Abstract
Mammalian apurinic/apyrimidinic endonuclease (APE1) initiates the repair of abasic sites (AP-sites), which are highly toxic, mutagenic, and implicated in carcinogenesis. Also, reducing the activity of APE1 protein in cancer cells and tumors sensitizes mammalian tumor cells to a variety of laboratory and clinical chemotherapeutic agents. In general, mouse models are used in studies of basic mechanisms of carcinogenesis, as well as pre-clinical studies before transitioning into humans. Human APE1 (hAPE1) has previously been cloned, expressed, and extensively characterized. However, the knowledge regarding the characterization of mouse APE1 (mAPE1) is very limited. Here we have expressed and purified full-length hAPE1 and mAPE1 in and from E. coli to near homogeneity. mAPE1 showed comparable fast reaction kinetics to its human counterpart. Steady-state enzyme kinetics showed an apparent Km of 91 nM and kcat of 4.2 s−1 of mAPE1 for the THF cleavage reaction. For hAPE1 apparent Km and kcat were 82 nM and 3.2 s−1, respectively, under similar reaction conditions. However, kcat/Km were in similar range for both APE1s. The optimum pH was in the range of 7.5–8 for both APE1s and had an optimal activity at 50–100 mM KCl, and they showed Mg2+ dependence and abrogation of activity at high salt. Circular dichroism spectroscopy revealed that increasing the Mg2+ concentration altered the ratio of “turns” to “β-strands” for both proteins, and this change may be associated with the conformational changes required to achieve an active state. Overall, compared to hAPE1, mAPE1 has higher Km and kcat values. However, overall results from this study suggest that human and mouse APE1s have mostly similar biochemical and biophysical properties. Thus, the conclusions of mouse studies to elucidate APE1 biology and its role in carcinogenesis may be extrapolated to apply to human biology. This includes the development and validation of effective APE1 inhibitors as chemosensitizers in clinical studies.
Keywords: APE1, Base excision repair, CD
Introduction
Cellular DNA is continuously exposed to endogenous or exogenous chemical or physical agents that induce DNA lesions. DNA-base damage threatens genomic stability and cellular viability. Multiple DNA repair pathways exist in all organisms—from bacteria to humans—to preserve the integrity of the genome [1]. Damaged bases, if not repaired, can be mutagenic [2], and/or trigger cell death [3].
Mammalian AP-endonuclease (APE1) initiates the repair of abasic sites (AP-sites), which are directly formed by free radicals during cellular metabolism, inflammatory diseases, carcinogenesis, and aging. AP-sites are alsoinduced by anti-tumoricidal agents or generated by excision of damaged bases by DNA glycosylases via the base excision repair (BER) pathway. As a BER enzyme, APE1 catalyzes the hydrolytic cleavage of the phosphodiester bond immediately 5′ to the AP-sites [4]. Being a multifunctional protein, APE1 also acts as an endoribonuclease (5) and a redox factor to activate various transcription factors and is involved in gene regulation [6, 7]. Moreover, the rRNA quality control process is known to be regulated by APE1 (8).
Elevated levels of APE1 have been linked to resistance to chemotherapy, poor prognosis, and poor survival. Reducing the activity of APE1 protein in cancer cells and tumors sensitizes mammalian tumor cells to a variety of laboratory and clinical chemotherapeutic agents [9-17].
To understand the biology of APE1 and its role in carcinogenesis, transgenic mice with conditional APE1 gene knock-out have been developed [18-20]. In general, the mouse model is used to study basic mechanisms of carcinogenesis. Moreover, most pre-clinical studies use the mouse model for translational experiments before transitioning into humans. Human APE1 (hAPE1) has previously been cloned, expressed, and extensively characterized [4-20]. Mouse APE1 (mAPE1) has 6% dissimilarity with the hAPE1 in amino acid sequence. Even a change in a single amino acid which is far from catalytic pocket can have a profound effect on the activity of APE1 [21]. Surprisingly, knowledge regarding the characteristics of mAPE1 is very limited. Therefore, it is important to know the basic properties of the mouse protein to complement animal studies and to provide necessary background information for full-scale animal and human translational studies. Here, we have purified and biochemically and biophysically characterized mAPE1, comparing with the hAPE1 under similar conditions. Overall, this study insures that mouse and human APE1s are mostly similar in respect to their basic physico-chemical properties.
