Abstract
Aflatoxin B 1 (AFB1) is a risk factor for hepatocellular carcinoma in humans. Infant, but not adult, mice are sensitive to AFB1-induced liver carcinogenesis; a single dose during the neonatal period leads to hepatocellular carcinoma in adulthood. Earlier work defined the mutational spectrum in the gpt gene of gpt delta B6C3F1 mice 3 weeks after exposure to aflatoxin. In the present study, we examined the gpt spectrum 10 weeks postdosing and expanded the study to examine, at 3 and 10 weeks, the spectrum at a second locus, the red/gam genes of the mouse λEG10 transgene. Whereas the gpt locus is typically used to define local base changes, the red/gam genes, via the Spi– assay, often are used to detect more global mutations such as large deletions and rearrangements. Three weeks after dosing with AFB1, there was a 10-fold increase over the control in the Spi– mutant fraction (MF) in liver DNA; after 10 weeks, a further increase was observed. The MF in the gpt gene was also increased at 10 weeks compared with the MF at 3 weeks. No gender-specific differences were found in the Spi– or gpt MFs. Whereas Spi– mutations often signal large genetic changes, they did not in this specific case. The Spi– spectrum was dominated by GC to TA transversions, with one exceptionally strong hotspot at position 314. Using two genetic loci, the data show a strong preference for the induction of GC to TA mutations in mice, which is the dominant mutation seen in people exposed to aflatoxin.
Key Words: aflatoxin B1, hepatocellular carcinoma;, mutation, infant mouse.
In mice, the ability of aflatoxin B1 (AFB1) to produce covalent adducts in liver DNA of infant and adult animals differs, respectively, by 20-fold (Shupe and Sell, 2004) with correspondingly different incidences of hepatocellular carcinoma (HCC) (Busby and Wogan, 1984). AFB1 is a potent hepatocarcinogen in infant mice, but older mice are refractory to its carcinogenic effects because of changes in AFB1 metabolism that reduce its ability to cause genotoxic damage (Dycaico et al., 1996; Hayes et al., 1991). In infant B6C3F1 mice, a single dose of AFB1 produces a high incidence of HCC in adult male mice but not in female mice (Vesselinovitch et al., 1972). This gender disparity in HCC is seen in other rodent models and also parallels human epidemiological data. In humans, there is an established relationship between exposure to AFB1, levels of AFB1-DNA adducts in urine, and the incidence of HCC (Groopman et al., 2008). Furthermore, the incidence of HCC in men is about two to four times higher than in women (El-Serag and Rudolph, 2007). Elucidating the molecular details underlying age, species, and gender differences in carcinogenesis is central to the development of effective preventive strategies.
Carcinogenesis is characterized by sequential alterations in gene expression involving genetic and epigenetic mechanisms that can be induced by endogenous and/or environmental factors. Mutations produced by genotoxicants are essential for initiation of the carcinogenic process by most chemical carcinogens. Several studies have characterized the number and types of mutations produced in animal models shortly after treatment with AFB1 and other genotoxic carcinogens (Chen et al., 2010; Woo et al., 2011). However, little is known about the numbers and types of mutations that are present in the liver over the extended period required for the development of HCC.
Integrated bacterial reporter genes in transgenic mice have enabled characterization of somatic mutations that occur spontaneously or are produced by genotoxins that can cause cancer. Using transgenic gpt delta B6C3F1 mice, we recently examined the number and types of mutations produced in the liver after treatment of newborn animals with AFB1 (Woo et al., 2011). The gpt assay detected base pair substitutions, single base insertions, and small deletions, all of which have been associated with tumor development. Three weeks after AFB1 administration, the gpt mutant fraction (MF) in the liver was increased approximately 20-fold in both males and females, indicating that shortly after exposure both sexes were at comparable risk of tumor-initiating mutations from AFB1-induced DNA damage.
The purpose of the present study was to assess the capability of AFB1 to generate other types of DNA sequence changes including those driven by nonhomologous recombination in vivo by analyzing the Spi– (Sensitive to P2 Inhibition) phenotype resulting from the disruption of red and gam genes in the λEG10 transgene of gpt delta B6C3F1 mice. We also extended our previous studies to demonstrate the persistence of base substitutions, insertions, and deletions in liver of both sexes following puberty. Point mutations were found to occur most frequently; however, AFB1 was also a potent inducer of Spi– mutants, which were molecularly characterized in this work.
MATERIALS AND METHODS
Chemicals. AFB1, DMSO and 6-thioguanine (6-TG) were obtained from Sigma Chemical Co. (St Louis, MO). All other chemicals and media were obtained from commercial suppliers and used as provided.
Animals and treatments. Female gpt delta C57BL/6J mice (Nohmi et al., 1996) were crossed with male C3H/HeJ mice to obtain an F1 that was hemizygous for the gpt transgene on chromosome 17. Four-day-old B6C3F1 animals were treated with AFB1 dissolved in DMSO administered via ip injection. Infant mice were weaned at 21 days. Animals were fed AIN93G diet ad libitum. All procedures involving animals were approved by the MIT Committee on Animal Care.
Analysis of covalent AFB 1 adducts in liver DNA. Liver DNA was isolated from AFB1-treated mice and AFB1-DNA adducts hydrolyzed using previously described procedures (Woo et al., 2011). Internal 15N5-guanine-derived standards for both AFB1-N7-guanine and AFB1-FAPY were added after hydrolysis of AFB1-DNA adducts to permit quantitative analysis by isotope dilution mass spectrometry. DNA adducts were separated by ultra-high performance liquid chromatography and injected for MS/MS analysis. The protonated parent ion of the AFB1-N7-guanine adduct (m/z 480.1) was selected and subjected to collision-induced fragmentation producing a m/z 152 product ion that was monitored to quantify adduct levels. The AFB1-FAPY adduct was quantified by selection of the m/z 498 parent ion and monitoring the collision-induced product ion m/z 452 (Egner et al., 2006).
