Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) is a mammalian enzyme that attaches long branching chains of ADP-ribose to specific nuclear proteins, including itself. Because its activity in vitro is dependent upon interaction with broken DNA, it has been postulated that PARP-1 plays an important role in DNA strand-break repair in vivo. The exact mechanism of binding to DNA and the structural determinants of binding remain to be defined, but regions of transition from single-stranded to double-strandedness may be important recognition sites. Here we employ surface plasmon resonance (SPR) to investigate this hypothesis. Oligodeoxynucleotide (ODN) substrates that mimic DNA with different degrees of single-strandedness were used for measurements of both PARP-1/DNA binding kinetics and PARP-1’s enzyme activities. We found that binding correlated with activity, but was unrelated to single-strandedness of the ODN. Instead, PARP-1 binding and activity were highest on ODNs that modeled a DNA double-strand break (DSB). These results provide support for PARP-1 recognizing and binding DSBs in a manner that is independent of single-stranded features, and demonstrate the usefulness of SPR for simultaneously investigating both PARP-1 binding and PARP-1 auto-poly(ADP-ribosyl)ation activities within the same in vitro system.
Keywords: poly(ADP-ribose) polymerase-1, PARP-1, DNA repair, DNA double-strand breaks, surface plasmon resonance, poly(ADP-ribosyl)ation, binding kinetics, non-homologous end joining
INTRODUCTION
Poly(ADP-ribose) polymerase-1 (PARP-1) is a major constituent of mammalian chromatin. Its sole known enzymatic activity is the attachment of long branching chains of ADP-ribose to specific nuclear proteins. NAD+ is the substrate, and binding to DNA strand breaks is required for enzymatic activation. Because of its DNA-strand-break-dependent activity, it has long been postulated that PARP-1 plays an important role in mammalian DNA strand-break repair. In recent years, PARP-1 has been associated most closely with the base excision repair (BER) pathway (Masson et al., 1998; Bernstein et al., 2002), which is responsible for repairing both single-strand breaks (SSBs) and oxidatively-damaged bases in DNA.
Upon binding to a DNA nick, PARP-1 enzymatic activity is stimulated. The newly synthesized poly(ADP-ribose) plays an important role in recruiting the BER protein, XRCC1, to the site (El-Khamisy et al., 2003; Okano et al., 2003). Then PARP-1 interacts with the BRCT I domain of XRCC1 (Masson et al., 1998; Schreiber et al., 2002; Horton et al., 2008). Multiple acceptor proteins, including XRCC1 (Masson et al., 1998), are rapidly poly(ADP-ribosyl)ated. However, the major acceptor is PARP-1 itself (Lindahl et al., 1995). Auto-poly(ADP-ribosyl)ation accounts for 90% of the intracellular poly(ADP-ribosyl)ated protein in oxidatively damaged cells (Huletsky et al., 1989; D’Amours et al., 1999). The auto-poly(ADP-ribosyl)ated PARP-1 loses its affinity for the DNA break and releases from the nicked DNA, allowing downstream DNA repair enzymes access to the damage site (D’Amours et al., 1999). The auto-poly(ADP-ribosyl)ated PARP-1 may also play a role in recruiting other DNA repair proteins, such as DNA ligase IIIalpha to the site (Leppard et al., 2003).
One strategy used to infer PARP-1’s role in DNA repair has been to measure its binding or activation by DNA strand breaks having different structures and chemistries. This work has shown that PARP-1 binds to double-strand breaks (DSBs), as well as SSBs. Yet, DSBs are not intermediate structures in the BER pathway, implying that PARP-1 may also play some role in DSB mediated pathways. There is evidence that PARP-1 may regulate homologous recombinational (HR) repair of DSBs (Schultz et al., 2003; Bryant and Helleday, 2006), and it may also participate in a poorly understood non-homologous end joining (NHEJ) DSB repair pathway that is separate from the well-characterized DNA-PK-dependent NHEJ (Audebert et al., 2004). It has further been hypothesized that the major determinant of the pathway utilized for DSB repair is the particular end-group structure of the DSB (Audebert et al., 2004). While other DSB binding proteins appear to have rather restrictive DNA termini constraints for successful loading at DSBs (e.g., DNA-PKCS) (Jovanovic and Dynan, 2006), PARP-1 binding can recognize more diverse terminal structures such as those produced through oxidative damage. However, the structural determinants of DSB binding for PARP-1 are poorly understood.