Methods
Materials
hAPE1 cDNA was bought from Open Biosystems (Huntsville, AL, USA). mAPE1 cDNA was a kind gift from Prof. Sankar Mitra of UTMB, Galveston, Texas.
Cloning and purification of hAPE1 and mAPE1
hAPE1 and mAPE1 were cloned, expressed as His-tagged protein, and purified as described previously [22] using an ion-exchange column with slight modification. We had 2 mM EDTA in all the buffers used for protein purification.
In-gel tryptic digestion and protein identification by mass spectrometry
In brief, the protein was resolved on a 4–12% NUPAGE gel (Invitrogen), and the protein band of interest was manually excised from the 1D SDS-PAGE-gel and transferred to a microcentrifuge tube. The gel slices were washed with 100 mM ammonium bicarbonate and incubated with 50 mM ammonium bicarbonate and 10 μl of 10 mM DTT at 60°C for 30 min. When tubes were cooled to room temp, 10 μl of 55 mM iodoacetamide was added, and they were incubated for another 30 min in the dark at room temperature. The solvent was discarded, and the gel slices were washed in 50% acetonitrile/100 mM ammonium bicarbonate. Subsequently, the gel slices were transferred onto a 96-well Montage plate (Millipore), destained with 50% acetonitrile in 25 mM ammonium bicarbonate, dehydrated with acetonitrile for 5 min, and vacuum dried. Gel pieces were then rehydrated with 15 ll of ammonium bicarbonate:acetonitrile (25 mM:10%) supplemented with trypsin (5 ng/ll, Promega, Madison, WI, USA) at 37°C for 16 h. Afterwards, tryptic peptides were extracted in 0.1%TFA/50% acetonitrile and mixed with equal volume of 5 mg/ml alpha cyano-4-hydroxycinnamic acid (Acros Organics, New Jersey, USA). Mass spectra (MS) were recorded with a matrix assisted laser desorption/ionization-time of flight, time of flight (MALDI-TOF-TOF) spectrometer (4800 Proteomics Analyzer, Framingham, MA, USA) set in reflector positive mode by spotting the samples onto a MALDI plate. The samples were ionized with a fixed LASER intensity of 3,800 J, and 1000 LASER shots were collected per subspectrum. Moreover, the samples were shot randomly with uniform bias. The detector voltage was 2.1 kV, the bin size was set at 0.5 ns, and the signal/noise threshold was set at 15. The spectra were collected with a specified mass range of 700-4,000 Da with a focus mass of 2,100 Da. Peptide masses were compared with the theoretical masses derived from the sequences contained in SWISS-PROT/NCBI databases using MASCOT. The search parameters were set as follows: cysteines as carbamidomethyl derivatives, allowed peptide mass error 50 ppm, at least four peptide mass hits required for a protein match, and up to one missed cleavage and methionine oxidized form.
Oligonucleotide substrate preparation
Tetrahydrofuran (THF), a synthetic stable AP-site analog, containing 50-mer oligonucleotide with the sequence 5′-TCGAGGATCCTGAGCTCGAGTCGACGXTCGCGAATTCTGCGGATCCAAGC-3′ (where X represents THF) was purchased from Gene Link (Hawthorne, NY). The complementary oligonucleotide containing T opposite THF was synthesized by the Recombinant DNA Laboratory Core Facility at the University of Texas Medical Branch (Galveston, TX). The oligonucleotides were purified on a sequencing gel. The THF oligonucleotide was labeled at the 5′ end using T4 polynucleotide kinase and γ[32P]ATP and annealed to the complementary oligonucleotide to prepare 32P-end-labeled duplex oligonucleotide as described previously [23].