Mutation assays. The gpt and Spi– assays were performed using genomic DNA isolated from mice injected with AFB1 or vehicle (DMSO). Genomic DNA was extracted from 25 mg liver tissue using a RecoverEase DNA Isolation Kit (Agilent Technologies, Santa Clara, CA); subsequently, λEG10 phages were packaged in vitro from the genomic DNA using Transpack Packaging Extract (Agilent Technologies) following the manufacturer’s instructions. The selection of 6-TG-resistant mutants and sequencing of the gpt gene were performed as described previously (Woo et al., 2011).
The Spi– assay was performed by infecting XL1 Blue MRA Escherichia coli with rescued phage to estimate the total number of phage and XL1 Blue MRA (P2) E. coli to enumerate phage with the Spi– phenotype. Mutants were confirmed by growth on WL95 (P2) and XL1 Blue MRA E. coli strains. The Spi– mutant fraction was calculated as the number of confirmed mutants divided by the total number of packaged phage (Nohmi et al., 1996).
Analysis of Spi – mutants. Spi– phage were used to infect E. coli LE392 and incubated until lysis. Phage DNA was isolated from the Spi– lysate and a 5 kb segment containing the red and gam genes was amplified by PCR using primers: (5′-CTCTCCTTTGATGCGAATGCCAGA-3′) and (5′-GGAGTAATTATGCGGAACAGAATCATGC-3′). The PCR products were analyzed by agarose gel electrophoresis and purified for sequencing (QIAquick PCR Purification Kit, Qiagen). Sequencing of the gam gene was performed at the Biopolymers Facility at Harvard Medical School (Boston, MA) using the primer (5′-CCACTTTTATTGGCGATGAAAAGATGTTC-3′). Sequences were aligned with the λ gam gene (NC_001416) using NCBI Nucleotide Blast.
Statistical analyses. Mutational spectra were compared using the Adams-Skopek test (Adams and Skopek, 1987; Cariello et al., 1994). The Mann-Whitney nonparametric test was used to determine statistical significance of differences in MF.
RESULTS
AFB 1 adducts in liver DNA 3 weeks after treatment. The experimental protocol used in this investigation is shown in Figure 1. Mice were injected ip with a single dose of 6 mg/kg AFB1 on day 4 after birth. AFB1-DNA adduct levels in liver were analyzed 3 weeks later as described earlier. Our previous investigation quantified adduct levels up to 48 h after AFB1 was administered (Woo et al., 2011). Here we extended the period of observation to determine whether the persistent and more mutagenic AFB1-FAPY adduct had the potential to increase the mutation frequency in liver cells over an extended period. The previous analyses showed that both the AFB1-N7-guanine adduct and the imidazole ring-opened form (AFB1-FAPY) were present 48 h after dosing (Woo et al., 2011). Three weeks after dosing, the AFB1-N7-guanine adduct was undetectable, whereas amounts of the highly mutagenic AFB1-FAPY persisted in liver DNA at a level of 1 adduct/107 nucleotides. This result suggested that additional mutations could be induced over the subsequent period of liver cell proliferation and prompted us to extend our original analyses of mutations in the gpt transgene.
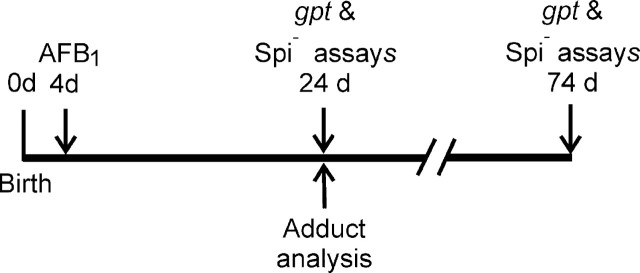
FIG. 1.
Experimental scheme for treatment of 4-day old gpt delta B6C3F1 mice with 6 mg/kg AFB1, analysis of DNA adducts and gpt and Spi– mutation assays.
The time point of 3 weeks in our previous study was chosen because by that age the liver had completed its initial phase of rapid growth during which mutations could be created through errors in repair or replication of DNA containing AFB1 adducts. In the present study, the “fixation/expression” period was extended to determine how the mutant fraction in liver changed between 3 and 10 weeks after dosing. We recorded liver weight as a measure of age-related changes in cell proliferation and growth. At the time of AFB1 administration on day 4 after birth, the liver in males and females weighed 54 ± 11 mg and 48 ± 13 mg, respectively; two days later (day 6), liver weights had increased significantly in both males (80 ± 23 mg) and females (74 ± 17 mg). At 25 days of age, which was 21 days after treatment, liver weights had increased by about 10-fold in both sexes (male 815 ± 73 mg; female 720 ± 50 mg). Thereafter, the rate of increase in liver weight slowed, nearly doubling during the subsequent 7 weeks and attaining an average weight of 1400 ± 374 mg in males and 970 ± 340 mg in females.
Mutation fraction in the gpt gene of AFB 1 -treated infant mice. Three and 10 weeks after treatment with a single dose of 6 mg/kg AFB1, λ transgenes were recovered from liver DNA by in vitro packaging. Growth of the λ phage on E. coli YG6020 expressing Cre recombinase produced chloramphenicol (CM)-resistant plasmids. Mutations in the gpt gene resulted in additional resistance to 6-TG. Tables 1 and 2 show the numbers of colonies resistant to CM and CM + 6-TG for each animal and the calculated gpt mutant fractions.
