pathways involving HIRA or CAF-1 (reviewed in (Amin et al. 2011; Ransom et al. 2010)).
A strength of the methods presented here is that they ought to be equally applicable to the analysis of other H3/H4 histone chaperones as well as post-translationally modified histones or histone chaperones. It is already known that histone modifications play an important role in the nucleosome assembly process ((Neumann et al. 2009; Somers and Owen-Hughes 2009; Williams et al. 2008) and reviewed in (Liu and Churchill 2012; Ransom et al. 2010)). We recently found using an acetylation mimicking residue substitution in H3, to give H3K56Q/H4*FM, that there is no effect on Asf1-mediated disome deposition (Donham et al. 2011), but this type of modification might alter the interactions of the histones with other histone chaperones. There are several other H3/H4 histone chaperones that are not yet well characterized, which we expect can also be used for fluorescence quenching of fluorophore-labeled H3/H4. As most histone chaperones are acidic proteins, they would also be amenable to non-denaturing electrophoresis under the appropriate buffer conditions. As preparation of modified proteins becomes common place, quantitative measurement of their contributions to binding and (dis)assembly processes will be an important addition to our general understanding of chromatin dynamics.
Acknowledgments
We are grateful to Karolin Luger for providing the histone gene plasmids and Jessica Tyler for helpful discussions. We thank David Jones for the use of the Horiba Fluorolog-3 spectrometer, and the Department of Cellular and Developmental Biology for the use of the Typhoon 9400 imager. We acknowledge funding from the NIH R01 GM079154 to M.E.A.C for the support of this work.
References
- Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13(1):6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Alilat M, Sivolob A, Revet B, Prunell A. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3–H4)2 tetramer-DNA particle. J Mol Biol. 1999;291(4):815–41. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin’. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37(6):834–42. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17(11):1343–51. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Das C, Tyler JK. Histone exchange and histone modifications during transcription and aging’. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Tyler JK, Churchill MEA. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–89. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donham DC, II, Scorgie JK, Churchill MEA. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4–DNA complexes’. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr097. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65(3):414–44. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3–H4 heterotetramer on DNA. Biochemistry. 2005;44(42):13673–82. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127(3):495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Preferential binding of histones H3 and H4 to highly positively coiled DNA. Biochemistry. 1995;34(33):10607–19. doi: 10.1021/bi00033a036. [DOI] [PubMed] [Google Scholar]
- Jorcano JL, Ruiz-Carrillo A. H3.H4 tetramer directs DNA and core histone octamer assembly in the nucleosome core particle. Biochemistry. 1979;18(5):768–74. doi: 10.1021/bi00572a005. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lavelle C, Prunell A. Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle. 2007;6(17):2113–9. doi: 10.4161/cc.6.17.4631. [DOI] [PubMed] [Google Scholar]
- Liu WH, Churchill MEA. Histone Transfer Among Chaperones’. Biochem Soc Trans. 2012 doi: 10.1042/BST20110737. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116(2):79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446(7133):338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36(1):153–63. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18(3):282–9. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279(23):24274–82. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15(9):957–64. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140(2):183–95. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J, Owen-Hughes T. Mutations to the histone H3 alpha N region selectively alter the outcome of ATP-dependent nucleosome-remodelling reactions. Nucleic Acids Res. 2009;37(8):2504–13. doi: 10.1093/nar/gkp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A. 2008;105(26):9000–5. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17(2):160–71. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]