Human and mouse APE1-mediated excision activity assay
5′-32P-labeled THF-containing duplex oligonucleotide substrate (3.25 nM) were individually incubated with varying hAPE1 and mAPE1 protein concentration (0–170 pM) for 7 min at 37°C for the protein dose kinetics experiment. For time kinetics experiment, we have used a fixed amount of proteins (27 pM) and different concentrations of the same substrate (1.75–3.5 nM) and incubations for up to 10 min at 37°C. The assay buffer used for both the experiments was 50 mM Tris–HCl, pH 7.5, 0.5 mM DTT, 100 μg/ml nuclease-free bovine serum albumin, 2 mM MgCl2, and 50 mM KCl in a total volume of 20 μl. Reactions were stopped by inactivating the enzyme at 70°C for 5 min. The reaction mixture was then mixed with 20 μl of loading buffer containing 1 × DNA dye (diluted from blue-orange 6 × loading dye; Promega, Madison, WI) and 85% formamide heated at 95°C for 2 min. The samples were resolved by electrophoresis at 60°C using Criterion gel (BioRad, Hercules, CA) containing 20% polyacrylamide and 7 M urea. Radioactivity in the incised oligonucleotides was quantified by exposing the gel to X-ray film and measuring the band intensities using an imager (Chemigenius Bioimaging System, Frederick, MD) with quantification software (Syngene Inc., San Diego, CA) [23].
pH, salt, and MgCl2 dependence of mouse and human APE1 activity
The APE1 proteins (27 pM) were incubated with 30 nM oligonucleotides in the presence of buffer that differed in pH (7–10), salt concentration (50–200 mM KCl), and MgCl2 concentration (0–20 mM). The reaction was stopped and the products were analyzed as described in the previous section.
Steady-state kinetic study
The enzymes (27 pM) were incubated with 5′-32P-labeled THF-containing duplex oligonucleotide (0–160 nM) substrates for 2 min at 37°C under the assay conditions similar to those described in the previous section. The reaction products were also analyzed and quantified as described for the activity assay. The steady-state parameters were calculated by non-linear regression using Michaelis-Menten equation and confirmed by double reciprocal plot.
Circular dichroism spectra analysis
Circular dichroism (CD) measurements were acquired on a J-810 spectropolarimeter (JASCO, Easton, MD), which was calibrated with and without camphorsulphonic acid before measurements. CD measurements were performed on APE1 samples with and without the addition of different amounts of Mg2+. Samples were equilibrated in 20 mM sodium phosphate (pH 7.6) in the presence of 1 mM DTT, 2 mM EDTA, and 150 mM NaCl. Far-UV spectra were recorded over 185-260 nm in a 0.1 cm cell. Measurements were performed with a bandwidth of 1 nm, response time of 0.5 s, and scan speed of 20 nm/min. Each spectrum (the average of five scans) was corrected for the appropriate solvent blank. Secondary structure composition was estimated on the basis of CD data using the CDSSTR program (available at http://public-1.cryst.bbk.ac.uk/cdweb/html/, by request) [24-27].