Table 1.
gpt Mutant Fractions in Liver of gpt Delta B6C3F1 Mice 3 Weeks After Treatment With DMSO or AFB1
| Treatment | Animal no. | Sex | CMR colonies (X106) | 6-TGR colonies | Mutant fraction (X10–5) | Mean ± SD (X10–6) |
|---|---|---|---|---|---|---|
| DMSO | 6 | M | 1.01 | 3 | 0.30 | 4.0 ± 2.3 |
| 9 | M | 1.41 | 9 | 0.66 | ||
| 12 | M | 1.97 | 10 | 0.51 | ||
| 15 | M | 1.47 | 2 | 0.14 | ||
| 7 | F | 1.94 | 10 | 0.52 | 7.7 ± 4.3 | |
| 8 | F | 0.31 | 3 | 1.10 | ||
| 10 | F | 1.50 | 17 | 1.17 | ||
| 20 | F | 1.15 | 3 | 0.29 | ||
| AFB1 | 26 | M | 1.27 | 177 | 13.4 | 136 ± 2 |
| 30 | M | 1.44 | 171 | 13.4 | ||
| 23 | M | 1.80 | 207 | 11.5 | ||
| 24 | M | 1.63 | 262 | 16.2 | ||
| 22 | F | 1.34 | 217 | 16.2 | 153 ± 3 | |
| 25 | F | 1.39 | 173 | 12.8 | ||
| 32 | F | 1.29 | 233 | 18.5 | ||
| 35 | F | 1.23 | 163 | 13.6 | ||
| Note. CMR = chloramphenicol resistant; 6-TGR = 6-thioguanine resistant. | ||||||
Table 2.
gpt Mutant Fractions in Liver of gpt Delta B6C3F1 Mice 10 Weeks After Treatment With DMSO or AFB1
| Treatment | Animal no. | Sex | CMR colonies (X106) | 6-TGR colonies | Mutant fraction (X10–5) | Mean ± SD (X10–6) |
|---|---|---|---|---|---|---|
| DMSO | 110 | M | 1.90 | 21 | 1.11 | 12.5 ± 2.3 |
| 111 | M | 1.07 | 12 | 1.12 | ||
| 112 | M | 1.32 | 20 | 1.51 | ||
| 113 | F | 1.18 | 17 | 1.44 | 10.0 ± 4.7 | |
| 114 | F | 3.18 | 16 | 0.50 | ||
| 115 | F | 1.40 | 17 | 1.22 | ||
| AFB1 | 103 | M | 0.71 | 121 | 17.0 | 183 ± 19 |
| 105 | M | 1.51 | 286 | 19.0 | ||
| 131 | M | 1.53 | 315 | 20.5 | ||
| 132 | M | 0.49 | 78 | 15.8 | ||
| 133 | M | 1.07 | 206 | 19.3 | ||
| 101 | F | 0.87 | 109 | 12.5 | 191 ± 40 | |
| 102 | F | 1.97 | 433 | 22.0 | ||
| 120 | F | 2.39 | 443 | 18.5 | ||
| 122 | F | 1.26 | 260 | 20.6 | ||
| 130 | F | 1.03 | 227 | 22.1 | ||
| Note. CMR = chloramphenicol resistant; 6-TGR = 6-thioguanine resistant. | ||||||
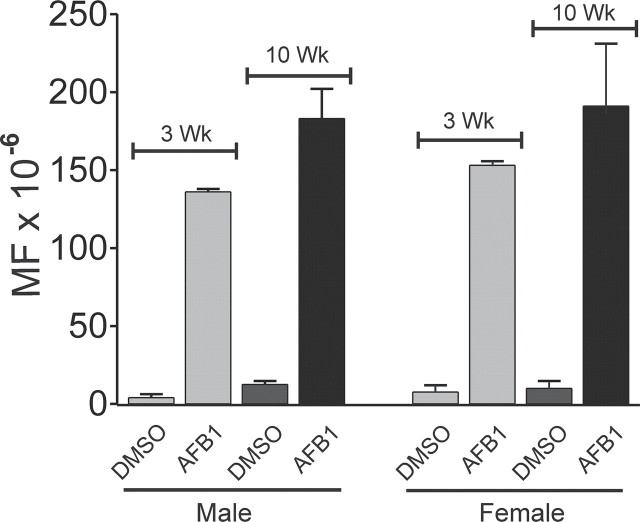
Tables 1 and 2 (also Fig. 2) depict that the average gpt MFs in the liver in AFB1-treated mice was greater at 10 weeks than at 3 weeks, with the increase in males (p = 0.02) during this 7-week period being greater than that found in females (p = 0.07). In DMSO-treated controls, the gpt MFs also increased, but the differences did not reach statistical significance (males p = 0.06; females p = 0.4); at 10 weeks, the gpt MF in the livers of males and females was the same (p = 0.55).
FIG. 2.
gpt MF in gpt delta B6C3F1 mouse liver 3 weeks or 10 weeks after treatment with 6 mg/kg AFB1 or vehicle (DMSO) at day 4 after birth.
Sequencing of the 456 bp gpt gene in 157 6-TG-resistant colonies revealed similar types and positions of mutations at 3 weeks and 10 weeks. Table 3 summarizes the types of mutations found in the gpt gene in livers of 17 AFB1-treated mice (males and females) 10 weeks after treatment. As we previously observed, 3 weeks after AFB1 treatment, the G:C to T:A transversion was the most frequent type of mutation present (55%) (Woo et al., 2011). Figure 3 shows the distribution of base pair changes, insertions, and small deletions in the gpt gene at 10 weeks. Monte Carlo-based analysis revealed no differences between the gpt mutational spectra at 3 or 10 weeks after treatment (p > 0.05) (Adams and Skopek, 1987).
FIG. 3.
