Abstract
In this study, we evaluated Cu(L1) in two xenografted tumor-bearing (U87MG and MDA-MB-435) animal models to prove the concept that Cu(II)-labeled rhodamine derivatives, Cu(L) (L = L1 - L4) are useful as selective fluorescent probes for tumor imaging. We found that both multidrug resistance (MDR) negative U87MG gliomas and MDR-positive MDA-MB-435 breast tumors could be visualized. Because of tissue attenuation, accurate quantification of tumor uptake was difficult by optical methods. Therefore, 64Cu(L) (L = L1 - L4) were evaluated to compare their biodistribution properties. It was found that all four 64Cu radiotracers had a high glioma uptake (64Cu(L1): 5.71± 1.43 %ID/g; 64Cu(L2): 5.98 ± 2.75 %ID/g; 64Cu(L3): 4.28 ± 1.45 %ID/g; and 64Cu(L4): 6.25 ± 3.42 %ID/g) with 64Cu(L1) showing the highest tumor/background ratios. In athymic nude mice bearing MDA-MB-435 breast cancer xenografts, 64Cu(L4) showed almost identical normal organ uptake to that in the glioma-bearing animals, but its breast tumor uptake (1.26 ± 0.10% ID/g) was significantly lower (p < 0.001) than that in the glioma (6.25 ± 3.42% ID/g) because of MDR Pgps (P-glycoproteins) and MRPs (multidrug resistance-associated proteins) overexpressed in the xenografted MDA-MB-435 breast tumors. Results from cellular staining assays showed that both Cu(L2) and Cu(L4) were able to localize in mitochondria of U87MG cells, and their tumor selectivity was caused by the elevated negative mitochondrial potential in U87MG glioma cells as compared to that in human fibroblast cells. On the basis of these results, it was concluded that Cu(L) (L = L1 - L4) are useful as selective fluorescent probes for cellular staining assays and optical tumor imaging while 64Cu(L) (L = L1 - L4) have the potential as PET radiotracers for tumor imaging. This study represents a good example of dual modality imaging (PET and optical) using two agents, 64Cu(L) and Cu(L), with identical chemical composition. Future research will focus on developing new fluorescent probes with longer wavelength and reduced liver uptake.
Keywords: Cu-labeled rhodamine derivatives, fluorescent probes, and optical imaging
INTRODUCTION
Cancer is the second leading cause of death worldwide 1. Alteration in mitochondrial potential (ΔΨm) is an important characteristic of cancer, and is caused by mitochondrial dysfunction, such as DNA mutation and oxidative stress 2-5. It was reported that the mitochondrial potential in carcinoma cells is significantly higher than that in normal epithelial cells 6-10. For example, the difference in ΔΨm between the colon carcinoma cell line CX-1 and the green monkey kidney epithelial cell line CV-1 was approximately 60 mV (-163 mV in tumor cells versus -104 mV in normal cells) 4, 5. The observation that enhanced mitochondrial potential is prevalent in tumor cell phenotype provides the conceptual basis to develop mitochondrion-targeted therapeutics and molecular imaging probes 11-16.
Rhodamines have been used as fluorescent probes for determination of plasma and mitochondrial potentials 6-9, 17-12, and for tumor imaging 23-29. Recently, we reported 64Cu(L1) (Figure 1) as a new radiotracer for tumor imaging by PET (positron emission tomography) 30. It was found that 64Cu(L1) had glioma uptake comparable to or higher than many radiolabeled cyclic RGD peptides in the same tumor-bearing animal model 30. These promising results inspired us to prepare three new DOTA-conjugated rhodamine derivatives (Figure 1: H3L2 - H3L4). These DOTA conjugates were designed in such a way that complexes Cu(L) (L = L1 - L4) are useful as fluorescent probes for cellular staining assays and tumor imaging by optical methods while complexes 64Cu(L) (L = L1 - L4) are used as radiotracers for biodistribution and PET imaging. The Cu-DOTA chelate is expected to improve their hydrophilicity and increase the tumor selectivity of Cu/64Cu-labeled rhodamine derivatives 30, 31.