Results
Purification of APE1s
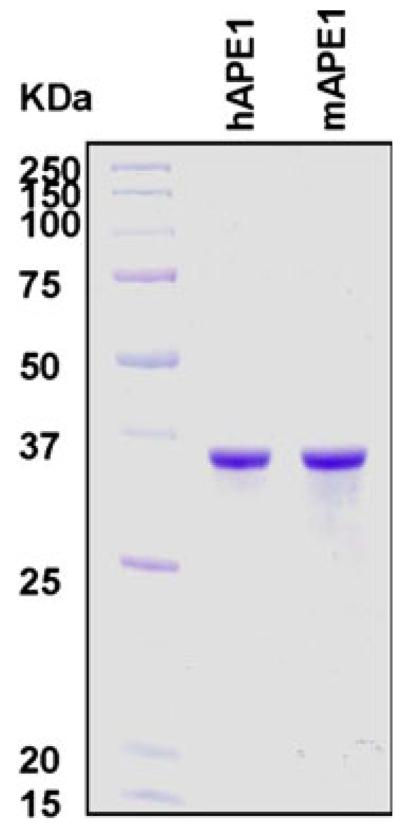
From 2 l of E. coli culture, we purified approximately 25 mg of human and mouse APE1 proteins with an apparent purity of ~99%, revealed by coomassie blue staining (Fig. 1).
Fig. 1.
Purification of apurinic/apyrimidinic endonucleases (APE1’s). APE1 proteins were purified to near homogeneity. The details of the purification are described in “Materials” section
Identification of proteins by mass spectrometry
The MALDI-TOF-TOF mass spectroscopic studies indicated that the purified single protein band on the SDS-PAGE was mouse or human APE1. At least four peptideswere identified for both APE1 sequences by MS, and the confidence score associated with the Proteins ID was 100%.
Comparison of activity of hAPE1 and mAPE1
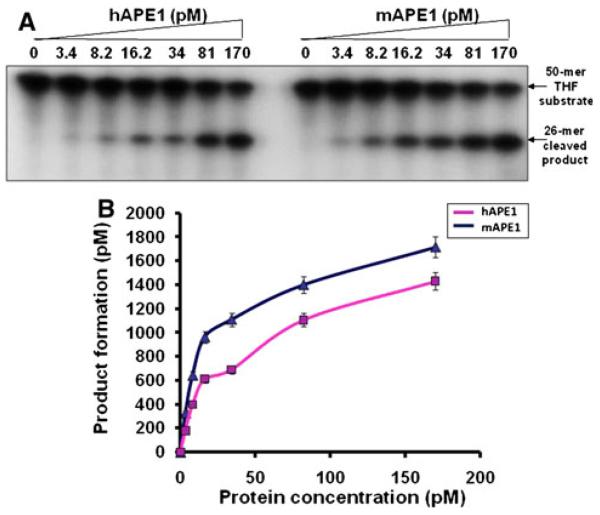
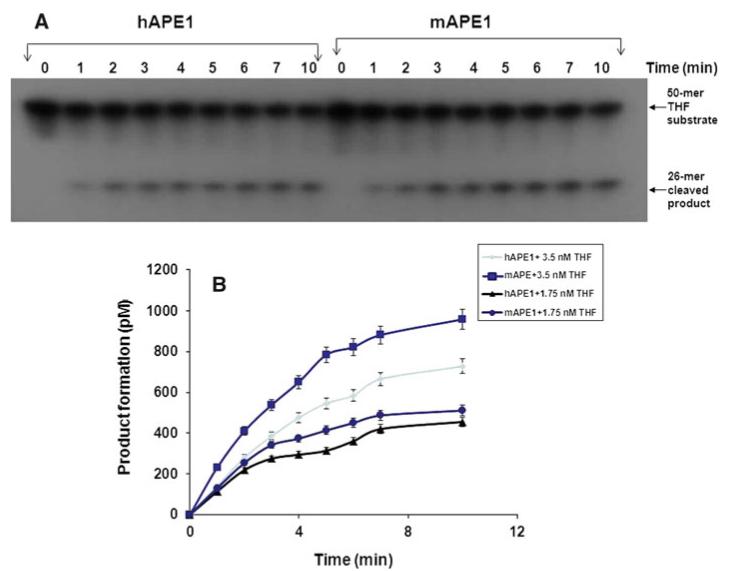
In order to compare the basic properties of human and mouse APE1s, we tested the activity of purified proteins under similar conditions (Fig. 2a, b). Both proteins showed fast kinetics, a typical signature of APE1. Also, both proteins showed a linear increase in product formation within a time period of 0–2 min (Fig. 3a, b). Of note, the results from our protein-dose dependence study (Fig. 2) showed a decent product formation at 16.2 and 34 pM concentrations. Thus, a protein concentration, 27 pM, within the 16.2 and 34 pM concentration range was chosen for further studies.