Although the exact structural requirements for PARP-1 to bind to DNA ends is not known, there is evidence that rather than having chemical end groups specificity PARP-1 differentially recognizes various helical junctures within DNA (Pion et al., 2003; Soldatenkov and Potaman, 2004; Chasovskikh et al., 2005; Lonskaya et al., 2005). Such structural junctures occur in oxidatively-induced DSBs. They also occur within DNA cruciform structures, which have likewise been reported to serve as binding substrates for PARP-1 (Sastry and Kun, 1990; Chasovskikh et al., 2005; Potaman et al., 2005). Localization by atomic force microscopy (AFM) of purified PARP-1 along various intact plasmid DNA substrates further suggests that single-strand/double-strand junctures may be determinants of recognition and binding (Chasovskikh et al., 2005). However, such studies have not measured the relative strength of PARP-1 binding to these different structures, nor has the stimulation of PARP-1s enzymatic activity by these different DNA binding substrates been well characterized.
In this study, we quantitated both the binding and enzymatic activities of PARP-1 on various oligodeoxynucleotide (ODN) substrates, that model different terminal DNA structures with different degrees of single-strandedness. We advanced the hypothesis that junctures transitioning from single- to double-strandedness in DNA were preferred sites for both PARP-1 binding and enzyme activation. We used surface plasmon resonance (SPR) technology and employed run conditions that mimic the intracellular environment, and we also measured PARP-1 binding kinetics and poly(ADP-ribose) chain growth to determine possible mechanistic functions based on PARP-1’s differential binding and activity on the ODNs.
We report here that PARP-1’s preferred binding substrate is a structure that mimics an oxidatively-produced DSB, and that this binding substrate also stimulated the greatest enzymatic activity. These findings support PARP-1 plays a significant role in DSB repair, and indicate that single-strandedness does not appear to be a significant determinant of binding.
MATERIAL AND METHODS
All chemicals and reagents were purchased form Sigma (St. Louis, MO). ODNs were purchased from Midland Certified Reagent Company, Inc. (Midland, TX). The ODN structures produced from the primary nucleotide sequences were characterized with web-based software written by N.R. Markham and M. Zuker at the Rensselaer Polytechnic Institute (Albany NY), which predicts secondary DNA structure based on primary base sequence (http://frontend.bioinfo.rpi.edu/applications/hybrid/quikfold.php).
All SPR data presented here were generated using recombinant human PARP-1 protein that was purchased from Trevigen Inc. (Gaithersburg, MD). Recombinant human PARP-1 protein from Axxora LLC (San Diego, CA) was also tested with SPR, and comparable results were obtained. SPR has been validated and shown to give reliable kinetics data comparable to established aqueous methods, such as isothermal titration calorimetry and stopped-flow fluorescence (Day et al., 2002).
SPR experiments were performed on a Biacore T-100 instrument (GE Healthcare, Piscataway NJ). For measurement of PARP-1/DNA interactions, streptavidin-coated chips were used (Series S Sensor Chip SA; GE Healthcare, Piscataway NJ).
Simple binding assays and enzymatic assays were performed at a flow rate of 20 μl/min. Kinetic analyses were performed at a flow rate of 75 μl/min. Buffer A (HBS-P) was used as the standard running buffer, which contained 10 mM Hepes (pH 7.4), 150 mM NaCl and 0.05% surfactant P-20. Measurements of enzymatic activities were determined by injection of NAD+ in Buffer B (50 mM Tris pH 8.0, 4 mM MgCl2 and 0.2 mM dithiothreitol).
Different regeneration conditions were evaluated. Best results were obtained with either two 30-second pulses of 50 mM NaOH/1.0 M NaCl or 0.08% SDS, which gave very comparable kinetics data. Regeneration between runs was done with 50 mM NaOH/1.0 M NaCl at 25°C.
Kinetics experiments required a low surface density chip, with a theoretical Rmax of 80–120 resonance units (RU). [1 RU = 0.0001 degree of change in reflected light angle.] Enzymatic activity studies required a high surface density chip with a theoretical Rmax of 8000–10 000 RU. Results were analyzed by using Biacore T-100 analysis software.
RESULTS
SPR was employed to measure the binding kinetics of purified human PARP-1 on three different ODN substrates. SPR is a technology that can measure direct molecular interactions without labeling (Fivash et al., 1998; Malmqvist, 1999). Experiments involve immobilization of a ligand on a microchip surface, and an analyte is then injected over the chip surface through a fluidics system. Interactions between the ligand and the analyte are measured based on total mass change on the chip surface. In our experiments, biotinylated ODN molecules were used as ligands immobilized on the chip surface. PARP-1 protein was the analyte.