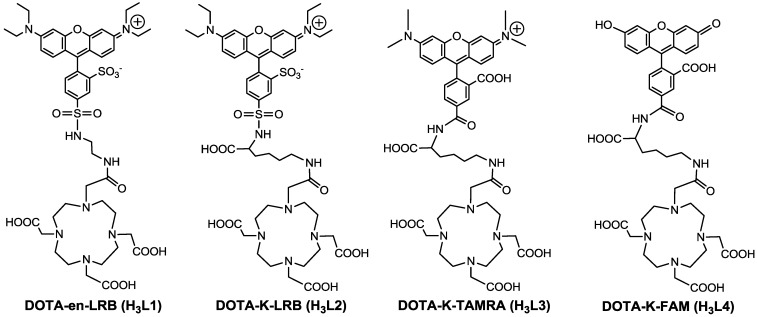
Figure 1.
Structures of DOTA-en-LRB (H3L1), DOTA-K-LRB (H3L2), DOTA-K-TAMRA (H3L3) and DOTA-K-FAM (H3L4). While complexes 64Cu(L) (L = L1 - L4) are useful as PET radiotracers, the corresponding Cu(II) complexes, Cu(L) (L = L1 - L4), have the potential as molecular probes for optical imaging of tumors.
In spite of its high sensitivity and capability of quantitation, PET suffers from several shortcomings, including poor clinical availability of tumor-specific radiotracers, requirement of an on-site cyclotron and a radiochemistry laboratory to produce the short-lived radionuclides and radiotracers, high cost, and radiation exposure to both nuclear medicine practitioners and cancer patients. Alternatively, optical imaging with bioluminescence/fluorescence has emerged as an attractive modality to study biological events at the molecular level in both cell culture and small animals 32-38. Optical imaging does not use ionizing radiation. It is inexpensive, highly sensitive and allows for a high throughput screening in both in vitro and in vivo assays 32, 33. Many fluorescent probes and optical imaging technologies have been developed for early tumor detection in tumor-bearing small animal models 23-29, 34-38.
As a continuation of our previous study 30, we now present the syntheses of three new DOTA-conjugated rhodamine derivatives (Figure 1: H3L2 - H3L4) and evaluations of Cu(L1) as a fluorescent probe for tumor optical imaging in two tumor-bearing models (U87MG glioma and MDA-MB-435 breast cancer). Cu(L1) was chosen due to its high tumor uptake 30. Cu(L2) and Cu(L4) were prepared to demonstrate their localization mechanism through a series of cellular staining assays. Radiotracers 64Cu(L) (Figure 1: L = L1 - L4) were prepared to compare their biodistribution properties using the same tumor-bearing animal models. The main objective of this study was to provide the proof-of-concept for the utility of complexes Cu(L) (L = L1 - L4) as selective molecular probes for optical imaging of tumors, where the negative mitochondrial transmembrane potential is significantly elevated in tumor cells as compared to that in the surrounding normal tissues.
EXPERIMENTAL PROCEDURES
Materials and Instruments. Chemicals were purchased from Sigma-Aldrich (St. Louis, MO), and were used without further purification. Fluorescein-5-carboxamide lysine (K-FAM, HCl salt), tetramethylrhodamine-5-carboxamide lysine (K-TAMRA), Lissamine rhodamine B ethylenediamine (en-LRB), Lissamine rhodamine B sulfonyl chloride (LRB-SC) were purchased from AnaSpec, Inc. (Fremont, CA). Lys(Boc)-OH was purchased from Bachem Americas, Inc. (Torrance, CA). DOTA-NHS and DOTA(OBu-t)3-NHS were purchased from Macrocyclics (Dallas, TX). The ESI (electrospray ionization) mass spectral data were collected on a Finnigan LCQ classic mass spectrometer, the School of Pharmacy, Purdue University. Complexes 64Cu(L1) and Cu(L1) were prepared using the method described in our previous report 30. 64Cu was produced using a CS-15 biomedical cyclotron at Washington University by the 64Ni(p,n)64Cu nuclear reaction.
HPLC Methods. The semi-prep HPLC method (Method 1) used a LabAlliance HPLC system equipped with a UV/vis detector (λ=254 nm) and Zorbax C18 semi-prep column (9.4 nm x 250 mm, 100 Å pore size; Agilent Technologies, Santa Clara, CA). For Method 1, the flow rate was 2.5 mL/min and the mobile phase was isocratic with 70% A (0.1% TFA in water) and 30% B (0.1% TFA in methanol) at 0 - 5 min, followed by a gradient mobile phase going from 30% B at 5 min to 80% B at 20 min. The radio-HPLC method (Method 2) used the LabAlliance HPLC system equipped with a β-ram IN/US detector (Tampa, FL) and Vydac protein and peptide C18 column (4.6 mm x 250 mm, 300 Å pore size; Grace Davison Discovery Sciences, Hesperia, CA). The flow rate was 1 mL/min with a gradient mobile phase being isocratic with 90% solvent A (25 mM NH4OAc, pH = 6.8) and 10% solvent B (acetonitrile) at 0 - 5 min, followed by a gradient mobile phase going from 10% B at 5 min to 90% B at 20 min.