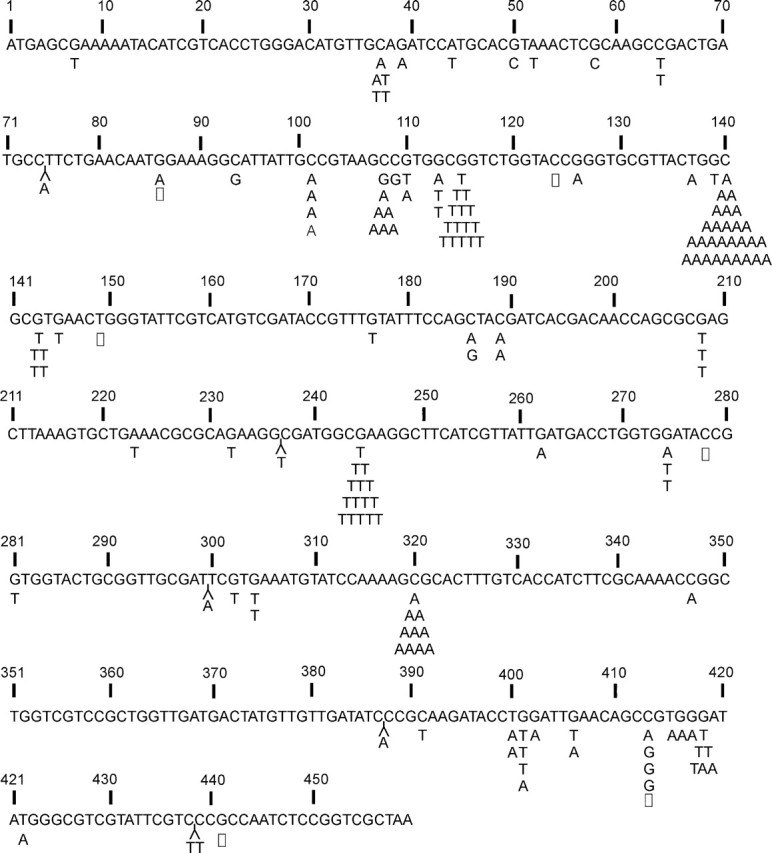
Distribution of mutations in the gpt gene in liver of gpt delta B6C3F1 mice 10 weeks after treatment with 6 mg/kg AFB1. Base substitutions are indicated by symbols Y = insertion; □ = deletion.
Table 3.
Summary of Mutation Types in the gpt Gene of Male and Female gpt Delta B6C3F1 Mice 10 Weeks After Treatment With a Single 6 mg/kg Dose of AFB1
| Mutation types | AFB1 | |
|---|---|---|
| Male | Female | |
| Transition: | ||
| G:C to A:T | 11 (13) | 9 (12) |
| A:T to G:C | 0 | 0 |
| Transversion: | ||
| G:C to T:A | 58 (71) | 50 (67) |
| G:C to C:G | 6 (7) | 4 (5) |
| A:T to T:A | 3 (4) | 5 (7) |
| A:T to C:G | 0 | 0 |
| Deletion: | ||
| 1 bp | 2 (2) | 4 (5) |
| > 1 bp | 0 | 0 |
| Insertion | 2 (2) | 3 (4) |
| Total number of mutants | 55 (100) | 75 (100) |
Note. The percentage of each type of mutation is shown in parentheses.
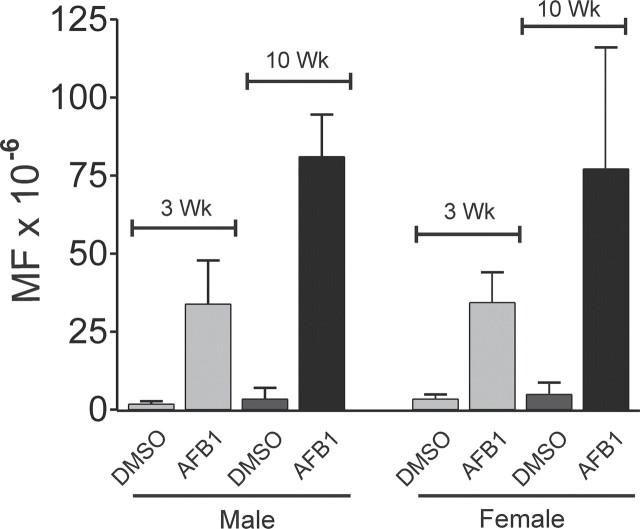
AFB 1 -Induced Spi – Mutations in Liver . The Spi– assay is sensitive to mutations in the red and gam genes of the λEG10 transgene (Masumura et al., 2000). Figure 4, along with the data in Tables 4 and 5, illustrates that at 3 and 10 weeks after AFB1 treatment there was a 10- to 20-fold increase in the Spi– MF in liver DNA compared with vehicle-treated control mice. Equal fractions of Spi– mutations were found in male and female mice at 3 weeks. At 10 weeks, the Spi– MF in AFB1-treated mice was increased significantly in both males (p = 0.004) and females (p = 0.02) compared with the 3-week MF.
FIG. 4.
Spi– MF in gpt delta B6C3F1 mouse liver 3 weeks or 10 weeks after treatment with 6 mg/kg AFB1 or vehicle (DMSO) at day 4 after birth.
Table 4.
Spi Mutant Fractions in Liver of gpt Delta B6C3F1 Mice 3 Weeks After Treatment With DMSO or AFB1
| Treatment | Animal no. | Sex | Plaques (X106) | Plaques (P2) (Spi–) | Mutant fraction (X10–5) | Mean ± SD (X10–6) |
|---|---|---|---|---|---|---|
| DMSO | 6 | M | 0.78 | 1 | 0.13 | 1.8 ± 0.9 |
| 9 | M | 0.75 | 2 | 0.28 | ||
| 12 | M | 0.88 | 2 | 0.24 | ||
| 15 | M | 1.09 | 1 | 0.09 | ||
| 7 | F | 1.03 | 2 | 0.19 | 3.4 ± 1.5 | |
| 8 | F | 1.15 | 4 | 0.35 | ||
| 10 | F | 1.10 | 6 | 0.55 | ||
| 20 | F | 1.12 | 3 | 0.27 | ||
| AFB1 | 23 | M | 1.03 | 30 | 2.96 | 33.8 ± 14 |
| 34 | M | 0.32 | 12 | 3.77 | ||
| 26 | M | 0.61 | 31 | 5.04 | ||
| 30 | M | 0.46 | 8 | 1.75 | ||
| 22 | F | 1.57 | 52 | 3.38 | 34.3 ± 9.7 | |
| 25 | F | 1.34 | 54 | 4.00 | ||
| 32 | F | 1.01 | 16 | 2.09 | ||
| 35 | F | 1.54 | 65 | 4.25 |
Table 5.