The ODN substrates were designed to represent various DNA end-group structures with substantially differing degrees of single-strandedness, in order to test the hypothesis that PARP-1 recognizes and binds to junctures at transitions from double- to single-stranded DNA. The specific nucleotide sequences were selected to be identical or similar to sequences that had been previously used by Dynan and coworkers to measure loading of DNA-PKCS on DNA ends (Jovanovic and Dynan, 2006).
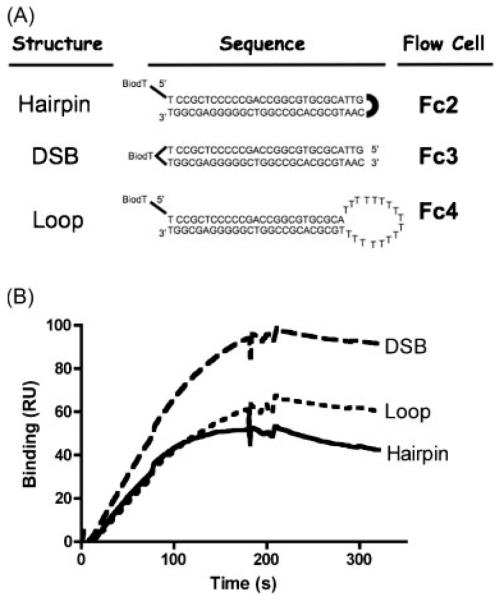
The ODNs were either terminally or internally biotinylated to allow immobilization on streptavidin-coated chips that were used to anchor ligands within SPR flow cells. A single chip had four flow cells (Fc), each of which could accommodate a different ODN. For these experiments, one flow cell (Fc1) had no bound ODN and was used as a reference for background subtraction. The other three flow cells contained one of the three ODNs. The sequence, structure, and flow cell location for each of the ODNs is shown in Figure 1A.
Figure 1.
PARP-1 binding to substrates. (A) Oligodeoxynucleotide sequences and structures. BioT allowed immobilization of the ODN on a Biacore-SA chip at the indicated flow cell (Fc) location. Location of a biotinylated thymidine (BioT) within each ODN sequence is shown. (Fc1 contained no ODN and was used as negative control for background subtraction.) (B) PARP-1 binding to different ODN stuctures. A solution of 10 nM recombinant PARP-1 protein was injected over all four flow cells of a Biacore T-100 chip. PARP-1 binding in each flow cell was detected in a Biacore T-100 instrument. Surface readings from an empty flow cell (Fc1) were subtracted from the flow cells containing different ODN structures (Fc2, Fc3, and Fc4), and the resulting binding quantity, in RU, was graphed over time post-injection. Injections started at 0 s and ended at 180 s, which constituted the association phase for PARP-1/ODN interaction. Dissociation phase data were collected from the end of injection (180 s) until 330 s.
Fc2 contained an ODN with a palindromic sequence that formed a tight hairpin. It had the same structure as the DSB ODN (Fc3) but lacked free 3′ or 5′ strand termini, so that there was no potential for it to unwind its helical structure and, thereby, induce single-strandedness. This ODN was designated the “hairpin.”
Fc4 contained the same base-pair sequence as the hairpin ODN, but was interrupted midway by a long sequence of thymine nucleotides (17 bases), which would not allow Watson-Crick base pairing and, therefore, formed an open loop of single stranded DNA. The loop ODN modeled DNA with a single- to double-stranded junction but, just as for the hairpin, it had no 5′ or 3′ termini. Preliminary imaging experiments of this ODN with AFM had suggested the possibility that PARP-1 might bind to ODNs containing such open loop structures (data not shown). This ODN was designated the “loop” structure. Together, the hairpin and the loop were considered to represent models at the extremes of double-strandedness and single-strandedness, respectively.
Fc3 contained an ODN with the same base-paired sequence as the hairpin, but with 5′ and 3′ hydroxyl termini. This ODN was considered to be a structural model of a DNA DSB that might be produced in a cell by DNA damaging agents or enzymatic cleavage. This structure was thought to have moderate potential for single-strandedness, since the termini are not covalently linked together, and can have a resonant single-stranded/double-stranded character. Thus, the hairpin, the DSB, and the loop ODNs were designed to represent progressively increasing single-strandedness at DSB termini. Further, the hairpin and DSB ODNs were thought to have natural counterparts in cruciform formations and oxidative DSBs, repectively, that PARP-1 might be expected to encounter intracellularly.