Fig. 2.
APE1-mediated THF cleavage reaction. mAPE1 or hAPE1 (0–170 pM) was reacted with a 50 mer 32P-labeled THF-containing oligonucleotide (3.25 nM) at 37°C for 7 min (a). The details of the reaction conditions are described in “Materials” section. b Data represent mean values with standard error derived from three independent experiments
Fig. 3.
Time kinetics of APE1-mediated THF cleavage reaction. mAPE1 or hAPE1 (27 pM) was reacted with a 50 mer 32P-labeled THF-containing oligonucleotide (1.75 nM) at 37°C for up to 10 min (a). The details of the reaction conditions are described in “Materials” section. b Data obtained in Panel A and a separate set of experiments using 3.5 nM substrate under similar reaction conditions as in Panel A were plotted. Data represent mean values with standard error derived from three independent experiments
Regulation of APE1s’ activity
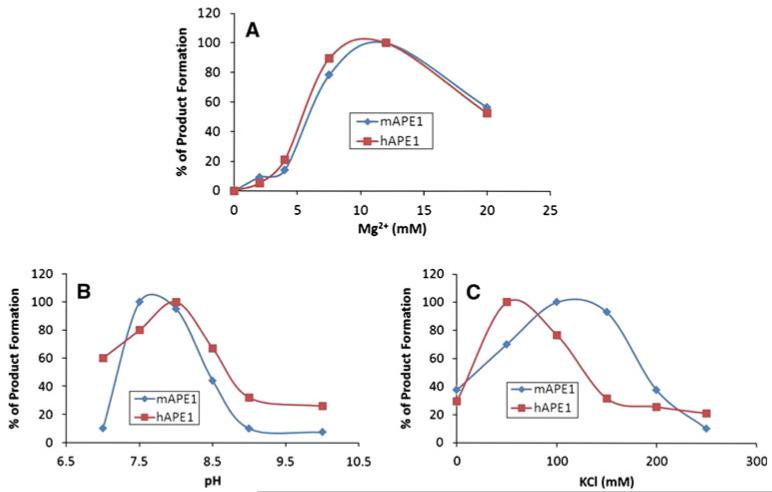
Both proteins required 12 mM MgCl2 as an optimum concentration under our reaction condition and even at 20 mM MgCl2, both APE1s retained significant portion of activity (Fig. 4a). Optimum activity was also obtained at pH 7.5–8 and 50–100 mM KCl (Fig. 4b, c). Interestingly, even 200 mM KCl abrogated the activity almost completely for both APE1s (Fig. 4c).
Fig. 4.
Comparison of basic enzymatic properties of the purified mAPE1 and hAPE1. Dependence of APE1 activity on a MgCl2 concentration, b pH and c KCl concentration. The relative activity is defined as the ratio of the THF cleavage activity at optimum pH, KCl, or MgCl2 concentrations to the similar activity at different pH and various concentrations of KCl or MgCl2. Data represent mean values with standard error derived from three independent experiments
Steady-state kinetic study
We further determined the steady-state parameters for both APE1s during the linear increase period (up to 2 min, Fig. 3) of product formation. The steady-state experiments showed an apparent Km of 91 nM and kcat of 4.2 s−1 for mAPE1 for the THF cleavage reaction. For hAPE1 Km and kcat were 82 nM and 3.2 s−1, respectively. The kcat/Km, an approximate measurement of enzyme efficiency, was similar for both species (2.3 min−1 nM−1 for hAPE1 and 2.8 min−1 nM−1 for mAPE1) under our reaction conditions, while the individual values of kcat and Km for mAPE1 were higher than for the hAPE1.