Spi Mutant Fractions in Liver of gpt Delta B6C3F1 Mice 10 Weeks After Treatment With DMSO or AFB1
| Treatment | Animal no. | Sex | Plaques (X106) | Plaques (P2) (Spi–) | Mutant fraction (X10–5) | Mean ± SD (X10–6) |
|---|---|---|---|---|---|---|
| DMSO | 110 | M | 2.26 | 2 | 0.09 | 3.4 ± 3.6 |
| 111 | M | 2.14 | 4 | 0.19 | ||
| 112 | M | 0.53 | 4 | 0.75 | ||
| 113 | F | 2.11 | 12 | 0.57 | 4.9 ± 3.8 | |
| 114 | F | 2.40 | 2 | 0.08 | ||
| 115 | F | 1.91 | 16 | 0.84 | ||
| AFB1 | 103 | M | 0.69 | 58 | 8.37 | 81.0 ± 13.5 |
| 105 | M | 0.57 | 59 | 10.30 | ||
| 131 | M | 1.45 | 112 | 7.71 | ||
| 132 | M | 1.02 | 71 | 6.94 | ||
| 133 | M | 0.82 | 59 | 7.18 | ||
| 101 | F | 0.97 | 42 | 4.35 | 77.1 ± 38.9 | |
| 102 | F | 0.62 | 22 | 3.58 | ||
| 120 | F | 3.14 | 160 | 5.09 | ||
| 122 | F | 0.98 | 72 | 7.32 | ||
| 130 | F | 0.72 | 95 | 13.2 |
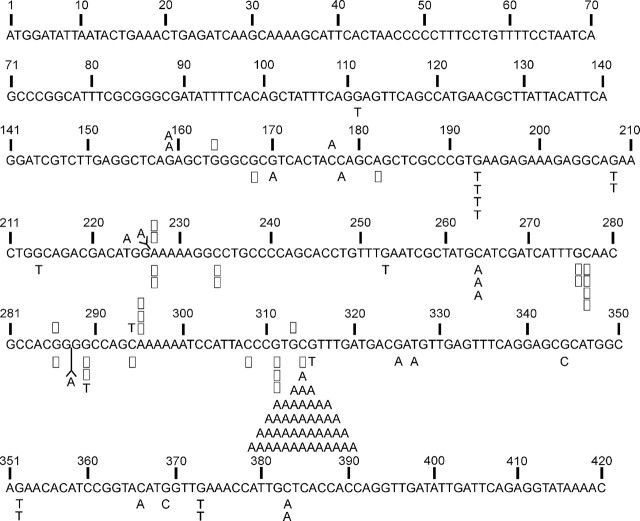
Spi– mutants from 10-week mice were first characterized by PCR amplification of a 5 kb segment of the λEG10 transgene containing both the gam and red genes. All of the mutants produced a full-length 5 kb PCR product indicating that there were no large deletions or other types of rearrangements in the gam and red gene coding sequences. Mutations in the gam gene alone can show the Spi– phenotype (Singer, 1971). Therefore, we decided to perform sequence analysis of gam to identify any base substitutions or small insertions/deletions characteristic of those produced by AFB1. Figure 5 shows the sequence changes that were identified in the gam gene of Spi– phage isolated from AFB1-treated mice and DMSO-treated controls. Ninety-six Spi– λEG10 phage were sequenced from 10 AFB1-treated mice. Only 14 mutants were available for sequence analysis from the 6 control mice treated with DMSO. Base pair substitutions were once again the predominant type of mutation, representing 88% of total mutations in AFB1-treated mice. There were no significant gender differences either in the type of mutation in the gam gene (Table 6). The C:G to A:T transversion mutation at nucleotide 314 is a hot spot that was found in all 10 AFB1-treated mice analyzed at 10 weeks, representing approximately 50% of the sequenced mutants. The most plausible explanation for this mutation is the formation of an AFB1-Gua adduct on the G-containing strand. Frameshifts resulting from single base pair deletions or insertions represented the next most abundant class of mutations (22% of total). Single base pair deletions predominated in limited number of mutants available for analysis in the 10-week control group.
FIG. 5.
Distribution of mutations in the gam gene in liver of gpt delta B6C3F1 mice 10 weeks after treatment with 6 mg/kg AFB1. Base substitutions are indicated by symbols Y = insertion; □ = deletion. Mutations in DMSO-treated mice are shown above the sequence; AFB1-induced mutations are shown below.
Table 6.
Summary of Mutation Types in the gam Gene of Male and Female gpt Delta B6C3F1 Mice 10 Weeks After Treatment With a Single 6 mg/kg Dose of AFB1
| Mutation types | AFB1 | DMSOa | |
|---|---|---|---|
| Male | Female | ||
| Transition: | |||
| G:C to A:T | 1 (3) | 1 (2) | 3 |
| A:T to G:C | 0 (0) | 0 (0) | 0 |
| Transversion: | |||
| G:C to T:A | 27 (67) | 40 (71) | 1 |
| G:C to C:G | 0 (0) | 2 (5) | 0 |
| A:T to T:A | 0 (0) | 1 (2) | 1 |
| A:T to C:G | 0 (0) | 0 (0) | 0 |
| Deletion | |||
| 1 bp | 11 (27) | 10 (18) | 8 |
| > 1 bp | 0 (0) | 0 (0) | 0 |
| Insertion | 1 (3) | 1 (2) | 1 |
| Total number of mutants | 40 (100) | 56 (100) | 14 |
Note. The percentage of each type of mutation is shown in parentheses.
aBecause few mutants were available for analysis, data from male and female mice were combined.