We first tested the binding of PARP-1 to all three ODNs by simultaneously injecting a 10 nM recombinant PARP-1 solution for 3 min (Figure 1B). The first flow cell of the chip, with no bound ODN, was used as a background control. PARP-1 bound to all three flow cells with immobilized ODN. However, PARP-1 binding to the DSB ODN showed a faster association compared to hairpin and loop ODNs. Dissociation of all three ODNs appeared to be similar.
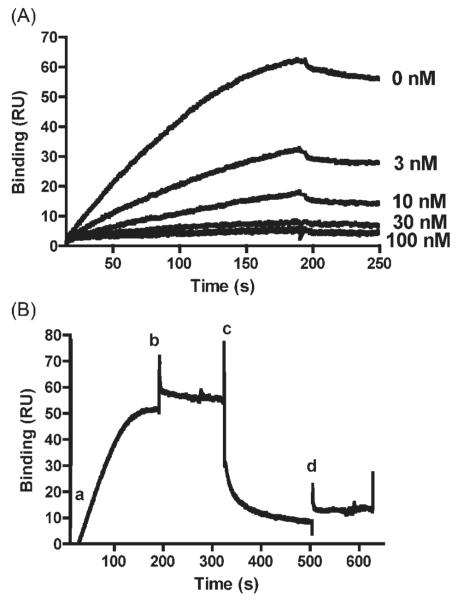
Before performing the kinetic analysis of PARP-1/ODN binding, assay validation was performed to ensure that specific protein-DNA interactions were being measured. The empty flow cell (Fc1) provided a reliable control for non-specific binding. Nevertheless, competition experiments were also conducted to increase confidence. Each ODNs was premixed with PARP-1 in solution before injection into the SPR instrument. (Since chip surfaces are coated with streptavidin, the same ODNs that were immobilized on the chip surface could not be used. Comparable ODNs with the same sequence but no biotin tag were, therefore, used for this experiment.) When the free ODN was premixed with PARP-1, it inhibited the PARP-1 binding to immobilized ODN on the chip surface. This effect could be titrated by changing the free ODN concentration (Figure 2A).
Figure 2.
Competition of PARP-1 binding to immobilized ODN by free ODN. (A) To determine whether free ODN could compete with bound ODN for PARP-1 binding, recombinant PARP-1 (8 nM) was premixed with increasing concentrations (3, 10, 30, 100 nM) of a free ODN that was similar in structure to the hairpin ODN, but lacked BioT. Premixing PARP-1 with free ODN inhibited PARP-1 binding to ODNs on the surface in an ODN concentration-dependent manner. Similar results were obtained with other immobilized ODN structures (data not shown). (B) To detemine whether prebound PARP-1 could be released by free ODN, PARP-1 (8 nM) was prebound to immobilized DSB ODN by injecting PARP-1 alone at time zero (a). PARP-1 injection was complete at 180 s (b). Free ODN was then injected starting at 320 s (c) and ending at 500 s (d). Results showed binding of PARP-1 to decrease rapidly upon injection of free ODN. The drop is due to free ODN competing with immobilized ODN on the surface for PARP-1 binding.
Another competition experiment was conducted by sequentially injecting PARP-1 followed by ODN. First, PARP-1 was allowed to bind to the chip surface, and then free ODN was injected (Figure 2B). Once PARP-1 binds to ODN on chip surface, the dissociation phase is very slow (see kinetics data below). Therefore, until a regeneration solution was injected, most of the PARP-1 stayed on the surface. Injection of free ODN however, was able to remove previously bound PARP-1 from the surface (Figure 2B). Thus, free ODN was able to compete for PARP-1 binding to ODN immobilized on the chip surface, both in premixed and sequential injection regimens. These results suggest that this experimental model can reliably measure reversible protein-DNA interactions.