Synthesis of 5-(N-(1-carboxy-5-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecan-1-yl)acetamido)pentyl)sulfamoyl)-2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)benzenesulfonate (H3L2). To a clean round-bottom flask were added K-FAM (2 mg, 3.5 µmol) and DOTA(OBu-t)3-NHS (28.26 mg, 34.6 µmol) anhydrous DMF (1 mL). To the mixture above was added excess triethylamine (Et3N: 35 µL, 250 µmol). The reaction mixture was stirred at room temperature for 37 h. Upon removal of volatiles under reduced pressure, the residue was dissolved in 1 mL of 12 N HCl. After stirring at room temperature for 30 min, the hydrolysis of t-butyl ester groups was completed. Volatiles were removed under reduced pressure on a rotary evaporator. The residue was dissolved in 2 mL of 50:50 (v:v) DMF/water mixture. The resulting solution was filtered to remove any foreign particles, and the filtrate was subjected to HPLC purification (Method 1). Fractions at 15 min were collected, combined and lyophilized to give deep yellow powder (1.3 mg, 42%). The HPLC purity was >95%. ESI-MS: m/z = 891.3 for [M + H]+ (891.9 calcd. for [C43H51N6O15]+).
Synthesis of N-(9-(2-carboxy-4-((1-carboxy-5-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecan-1-yl)acetamido)pentyl)carbamoyl)phenyl)-6-(dimethylamino)-3H-xanthen-3-ylidene)-N-methylmethanaminium (H3L3). K-TAMRA (1 mg, 1.79 µmol) and DOTA-NHS (7.17 mg, 14.31 µmol) were dissolved in anhydrous DMF (0.5 mL). To the mixture was added excess Et3N (35 µL, 250 µmol). The resulting reaction mixture was stirred at room temperature overnight. After complete removal of volatiles, the residue was dissolved in 1 mL of water. The product was separated from the reaction mixture by HPLC purification (Method 1). Fractions at 13.8 min were collected, combined and lyophilized to give deep purple powder (1.3 mg, ~77%). The HPLC purity was >95%. ESI-MS: m/z = 946.3 for [M+ H]+ (946.03 calcd. for [C47H61N8O13]+).
Lissamine Rhodamine B Sulfamide Lysine (K-LRB). Lys(Boc)-OH (9 mg, 36.54 µmol) and LRB-SC (10 mg, 17.3 µmol) were dissolved in 1.5 mL of DMF. To the mixture was added excess Et3N (0.6 mL). The reaction mixture was stirred at room temperature for 20 h. Upon removal of volatiles, the residue was dissolved in 1 mL of 12 N HCl to completely remove the Boc-protecting groups. After being stirred at room temperature for 30 min, volatiles were removed completely. The residue was dissolve in water (2 mL). The solution was subjected to HPLC purification (Method 1). Fractions at 18 min were collected, combined and lyophilized to give deep purple powder (3.8 mg, ~33%). The HPLC purity was >95%. ESI-MS: m/z = 687.2 for [M + H]+(687.84 calcd. for [C33H42N4O8S2]+).
Synthesis of 2,2',2''-(10-(2-((5-carboxy-5-(3-carboxy-4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzamido)pentyl)amino)-2-oxoethyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (H3L4). DOTA(OBu-t)3-NHS (16.63 mg, 20.4 µmol) and K-LRB (1.4 mg, 2.04 µmol) were dissolved in 0.5 mL of anhydrous DMF. To the mixture was added excess Et3N (35 µL, 250 µmol). The reaction mixture was stirred at room temperature for 10 h. Upon removal of volatiles, the residue was dissolved in 12 N HCl (1 mL) to completely hydrolyze the t-butyl ester groups. After being stirred at room temperature for 30 min, volatiles were removed under the reduced pressure. The residue was dissolve in water (2 mL). The product separated by HPLC purification (Method 1). Fraction at 13 min was collected, combined and lyophilized to give deep purple powder (0.8 mg, ~37%). The HPLC purity was >95%. ESI-MS: m/z = 1073.5 for [M + H]+ (1073 calcd. for [C49H69N8O15 S2]+).