DISCUSSION
Cancer development involves the acquisition of proliferative and invasive phenotypes through the accumulation of genetic variation spontaneously or through exposures to environmental carcinogens. The multiple genetic changes associated with carcinogenesis include point mutations, chromosomal rearrangements, and changes in gene copy number (Futreal et al., 2004). AFB1, one of the most potent known liver carcinogens in humans and animal models, has been shown to be a potent mutagen in bacterial and mammalian cells. Previous work using bacterial reporter genes in transgenic mice established that neonatal exposure to AFB1 leads to a 20-fold increase in the frequency of point mutations in the gpt gene in the liver (Chen et al., 2010; Woo et al., 2011). In the present investigation, we studied the ability of AFB1 to induce mutations in the gam and red genes of a λEG10 reporter in mouse liver. Mutations in these genes produce the Spi– phenotype that can be identified by the ability to grow on P2 lysogenic E. coli strains. This assay is sensitive to large deletions and rearrangements in the gam and red genes as well as small deletions, insertions, and base pair substitutions that disrupt transcription and translation of these genes.
In conjunction with the results of our previous study, the findings in the present study establish the ability of AFB1 to induce a broad range of mutation types in the liver DNA, including base pair substitutions and frameshifts resulting from small deletions and insertions. Earlier analyses showed a 13- to 15-fold increase in the MF of the gpt transgene at 3 weeks after treatment of 4-day-old mice (Woo et al., 2011); our present findings show that the MF continued to increase up to 20-fold over DMSO-treated controls at 10 weeks thereafter. These results are in agreement with another study that found a 22-fold increase in base substitution and frameshift mutations in the cII transgene of neonatal Big Blue B6C3F1 mice 6 weeks after treatment with AFB1 (Chen et al., 2010). The slower increase in MF in the liver from 3 to 10 weeks was very likely due to the decrease in growth rate of the liver after 3 weeks. Liver weight increased approximately 10-fold during the initial 3-week period, and subsequently doubled in size over the next 7 weeks. Cell proliferation provides opportunities for fixation of mutations at damaged sites in DNA through replication errors. Reynolds et al. (2004) reported a 27-fold difference in the proliferation index between 4-week and 12-week-old B6C3F1 mice. Thus, during the initial 3-week period, a greater number of AFB1-DNA adducts are present along with greater opportunity for them to cause mutations when cells are in S-phase during this period of rapid liver growth.
The strong mutagenic signal in the Spi– assay from the liver of AFB1-treated animals is particularly noteworthy. The sequence characteristics of deletions found in gpt delta mice after treatment with various agents include –1 bp frameshifts and larger deletions (> 10 kb) containing junction sequences of limited homology that could result from end joining of double strand breaks (Horiguchi et al., 2001; Masumura et al., 2000; Nohmi et al., 1999). The molecular characteristics of the Spi– mutants from the liver of AFB1-treated mice differ from those found in the aforementioned studies. In the present work on aflatoxin, there were some small deletions and frameshifts observed, but point mutations were more common.
A significant number of base substitutions resulted in the appearance of a stop codon in the gam gene, which presumably resulted in a halt in translation in both the gam and the contiguous redAB genes. Approximately 50% of all base substitution mutations (44 out of 98 sequenced mutations) were found at a single site: the C:G to A:T transversion that occurred at base pair 314. This mutation was not seen among the 15 Spi– mutants analyzed from DMSO-treated animals, which would indicate that it is a hot spot for AFB1-induced mutations. This transversion mutation in the gam gene results in the substitution of glutamate for an alanine residue that is located in one of the two small helices of the gam protein that functions to anchor larger helical regions for proper binding to the RecBCD complex (Court et al., 2007). The charged glutamate residue may prevent this interaction. Inhibition of the RecBC complex by gam is required for initiation of the rolling circle mode of lambda replication that generates linear concatemers of DNA that can be packaged into infectious phage (Furth and Wickner, 1983). Gam– phage are confined to the closed-circular form of replication, producing single circular genomes that are not substrates for packaging and consequently are unable to produce plaques. Recombination between circles, however, produces dimers that can be packaged to form infectious progeny. The λEG10 genome contains several Chi sequences that promote recombination by RecBC, facilitating the concatenation of circular genomes and formation of infections particles by gam– phage (Smith, 1983). This could partly explain our observation of the Spi– phenotype resulting from the transversion at base pair 314. However, a full explanation will require a better understanding of the basis for Spi– selection on P2 lysogenic strains. The strong mutagenicity of AFB1 at base pair 314 may result from its preferential reaction with AFB1-8,9-oxide, lack of repair, or susceptibility to replication errors. It is also possible, however, that unexpected variables in the conditions used for selection of the Spi– phenotype are responsible for the apparent prevalence of this mutant in AFB1-treated animals.
Aflatoxin DNA adducts that could be responsible for these mutations include AFB1-N7-Gua and the multiple AFB1-FAPY adducts. These adducts block replication in in vitro assays (Johnston and Stone, 2000; Levy et al., 1992; Refolo et al., 1985). AFB1-FAPY exists in two rotameric forms (major and minor), which differ substantially with respect to lethal and mutagenic potencies in a bacterial reporter system. The major adduct is a particularly strong block for replication by both normal and bypass DNA polymerases, whereas the minor adduct is most potent at causing base substitutions (Smela et al., 2002), specifically G to T transversions. The presence of AFB1-FAPY in liver DNA 3 weeks after dosing indicates that this highly persistent adduct has multiple opportunities to cause genetic alterations over an extended period of time during hepatocarcinogenesis.