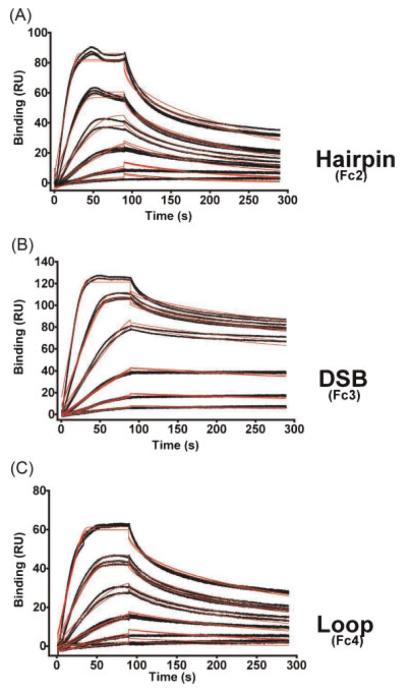
To calculate kinetics of PARP-1/ODN binding interactions, runs were repeated with various concentrations of PARP-1 (Figure 3). Association rate constants (kON), dissociation rate constants (kOFF) and equilibrium dissociation constants (KD) were calculated (Table 1). The DSB had the strongest binding (KD=0.49 nM), while the hairpin and loop ODNs bound more weakly (KD=4.31 and 3.50 nM, respectively). The difference in KD was mainly due to differences in kON rates. kOFF rates were comparable among three ODNs (Table 1). Repetitive runs at different temperatures (25 vs. 37°C), or using different regeneration conditions, gave similar KD values, consistently showing the DSB ODN to be the preferred PARP-1 binding substrate, regardless of the run conditions. Representative Biacore statistical parameters for the three ODNs are shown in Table 2.
Figure 3.
Kinetics of PARP-1 binding to ODN. To analyze binding kinetics of PARP-1 ODN interactions, recombinant PARP-1 protein was injected at six different concentrations (1, 2, 4, 8, 15, 30 nM) in duplicates or triplicates. PARP-1 injection was done for 90 s and dissociation data was collected for 200 s. Data were fitted to a Langmuir 1:1 interaction plot to calculate rate constants (Table 1). (Actual data in black lines and curve fits in red lines).
Table 1.
PARP-1 binding constants and enzymatic activity for different ODN structures
| kON (1/Ms) | kOFF (1/s) | KD (M) | Enzymatic activity (units) | |
|---|---|---|---|---|
| Hairpin | 1.51±1.27×106 | 4.72±1.67×10−3 | 4.31±2.48×10−9 | 1.16±0.05×10−2 ** |
| DSB | 1.23±1.35×107 * | 3.42±2.57×10−3 | 4.91±2.96×10−10 * | 2.60±0.13×10−2 ** |
| Loop | 1.99±0.96×106 | 6.51±2.81×10−3 | 3.50±1.52×10−9 | 0.54±0.06×10−2 ** |
Binding constants (kON, kOFF, and KD)and enzymatic activity units (incorporated ADP-ribose RU per minute per immobilized PARP-1 RU) are shown, plus/minus one standard deviation. KD values were determined by fitting SPR run data to a kinetic model (see experimental procedures), and the standard deviations were determined from KD values calculated from multiple runs (n=8). Activity units were calculated by dividing the slopes of the fitted lines, in Figure 4D, by the NAD+ injection time for the runs (i.e., 5 min). The standard deviation of the slope parameter for equation of the fitted line was used as the standard deviation for the enzymatic activity.
kON and KD for DSB were significantly different (p≤0.05) from those values for the loop and hairpin structures (one-way ANOVA).
There were no significant differences between any of the kOFF values. Hairpin and loop values were not significantly different from one another for any of the binding constants.
Enzymatic activities for all of the structures were significantly different from one another at p<0.001 (one-way ANOVA).
Table 2.
Representative Biacore statistical parameters for oligonucleotides
| Hairpin (Fc2) | DSB (Fc3) | Loop (Fc4) | |
|---|---|---|---|
| kA (1/Ms) | 1.20E×106 | 3.66×107 | 1.45×106 |
| SE(kA) | 1.30×104 | 1.90×106 | 2.20×104 |
| kD (1/s) | 0.005581 | 0.007734 | 0.005861 |
| SE(kD) | 4.90×10−5 | 4.00×10−4 | 7.20×10−5 |
| Rmax (RU) | 77.2 | 101 | 60.2 |
| SE(Rmax) | 0.25 | 0.15 | 0.15 |
| tc | 3.71×108 | 2.70×107 | 3.04×107 |
| SE(tc) | 8.90×107 | 8.50×104 | 1.00×106 |
| χ2 (RU2) | 12.0 | 6.67 | 3.87 |
Statistical parameters for a representative experiment are presented. Values are provided with no further manipulation as they are calculated by the BIAevaluation™ software. Relatively small values of standard error (SE) in each category suggests reliable data analysis and low χ2 indicates good curve fit to 1:1 Langmuir model.