Cu(L2). To a clean vial containing H3L2 and one equivalent of Cu(OAc)2·H2O was added 2.5 mL of NH4OAc buffer (0.5 M, pH = 5.5). The resulting solution was heated at 100 oC in a water bath for 20 min. The product was isolated by HPLC purification (Method 1). The fractions at 14.8 min were collected. Lyophilization of the collected fractions afforded Cu(L2) as a purple powder (0.3 mg, 76%). The HPLC purity was >95%. ESI-MS: m/z = 1012 for [M + H]+ (1012.14 calcd. for [C49H67N8O15Cu]+).
Cu(L4). To a clean vial containing H3L4 and one equivalent of Cu(OAc)2·H2O was added 2.5 mL of NH4OAc buffer (0.5 M, pH = 5.5). The resulting solution was heated at 100 oC for 30 min. The product was isolated by HPLC (Method 1). The fractions at 15.7 min were collected. Lyophilization of the collected fractions yielded Cu(L4) as a bright yellow powder (0.6 mg, 80%). The HPLC purity was >95%. ESI-MS: m/z = 952 for [M + H]+ (953.26 calcd. for [C43H49N6O15Cu]+).
64Cu-Labeling and Dose Preparation. To a clean Eppendorf tube was added 40 µg of the DOTA conjugate in 0.3 mL of 0.1 M NaOAc buffer (pH = 5.5) and 20 µL of 64CuCl2 solution (~ 520 µCi) in 0.05 N HCl. The reaction mixture was then heated at 100 oC for 20 min in water bath. After heating, the vial was allowed to stand at room temperature for ~ 5 min. A sample of the resulting solution was analyzed by HPLC (Method 2). For biodistribution studies, 64Cu radiotracers were prepared and purified by HPLC. Volatiles in HPLC mobile phases were removed on a rotary evaporator. Doses were prepared by dissolving the purified 64Cu radiotracer in saline to ~20 µCi/mL. The resulting solution was filtered with a 0.20 micron Millex-LG filter before being injected into animals. Log P values were determined by following literature procedure 13-16, 30, and are reported as an average of three independent measurements plus the standard deviation.
Animal Model. Biodistribution and optical imaging studies were performed in compliance with the NIH animal experimentation guidelines (Principles of Laboratory Animal Care, NIH Publication No. 86-23, revised 1985). The protocol was approved by the Purdue University Animal Care and Use Committee (PACUC). U87MG and MDA-MB-435 cell lines were obtained from ATCC (Manassas, VA). The U87MG cells were cultured in the Minimum Essential Medium, Eagle with Earle's Balanced Salt Solution (non-essential amino acids sodium pyruvate) (American Type Culture Collection or ATCC, Manassas, VA). MDA-MB-435 cells were grown in the RPMI Medium 1640 with L-Glutamine (GIBCO Industries Inc., Langley, OK). Both cell lines were supplemented with 10% fetal bovine serum (FBS, ATCC) and 1% penicillin and streptomycin (GIBCO) solution, and grown at 37 °C in a humidified atmosphere of 5% CO2 in air. Cells were grown as monolayers and were harvested or split when they reached 90% confluence to maintain exponential growth. Female nu/nu mice (5 - 6 weeks of age) were purchased from Harlan (Indianapolis, IN), and were implanted with 5 × 106 tumor cells into mammary fat pads (MDA-MB-435) or shoulder flanks (U87MG). All procedures were performed in a laminar flow cabinet using aseptic techniques. Four weeks after inoculation, the tumor size was 0.1 - 0.4 g, and animals were used for biodistribution and optical imaging studies.
Optical Imaging. Each tumor-bearing mouse was administered with 0.5 mg Cu(L1) in 100 μL PBS via tail vein injection. Animals without the tumor or administration of Cu(L1) were also imaged as the “negative” control. The animal was anesthetized by inhalation of 3% isofluorane and 97% O2 at 4 h post-injection (p.i.), and then placed in supine position in the imaging chamber, which was maintained at 37 °C and equipped with a controlled flow of 1.5% isofluorane and 98.5% O2. Images were acquired with a Kodak Imaging Station (In-Vivo FX, Eastman Kodak Company) using excitation at 555 nm and emission at 600 nm in combination with CCD camera and Kodak molecular imaging software (version 4.0).