The increase in the Spi– MF (10-fold) was a little less than half that of the gpt MF (20-fold) measured at 10 weeks of age (see Figs. 2 and 4). It is difficult to compare mutational potencies in different loci, especially in loci that encode such disparate functions as those of the gpt and gam genes. Nevertheless, the data from these two loci support the view that aflatoxin is a good point mutagen. If it induced frequent more global genetic changes, we would have expected to have seen such mutagenic events among the sequenced Spi– mutants. In this respect, the types of Spi– mutants induced by AFB1 were similar to those reported for 2-amino-1-methyl-6-phenylinidazo[4,5-b]pyrine (PhIP) and aminophenylnorharman, which were dominated by small deletions and single base substitutions (Masumura et al. 1999, 2003). The absence of large deletions and rearrangements in the gam and red genes of Spi– phage in our experiments is in contrast to the reported ability of AFB1 to cause these types of recombinant-driven changes in yeast (Sengstag et al., 1996) and lymphoid cells in culture (Preisler et al., 2000; Stettler and Sengstag, 2001) as well as reports of its ability to cause micronuclei and sister chromatid exchanges (Busby and Wogan, 1984). Further studies will be required to clarify these apparent discrepancies to understand the role of clastogenic activities of AFB1 in HCC.
The infant mouse model provides a unique environment in which to investigate the extended process of carcinogenesis following a single exposure to a carcinogen such as AFB1. A significant finding in this study is the absence of gender differences in gpt or Spi– MFs in liver DNA up to 10 weeks after treatment. On this basis, it would appear that the potential for initiation of HCC development exists in animals of both sexes; eventually, however, female mice develop a much lower incidence of HCC than do males (Vesselinovitch et al., 1972). If formation of mutations and adducts are central to carcinogenesis, this study helps to identify periods later than 10 weeks as the critical periods in defining the sex differences in carcinogenic outcome. In rodent models of HCC, development of premalignant lesions and their rate of progression to malignancy can be modulated by sex hormones and tumor-promoting cytokines (Maeda et al., 2005). Cytokine expression by injured hepatocytes results in the recruitment of inflammatory cells that aid in the process of tumor development (He et al., 2010). The discovery that cytokine expression can be suppressed by estrogenic hormones provides one possible mechanism responsible for the gender difference in incidence of chemically induced HCC in rodents (Naugler et al., 2007). Despite a wealth of information, it is still unclear how the inflammatory process promotes the genetic and/or epigenetic changes required to complete the carcinogenic process. One possibility is that chronic exposure to reactive oxygen species (ROS) or other reactive products of inflammatory cells promotes further mutagenesis and genomic instability in initiated hepatocytes. Further evaluation of the genetic changes that occur in cells as they progress to malignancy will help to clarify the biochemical mechanisms responsible for tumor progression and identify targets for preventive strategies.
Funding
National Institutes of Health (R01 ES016313, P30 ES002109, P01 ES006052, P30 ES003819); The Center of Excellence on Environmental Health, Toxicology and Management of Chemicals, Thailand (R.W.).
References
- Adams W. T., Skopek T. R. (1987). Statistical test for the comparison of samples from mutational spectra J. Mol. Biol. 194 391–396 [DOI] [PubMed] [Google Scholar]
- Busby W. F., , Jr., Wogan G. N. (1984). Aflatoxins. In Chemical Carcinogens, 2nd ed. (Searle C. E. , Ed.), pp. 945–1136 American Chemical Society; Washington, DC. [Google Scholar]
- Cariello N. F., Piegorsch W. W., Adams W. T., Skopek T. R. (1994). Computer program for the analysis of mutational spectra: Application to p53 mutations Carcinogenesis 15 2281–2285 [DOI] [PubMed] [Google Scholar]
- Chen T., Heflich R. H., Moore M. M., Mei N. (2010). Differential mutagenicity of aflatoxin B1 in the liver of neonatal and adult mice Environ. Mol. Mutagen. 51 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court R., Cook N., Saikrishnan K., Wigley D. (2007). The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition J. Mol. Biol. 371 25–33 [DOI] [PubMed] [Google Scholar]
- Dycaico M. J., Stuart G. R., Tobal G. M., de Boer J. G., Glickman B. W., Provost G. S. (1996). Species-specific differences in hepatic mutant frequency and mutational spectrum among lambda/lacI transgenic rats and mice following exposure to aflatoxin B1 Carcinogenesis 17 2347–2356 [DOI] [PubMed] [Google Scholar]
- Egner P. A., Groopman J. D., Wang J. S., Kensler T. W., Friesen M. D. (2006). Quantification of aflatoxin-B1-N7-Guanine in human urine by high-performance liquid chromatography and isotope dilution tandem mass spectrometry Chem. Res. Toxicol. 19 1191–1195 [DOI] [PubMed] [Google Scholar]
- El-Serag H. B., Rudolph K. L. (2007). Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis Gastroenterology 132 2557–2576 [DOI] [PubMed] [Google Scholar]
- Furth M. E., Wickner S. H. (1983). Lambda DNA replication. In Lambda II (Hendrix R. W. , Roberts J. W. , Stahl F. W. , Weisberg R. A. , Eds.), pp 145–173 Cold Spring Harbor Press; Cold Spring Harbor, NY. [Google Scholar]