These experiments suggested that a DSB was the preferred substrate for stimulation of PARP-1activity. We, therefore, designed an experiment to measure auto-poly(ADP-ribosyl)ation activity of PARP-1by SPR technology.
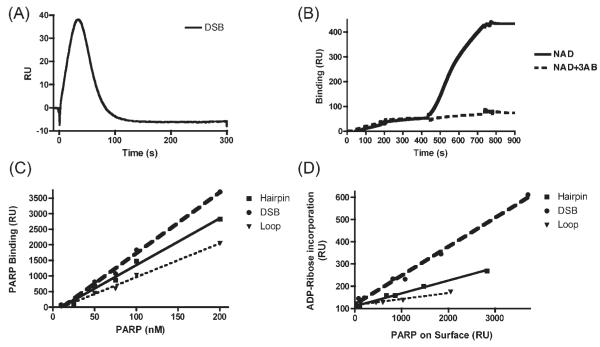
Detection of the total mass on the chip surface allowed us a means to monitor polymerization of ADP-ribose on PARP-1 itself (i.e., auto-poly(ADP-ribosyl)ation). However, increases in surface mass were transient, as auto-poly(ADP-ribosyl)ated PARP-1 was quickly released from its immobilized ODN binding substrate (Figure 4A). This is consistent with the proposition that auto-poly(ADP-ribosyl)ated PARP-1 is released from its ligand DNA due to the accumulation of negatively charged ADP-ribose polymer, which electrostatically repels the poly(ADP-ribosyl)ated protein from the likewise negatively charged DNA phosphodiester backbone.
Figure 4.
Measurement of PARP-1 enzymatic auto-poly(ADP-ribosyl)ation activity. SPR was used to measure auto-poly(ADP-ribosyl)ation activity of PARP-1 bound to ODNs immobilized on a high density Biacore chip surface. (A) When NAD is injected over prebound PARP-1 in the presence of 150 mM NaCl, surface mass increases were transient, due to release of auto-poly(ADP-ribosyl)ated PARP-1 from the ODN substrate. Representative data for the DSB ODN are shown. (B) Use of NAD running buffer without NaCl allowed auto-poly(ADP-ribosyl)ated PARP-1 to accumulate on the chip surface. Recombinant PARP-1 protein (30 nM) was first injected and allowed to bind to ODN surfaces for 200 s in Buffer A. Starting at 440 s 100 μM NAD+ solution was injected for 300 s in Buffer B without NaCl. An increase in total surface mass was detected. When 100 μM NAD+ was premixed with 100 μM 3-AB, mass increases were inhibited (dashed line). (C) Increasing concentrations of recombinant PARP-1 were injected over all flow cells and binding was plotted against PARP-1 concentrations. (D) Specific concentrations of PARP-1 were injected in each flow cell to achieve varying PARP-1 binding in each, based on the data from panel C. Then, enzymatic activity for the different PARP-1/ODN substrates was quatitated by injecting 100 μM NAD+ in Buffer B, without NaCl. The amount of PARP-1 bound was plotted in horizontal axis, and the amount of ADP-ribose polymerization for each ODN was plotted in vertical axis. The graph, therefore, displays enzymatic activity of PARP-1 on each ODN surface, independent of PARP-1/ODN binding affinity. The slopes of the plotted lines are the specific activities for PARP-1 on the different ODN substrates (Table 1).
Release of auto-poly(ADP-ribosyl)ated PARP-1 from the chip prevented us from directly measuring auto-poly(ADP-ribosyl)ation levels of ODN-bound PARP-1. Nevertheless, we found that by lowering the salt concentration of the NAD+ running buffer, auto-poly(ADP-ribosyl)ated PARP-1 remained associated with the chip surface through non-specific binding interactions. We exploited this phenomenon to measure enzymatic activity using a dual buffer system.
The dual buffer approach allowed us to measure both the rate and level of PARP-1 auto-poly(ADP-ribosyl)ation for the various ODNs. This was achieved by allowing PARP-1 to specifically bind to the ODNs in Buffer A, which contained 150 mM NaCl, then switching the Buffer B, which lacked NaCl, for injection of NAD+ (Figure 4B). Switching from high to low ionic strength permitted us to trap auto-(ADP-ribosyl)ated PARP-1 non-specifically on the chip surface of the ODN flow cells, yet without any non-specific binding of PARP-1 to the reference flow cell (Fc1). This strategy enabled us to observe a continuous accumulation of surface mass during NAD+ injection. In this way, enzymatic activity was measured in terms of an increase in molecular mass on a given flow-cell containing PARP-1 prebound to different ODN substrates. (Comparable results were obtained when the PARP-1 binding step was done with either Buffer A, or Buffer B supplemented with 150 mM NaCl.)