Biodistribution Protocol. Five tumor-bearing mice (20 - 25 g) were randomly selected. Each animal was administered with ~3 µCi of 64Cu radiotracer by tail vein injection. Animals were sacrificed by sodium pentobarbital overdose (~200 mg/kg) at 4 h p.i. Blood samples were withdrawn from the heart. Tumor, brain, eyes, heart, spleen, lungs, liver, kidneys, muscle and intestine were harvested, dried with absorbent tissue, weighed, and counted on a γ-counter (Perkin Elmer Wizard - 1480, Shelton, CT). The organ uptake was calculated as the percentage of injected dose per gram of organ mass (%ID/g). Biodistribution data and T/B ratios were reported as an average plus the standard deviation based on the results from 5 animals. Comparison between two radiotracers was made using the one-way ANOVA test (GraphPad Prim 5.0, San Diego, CA). The level of significance was set at p < 0.05.
Cellular Staining Assay. Cells (U87MG glioma and human fibroblasts cells) were cultured in Lab-TekTM 8-well glass chamber slides for at least 24 h before being used for the cellular staining assay. The fluorescent probe in saline was added into the culture media to achieve a concentration of 10 µM for Cu(L2) and 20 µM for Cu(L4). After incubation for 12 h, the tumor cells were washed three times with phenol red-free Minimum Essential Medium. The fluorescence was immediately visualized with an Olympic BX51 fluorescence microscope (Olympus America Inc., Center Valley, PA) under 400x magnifications. Images were acquired using a Hamamatsu digital CCD camera ORCA-R2 (Hamamatsu Photonics K.K., Japan) with the Olympus MetaMorph software.
Uncoupling Experiments. These experiments were carried out according to the literature methods 39, 40 with slight modification. In the dose-response experiment, the cultured U87MG glioma cells were pretreated with carbonylcyanide-m-chlorophenylhydrazone (CCCP) at 0, 50, 100 and 150 µM. After incubation with 10 μM Cu(L2) and 20 μM Cu(L4), respectively, for 1 h, the U87MG glioma cells were washed with the phenol red-free Minimum Essential Medium. In the time-response experiment, U87MG glioma cells were incubated with 10 μM Cu(L2) and 20 μM Cu(L4), respectively, in the presence of 50 μM CCCP for specified period of time (0, 1, 4 and 8 h). After incubation, the U87MG cells were washed with phenol red-free Minimum Essential Medium. After washing, the fluorescence was visualized immediately with an Olympic BX51 fluorescence microscope. All images were acquired using a Hamamatsu digital CCD camera ORCA-R2 (Hamamatsu Photonics K.K., Japan).
RESULTS
DOTA Conjugates. For H3L2, LRB-K(Boc) was first prepared by reacting LRB-SC with Lys(Boc) in anhydrous DMF. Removal of the Boc group afforded K-LRB, which was then allowed to react with DOTA(OBu-t)3-NHS in presence of excess Et3N to yield L2(OBu-t)3. Hydrolysis of L2(OBu-t)3 produced H3L2. H3L3 was prepared by conjugation of K-TAMRA with DOTA-NHS in DMF under the basic conditions. L4(OBu-t)3 was prepared from the reaction of K-FAM with DOTA(OBu-t)3-NHS in DMF in the presence of excess Et3N. Hydrolysis of L4(OBu-t)3 gave the expected product H3L4. All three new DOTA conjugates were purified by HPLC, and their HPLC purity was > 95% before being used for radiochemistry. Their chemical compositions were confirmed by ESI-MS.
Cu(II) Complexes. Cu(L2) and Cu(L4) were prepared by reacting one equivalent of copper(II) acetate with H3L2 and H3L4, respectively, in 0.5 M NH4OAc. Both Cu(L2) and Cu(L4) were purified by semi-prep HPLC, and analyzed by ESI- MS. Supplementary Material: Figure SI1 shows HPLC chromatograms of Cu(L2) and Cu(L4). The ESI-MS spectra displayed molecular ions at m/z = 1012 of Cu(L2) (1012.14 calcd. for [C49H67N8O15Cu]+), and m/z = 952 of Cu(L4) (953.26 calcd. for [C43H49N6O15Cu]+).
Radiochemistry. All three new 64Cu radiotracers were prepared by reacting DOTA conjugates with 64CuCl2 in 0.1 M NaOAc buffer (pH = 5.5) at 100 oC for 10 min. Their radiochemical purity was >95% without further HPLC purification. The specific activity was ~50 Ci/mmol. No further optimization was made to improve their specific activity. Their HPLC retention times and log P values were listed in Table 1. As an example, Supplementary Material: Figure SI2 illustrated the radio-HPLC chromatograms of 64Cu(L3) and 64Cu(L4). All three new 64Cu radiotracers were stable in the kit matrix for >24 h post-labeling.