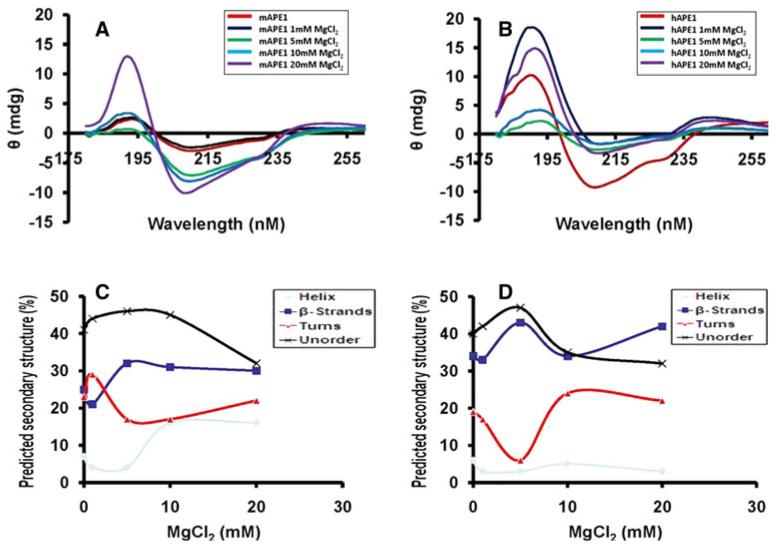
Secondary structure of mAPE1 and its modulation by Mg2+
Next, we performed circular dichroism (CD) analysis to test the effect of Mg2+ on the secondary structure of APE1. The changes in the far-UV CD spectra of native APE1 with increasing Mg2+ concentration are shown in Fig. 5a, b. Increasing Mg2+ concentrations could alter the secondary structure compositions (Fig. 5c, d). Interestingly, “turns” and “β-strands” showed “mirror image” changes in both APE1s. Also, overall CD structures seem similar for both APE1s.
Fig. 5.
Far UV-circular dichroism spectroscopy. The CD spectrums were measured in presence of different MgCl2 concentrations (0–20 mM) and 2 mM EDTA for mAPE1 (a) and hAPE1 (b). Panels C and D correspond to predicted secondary structure composition based on the CD data available from panels A and B, respectively
Overall, our study indicates that hAPE1 and mAPE1 have similar biochemical and biophysical properties. Moreover, our results suggest that it is safe to extrapolate preclinical mouse data to human biology and that the mouse data may also be used for proper design of human trials in translational studies.
Discussion
Various DNA repair mechanisms and pathways have evolved and are evolutionarily conserved from bacteria to humans for the repair of toxic and mutagenic damaged bases in DNA to prevent various diseases, including cancer (1–4). In addition, the damage produced by therapeutic agents can often be repaired by the BER proteins, which, in effect, confers therapeutic resistance. Efficient inhibition of a particular BER protein(s) may increase the efficacy of current chemotherapeutic regimens, which minimizes resistance and ultimately decreases the possibility of side effects. Therefore, pharmacological inhibition of DNA damage repair pathways has been explored as a useful strategy to enhance chemosensitivity. In the case of BER proteins, inhibition of APE1 increased sensitivity of cancer cells to alkylating chemotherapeutics. Methoxyamine is a small molecule that specifically targets BER by binding directly to AP-sites and preventing their processing by APE1, potentiating cytotoxicity of a wide range of DNA-damaging agents in both in vitro and tumor xenograft studies [11-13]. Preliminary studies also suggest that direct APE1 inhibitors for APE1’s endonuclease as well as redoxactivities potentiate the cytotoxicity of alkylating agents [13-17, 28].