We found that there were substantial increases in mass to the PARP-1 when NAD+ was presented to PARP-1 prebound to the DSB ODN (Figure 4B). This increase in mass was suppressed when an inhibitor of PARP-1 activity, 3-aminobenzamide (3-AB), was co-injected with the NAD+. This inhibition supported the contention that the observed increase in mass was dependent upon PARP-1’s enzymatic activity.
Since SPR technology measures total mass on the chip surface, it is possible to estimate the size of the poly-ADP-ribose chain on each PARP-1 molecule. The molecular weight ratio of PARP-1 to ADP-ribose is approximately 200:1. Therefore, 200 ADP-ribose residues will give the same RU response as one PARP-1 molecule. In Figure 4B, initial injection of PARP-1 resulted in a 45 RU signal. Following injection of NAD+ resulted in an additional 405 RU mass accumulation on the surface (i.e., nine times the initial PARP-1 immobilized on the chip surface). Hence, The bound mass increase corresponded to ADP-ribose chains with an average of 1800 (i.e., nine times 200) ADP-ribose residues per PARP-1 molecule.
The experiments described above (Figure 4A and 4B) used the DSB ODN to establish SPR technology as a reliable tool for measuring PARP-1 enzymatic activity in terms of auto-poly(ADP-ribosyl)ation. We then compared the enzymatic activity of PARP-1 immobilized on the different ODN structures. PARP-1 was injected at different concentrations in Buffer A into all flow cells (Figure 4C). Following PARP-1 binding, 100 μM NAD+ was injected in Buffer B and ADP-ribose polymerization was measured in each flow cell (Figure 4D).
Since PARP-1 bound to the three ODN surfaces at different rates, injection of PARP-1 at different concentrations allowed us to make comparisons between flow cells containing the same amount of bound PARP-1. Equal amounts of PARP-1 immobilized on the different ODN surfaces showed different enzymatic activities (Table 1). The highest activity was observed on the PARP-1/DSB complex. Therefore, even after controlling for the amount of PARP-1 binding to the chip surface, DSB ODN induced the highest enzymatic activity per PARP-1 molecule bound. This suggested that binding strength drives auto-poly(ADP-ribosyl)ation activity.
DISCUSSION
PARP-1 is known to bind to both oxidatively- and enzymatically-induced SSBs. In the case of BER, PARP-1 interacts with the base-damaged DNA only after the sequential actions of a base glycosylase and an AP endonuclease have nicked the phosphodiester backbone to produce a SSB. Thus, PARP-1 appears to have an important role in repairing SSBs regardless of their end group chemistries or mechanism of origin.
In the case of DSBs, however, PARP-1 has not been shown to be an integral component in either of the two known repair pathways—homologous recombination (HR) and non-homologous end joining (NHEJ). This is curious, since PARP-1 has long been known to have DSB binding capabilities and to be enzymatically activated by DSBs. However, a recent study suggests that PARP-1 may have a synaptic function in a NHEJ pathway that is separate from the well-characterized DNA-PK-dependent NHEJ (Audebert et al., 2004). In the putative PARP-1-dependent NHEJ, DSBs with various end structures are recognized and bound by PARP-1, which protects the ends and brings them together for synapsis.
Little is currently known about the mechanism of PARP-1’s recognition and binding to DSBs, but it has been suggested that the single-strand/double-strand junction regions, near DSB termini, allow PARP-1 to recognize DSBs without regard to their exact end-group chemistries. This type of recognition would also explain its binding to SSBs via the same DNA binding site, since SSBs also have transitions between single- and double-strandedness.
In this study, we quantitated PARP-1 binding to ODN substrates with very different DNA single-stranded to double-stranded transitions, to test the hypothesis that this parameter is an important determinant of PARP-1 binding. We measured the binding and activity using the SPR technology, which allowed us to measure binding constants and enzymatic activity within the same experimental system in real time and label-free. Results suggested that DSBs with free termini were preferred substrates. Furthermore, PARP-1 bound to DSB ODN showed higher activity compared to hairpin and loop ODNs. There was no indication that single-strandedness correlated with binding affinity.