- Futreal P. A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M. R. (2004). A census of human cancer genes Nat. Rev. Cancer 4 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groopman J.D., Kensler T.W., Wild C.P. (2008). Protective Interventions to prevent aflatoxin-induced carcinogenesis in developing countries Annu. Rev. Public Health 29 187–203 [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Judah D. J., McLellan L. I., Neal G. E. (1991). Contribution of the glutathione S-transferases to the mechanisms of resistance to aflatoxin B1 Pharmacol. Ther. 50 443–472 [DOI] [PubMed] [Google Scholar]
- He G., Yu G. Y., Temkin V., Ogata H., Kuntzen C., Sakurai T., Sieghart W., Peck-Radosavljevic M., Leffert H. L., Karin M. (2010). Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation Cancer Cell 17 286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi M., Masumura K. I., Ikehata H., Ono T., Kanke Y., Nohmi T. (2001). Molecular nature of ultraviolet B light-induced deletions in the murine epidermis Cancer Res. 61 3913–3918 [PubMed] [Google Scholar]
- Johnston D. S., Stone M. P. (2000). Replication of a site-specific trans-8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B(1) adduct by the exonuclease deficient Klenow fragment of DNA polymerase I Chem. Res. Toxicol. 13 1158–1164 [DOI] [PubMed] [Google Scholar]
- Levy D. D., Groopman J. D., Lim S. E., Seidman M. M., Kraemer K. H. (1992). Sequence specificity of aflatoxin B1-induced mutations in a plasmid replicated in xeroderma pigmentosum and DNA repair proficient human cells Cancer Res. 52 5668–5673 [PubMed] [Google Scholar]
- Maeda S., Kamata H., Luo J. L., Leffert H., Karin M. (2005). IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis Cell 121 977–990 [DOI] [PubMed] [Google Scholar]
- Masumura K., Matsui K., Yamada M., Horiguchi M., Ishida K., Watanabe M., Ueda O., Suzuki H., Kanke Y., Tindall K. R., Wakabayashi K., Sofuni T., Nohmi T. (1999). Mutagenicity of 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine (PhIP) in the new gpt delta transgenic mouse Cancer Lett. 143 241–244 [DOI] [PubMed] [Google Scholar]
- Masumura K., Matsui K., Yamada M., Horiguchi M., Ishida K., Watanabe M., Wakabayashi K., Nohmi T. (2000). Characterization of mutations induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in the colon of gpt delta transgenic mouse: Novel G:C deletions beside runs of identical bases Carcinogenesis 21 2049–2056 [DOI] [PubMed] [Google Scholar]
- Masumura K., Totsuka Y., Wakabayashi K., Nohmi T. (2003). Potent genotoxicity of aminophenylnorharman, formed from non-mutagenic norharman and aniline, in the liver of gpt delta transgenic mouse Carcinogenesis 24 1985–1993 [DOI] [PubMed] [Google Scholar]
- Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., Karin M. (2007). Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production Science 317 121–124 [DOI] [PubMed] [Google Scholar]
- Nohmi T., Katoh M., Suzuki H., Matsui M., Yamada M., Watanabe M., Suzuki M., Horiya N., Ueda O., Shibuya T., Ikeda H., Sofuni T. (1996). A new transgenic mouse mutagenesis test system using Spi- and 6-thioguanine selections Environ. Mol. Mutagen. 28 465–470 [DOI] [PubMed] [Google Scholar]
- Nohmi T., Suzuki M., Masumura K., Yamada M., Matsui K., Ueda O., Suzuki H., Katoh M., Ikeda H., Sofuni T. (1999). Spi(-) selection: An efficient method to detect gamma-ray-induced deletions in transgenic mice Environ. Mol. Mutagen. 34 9–15 [DOI] [PubMed] [Google Scholar]
- Preisler V., Caspary W. J., Hoppe F., Hagen R., Stopper H. (2000). Aflatoxin B1-induced mitotic recombination in L5178Y mouse lymphoma cells Mutagenesis 15 91–97 [DOI] [PubMed] [Google Scholar]
- Refolo L. M., Conley M. P., Sambamurti K., Jacobsen J. S., Humayun M. Z. (1985). Sequence context effects in DNA replication blocks induced by aflatoxin B1 Proc. Natl. Acad. Sci. U.S.A. 82 3096–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R., Witherspoon S., Fox T. (2004). The infant mouse as an in vivo model for the detection and study of DNA damage-induced changes in the liver Mol. Carcinog. 40 62–72 [DOI] [PubMed] [Google Scholar]
- Sengstag C., Weibel B., Fasullo M. (1996). Genotoxicity of aflatoxin B1: Evidence for a recombination-mediated mechanism in Saccharomyces cerevisiae Cancer Res. 56 5457–5465 [PMC free article] [PubMed] [Google Scholar]
- Shupe T., Sell S. (2004). Low hepatic glutathione S-transferase and increased hepatic DNA adduction contribute to increased tumorigenicity of aflatoxin B1 in newborn and partially hepatectomized mice Toxicol. Lett. 148 1–9 [DOI] [PubMed] [Google Scholar]
- Singer E. (1971). General recombination. In The Bacteriophage Lambda (Hershey A. D. , Ed.), pp. 139–174 Cold Spring Harbor Laboratory; Cold Spring Harbor, NY. [Google Scholar]
- Smela M. E., Hamm M. L., Henderson P. T., Harris C. M., Harris T. M., Essigmann J. M. (2002). The aflatoxin B(1) formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma Proc. Natl. Acad. Sci. U.S.A. 99 6655–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. (1983). General Recombination (Hendrix R. W. , Roberts J. W. , Stahl F. W. , Weisberg R.A. , Eds.), pp. 175–209 Cold Spring Harbor Press; Cold Spring Harbor, NY. [Google Scholar]
- Stettler P. M., Sengstag C. (2001). Liver carcinogen aflatoxin B1 as an inducer of mitotic recombination in a human cell line Mol. Carcinog. 31 125–138 [DOI] [PubMed] [Google Scholar]
- Vesselinovitch S. D., Mihailovich N., Wogan G. N., Lombard L. S., Rao K. V. (1972). Aflatoxin B1, a hepatocarcinogen in the infant mouse Cancer Res. 32 2289–2291 [PubMed] [Google Scholar]
- Woo L. L., Egner P. A., Belanger C. L., Wattanawaraporn R., Trudel L. J., Croy R. G., Groopman J. D., Essigmann J. M., Wogan G. N. (2011). Aflatoxin B1-DNA adduct formation and mutagenicity in livers of neonatal male and female B6C3F1 mice Toxicol. Sci. 122 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]