To understand the biology of APE1 and its role in carcinogenesis, transgenic mice with conditional APE1 gene knock-out have been developed [18], as direct APE1 gene knock-out showed embryonic lethality [19, 20]. APE1-null mice died because of their inability to repair toxic endogenous AP-sites [18]. Moreover, APE1 inhibitors are en route from preclinical to clinical studies. Thus, it is important to know the basic characteristics of mAPE1 and how it compares to its human counterpart. In addition, understanding the mAPE1 in isolation can offer better interpretation and complementation of animal biology research.
In this study, we purified recombinant mouse and human APE1s to near homogeneity. hAPE1 is known for its very fast kinetics [29], and we also observed fast kinetics for mouse protein. In our previous study, we found that low activation energy and the enthalpy of activation, which are perhaps a result of dipole-dipole interactions, play criticalroles in THF binding of hAPE1. Typically, the binding was abrogated significantly at 200 mM KCl [30]. In this study, we observed similar salt dependence for the activity of both APE1 enzymes (Fig. 4d) indicating, a conserved reaction mechanism of APE1 in evolution. However, the salt optimum for mAPE1 and hAPE1 are slightly different. hAPE1 and mAPE1 had maximum activity at 50 and 100 mM KCl, respectively. We have determined the enzyme efficiency of both APE1s under similar conditions for comparisons.
APE1s from both species showed similar kcat/Km, an approximate measurement of enzyme efficiency, under our reaction conditions, while the kcat and Km for mAPE1 were slightly higher than for the hAPE1. Of note, there are a number of different published values found in the literature for APE1 because most likely the reaction conditions and sequence context surrounding an abasic site can affect the local structure of the DNA duplex and hence APE1’s activity. For example, even oligonucleotides of the same size (26 mer) and sequence, except for the five nucleotides, showed threefold difference in activity [31, 32]. Therefore, it may not be scientifically correct to compare our results with those in the literature. To avoid this apparent confusion, we preferred to compare two proteins under similar conditions.
MgCl2 is a typical cofactor for APE1 activity. Interestingly, both APE1’s showed a typical bell-shaped curve with an optimum activity at 12 mM Mg2+. CD analysis showed that Mg2+ affects the secondary structure of APE1 and can convert some β-strand structures to turns. Increasing the Mg2+ concentration, however, altered the ratio of “turns” to “β-strands”, and this change may be attributed to the conformational changes required to achieve the active state. Increasing Mg2+ concentration from 10 to 20 mM does not have much effect on secondary structures, indicating that Mg2+-induced changes of secondary structures of APE1’s attain its optimum within 10 mM Mg2+ (Fig. 5c, d).
APE1 inhibitors can potentiate the cytotoxic effects of different DNA-damaging drugs, including alkylating agents such as methyl methane sulfonate and temozolomide (28). The latter is used regularly in clinics for treatment of brain cancer and melanoma (28). Preliminary data provided the proof of principle that APE1 can be targeted, even if further development is required to identify a clinical lead compound. In this study we found that human and mouse enzymes are similar, and thus, it may be safe to extrapolate preclinical mouse data to human biology. Overall, mAPE1 has 94% similarity with the hAPE1 amino acid sequence, and they have similar, if not the same, biochemical and biophysical properties. This is important to note because even a change in a single amino acid which is far from catalytic pocket can have a profound effect on the activity of APE1 [21].
mAPE1 has higher Km and kcat values, albeit the overall enzyme efficiency (kcat/Km) of mAPE1 is very similar to hAPE1. Overall, this study validates the idea that human and mouse APE1’s have mostly similar biochemical and biophysical properties. Thus, these in vitro data also complement the currently available and future findings from animal studies to elucidate APE1 biology and its role in carcinogenesis, as well as in developing and validating effective APE1 inhibitors as chemosensitizers to be used with chemotherapeutic agents.