We have employed a strategy similar to Dynan and coworkers, who used SPR to evaluate the binding kinetics of DNA-PKCS to different double-stranded ODN structures (Jovanovic and Dynan, 2006). The hairpin and DSB ODN sequences used here were identical to the ones used by them. Dynan’s group reported that DNA-PKcs binds more strongly to the DSB compared to the hairpin, and that the DSB stimulated more auto-phosphorylation than the hairpin. They postulated that a free termini from as single DNA strand was a requirement for binding. In this respect, it is intriguing that, of the three substrates used here, the DSB ODN was the only one with free termini available for binding, and it was the ODN with the greatest PARP-1 binding. This suggests that PARP-1 and DNA-PKCS may share a common DNA binding target, and may even be competitors for DSB binding in vivo.
The amino-terminal domain of PARP-1 contains three zinc fingers essential for DNA binding (Langelier et al., 2007). Pion, Bombarda, and coworkers (Pion et al., 2003) used a fragment of PARP-1 (residues 1-234), encompassing the zinc-finger domain, and measured its interaction with various DNA termini by fluorescence anisotropy. They concluded that PARP-1’s zinc finger domain preferentially interacted with 5′-recessed ends. They postulated that binding to 5′-ends allowed 3′-ends to remain exposed for processing by the 3′->5′exonucleases of BER or other repair pathways. Their findings are consistent with those presented here, in that PARP-1 binding to our ODN structures was weak in the cases where no 5′-terminus was available (i.e., hairpin and loop). Taken together, the data support a model where PARP-1 binds to oxidatively-induced DNA breaks through recognition and binding of 5′ termini.
SPR is a useful tool to explore the parameters important to PARP-1 binding to model ODN substrates. A better mechanistic understanding of PARP-1 binding may allow rational design of agents that target PARP-1 for therapeutic gain (Soldatenkov and Potaman, 2004). This technology may also prove valuable for screening and optimization of newly-designed inhibitors of PARP-1 for potential therapeutic activity.
Acknowledgements
SPR experiments were performed at the Biacore Molecular Interactions Shared Resource (BMISR) of Lombardi Comprehensive Cancer Center, which is supported by a grant from the National Cancer Institute (NIH P30 CA51008). The authors thank Ms Linshan Yuan for her excellent technical help.
REFERENCES
- Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat. Res. 2002;511(2):145–178. doi: 10.1016/s1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34(6):1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasovskikh S, Dimtchev A, Smulson M, Dritschilo A. DNA transitions induced by binding of PARP-1 to cruciform structures in supercoiled plasmids. Cytometry A. 2005;68(1):21–27. doi: 10.1002/cyto.a.20187. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(AD-P-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- Day YS, Baird CL, Rich RL, Myszka DG. Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods. Protein Sci. 2002;11(5):1017–1025. doi: 10.1110/ps.4330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31(19):5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivash M, Towler EM, Fisher RJ. BIAcore for macromolecular interaction. Curr. Opin. Biotechnol. 1998;9(1):97–101. doi: 10.1016/s0958-1669(98)80091-8. [DOI] [PubMed] [Google Scholar]
- Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18(1):48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huletsky A, De Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J. Biol. Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- Jovanovic M, Dynan WS. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res. 2006;34(4):1112–1120. doi: 10.1093/nar/gkj504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human PARP-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2007;283(7):4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(AD-P-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003;23(16):5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 2005;280(17):17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- Malmqvist M. BIACORE: an affinity biosensor system for characterization of biomolecular interactions. Biochem. Soc. Trans. 1999;27(2):335–340. doi: 10.1042/bst0270335. [DOI] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, De Murcia G. XRCC1 is specifically associated with poly(AD-P-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23(11):3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion E, Bombarda E, Stiegler P, Ullmann GM, Mely Y, de Murcia G, Gerard D. Poly(ADP-ribose) polymerase-1 dimerizes at a 5′ recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42(42):12409–12417. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J. Mol. Biol. 2005;348(3):609–615. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sastry SS, Kun E. The interaction of adenosine diphosphoribosyl transferase (ADPRT) with a cruciform DNA. Biochem. Biophys. Res. Commun. 1990;167(2):842–847. doi: 10.1016/0006-291x(90)92102-6. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277(25):23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31(17):4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatenkov VA, Potaman VN. DNA-binding properties of poly(ADP-ribose) polymerase: a target for anticancer therapy. Curr. Drug Targets. 2004;5(4):357–365. doi: 10.2174/1389450043345498. [DOI] [PubMed] [Google Scholar]