Acknowledgments
The authors thank Prof. Sankar Mitra of UTMB, Galveston, Texas for providing the mAPE1 cDNA. The authors thank Dr. Amrita Cheema for proteomics experiments performed at the Proteomics and Metabolomics Shared Resource of the Lombardi Comprehensive Cancer Center. The authors also thank Mrs. Jordan Woodrick for critically reading the paper. The study was supported by NIH grants RO1 CA 92306 (RR), RO1 CA 113447 (RR) and ACS grant ACS-IRG-92-152-17 (SA).
Abbreviations
- BER
Base excision repair
- AP
Apurinic/apyrimidinic
- mAPE1
Mouse apurinic/apyrimidinic endonuclease 1
- hAPE1
Human apurinic/apyrimidinic endonuclease 1
- THF
Tetrahydrofuran
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Clancy S. DNA damage & repair: mechanisms for maintaining DNA integrity. Nat Educ. 2008;1:1. [Google Scholar]
- 3.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 5.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/REF-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE11/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D’Ambrosio C, Scaloni A, Quadrifoglio F, Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., III Impairment of APE11 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res. 2009;7:897–906. doi: 10.1158/1541-7786.MCR-08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Gerson SL. Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs. 2004;5:623–627. [PubMed] [Google Scholar]
- 11.Taverna P, Liu L, Hwang HS, Hanson AJ, Kinsella TJ, Gerson SL. Methoxyamine potentiates DNA single strand breaks and double strand breaks induced by temozolomide in colon cancer cells. Mutat Res. 2001;485:269–281. doi: 10.1016/s0921-8777(01)00076-3. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 13.Madhusudan S, Hickson ID. DNA repair inhibition: a selective tumour targeting strategy. Trends Mol Med. 2005;11:503–511. doi: 10.1016/j.molmed.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Lou M, Kelly MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 15.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM., 3rd Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS One. 2009;4:e5740. doi: 10.1371/journal.pone.0005740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damia G, D’Incalci M. Targeting DNA repair as a promising approach in cancer therapy. Eur J Cancer. 2007;43:1791–1801. doi: 10.1016/j.ejca.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 21.Mantha AK, Oezguen N, Bhakat KK, Izumi T, Braun W, Mitra S. Unusual role of a cysteine residue in substrate binding and activity of human AP-endonuclease 1. J Mol Biol. 2008;379:28–37. doi: 10.1016/j.jmb.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adhikari S, Manthena PV, Sajwan K, Kota KK, Roy R. A unified method for purification of basic proteins. Anal Biochem. 2010;400:203–206. doi: 10.1016/j.ab.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adhikari S, Toretsky JA, Yuan L, Roy R. Magnesium, essential for base excision repair enzymes, inhibits substrate binding of N-methylpurine-DNA glycosylase. J Biol Chem. 2006;281:29525–29532. doi: 10.1074/jbc.M602673200. [DOI] [PubMed] [Google Scholar]
- 24.Compton LA, Johnson WC., Jr Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 25.Manavalan P, Johnson WC., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 26.Sreerama N, Woody RW. Estimation of protein secondary structure from CD spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 27.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adhikari S, Choudhury S, Mitra PS, Dubash JJ, Sajankila SP, Roy R. Targeting base excision repair for chemosensitization. Anticancer Agents Med Chem. 2008;8:351–357. doi: 10.2174/187152008784220366. [DOI] [PubMed] [Google Scholar]
- 29.Strauss PR, Beard WA, Patterson TA, Wilson SH. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J Biol Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 30.Adhikari S, Üren A, Roy R. Dipole-dipole interaction stabilizes the transition state of apurinic/apyrimidinic endonuclease—abasic site interaction. J Biol Chem. 2008;283:1334–1339. doi: 10.1074/jbc.M704594200. [DOI] [PubMed] [Google Scholar]
- 31.Berquist BR, McNeill DR, Wilson DM., 3rd Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson DM., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]