Table 1.
Radiochemical purity (RCP), retention time and log P values for 64Cu radiotracers.
| Radiotracer | RCP (%) | Retention Time (min)* | Log P Value |
|---|---|---|---|
| 64Cu(L1) | > 95 | 14.6 | -1.4 ± 0.1 |
| 64Cu(L2) | > 95 | 14.8 | -1.5 ± 0.1 |
| 64Cu(L3) | > 95 | 13.4 | -1.7 ± 0.2 |
| 64Cu(L4) | > 95 | 12.8 | -1.8 ± 0.1 |
*: Obtained using radio-HPLC (Method 2).
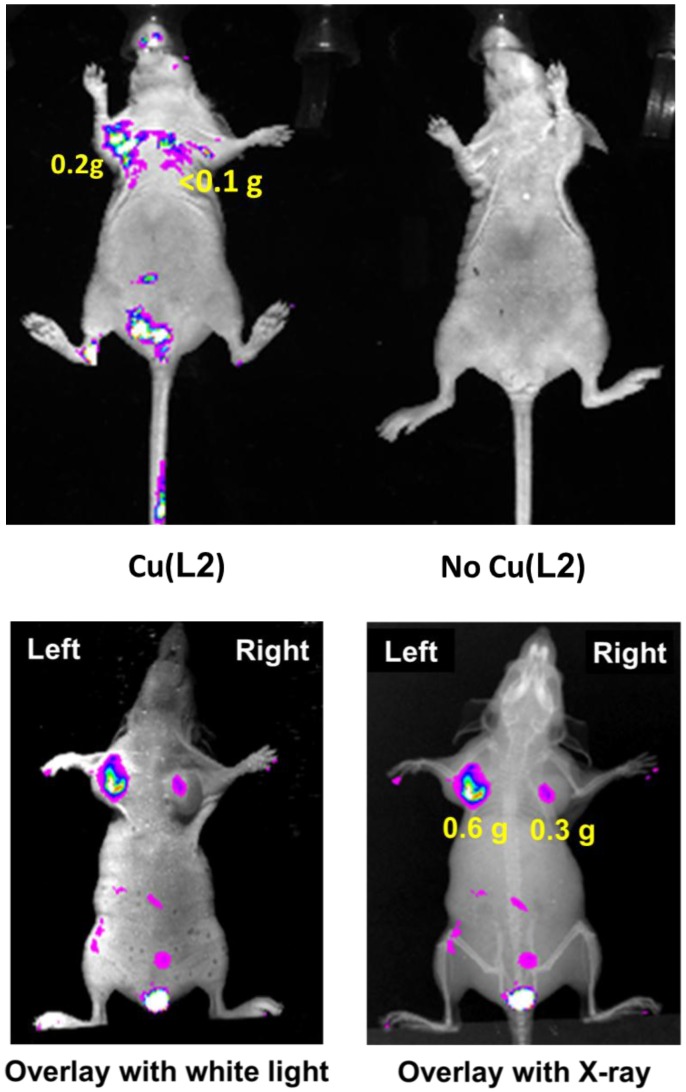
Optical Imaging. Optical imaging was performed using Cu(L1) in the U87MG and MDA-MB-435 models to demonstrate the utility of Cu(II)-labeled rhodamine derivatives as fluorescent probes for tumor imaging. Cu(L1) was chosen due to its high tumor uptake and longer wavelength than that of Cu(L4). As shown in Figure 2 (top), the tumors could be visualized with good contrast at 4 h p.i. Without Cu(L1), these tumors were not seen. We also found that Cu(L1) was able to retain in tumors for >24 h (data not shown). Because of tissue attenuation, accurate quantification of the tumor uptake was difficult. This was further supported by the fact that fluorescent signals were detected only in body parts closest to camera. Attempts were also made to quantify the fluorescent intensity in the extracted organ tissues (Supplementary Material: Figure SI3), but it always fluctuated depending on the orientation of organs to be imaged. We were surprised that Cu(L1) was able to localize and retain in the xenografted MDA-MB-435 breast tumors (Figure 2: bottom) since our previous study showed that the xenografted MDA-MB-435 tumors had high expression levels of MDR Pgps and MRPs 13.
Figure 2.
Top: The overlay optical images of the athymic nude mice (n = 3) bearing U87MG glioma xenografts (tumor size: 80 - 200 mg) before (right) and 4 h after (left) injection of Cu(L2). Bottom: Representative overlay optical images of athymic nude mice (n = 3) bearing MDA-MB-435 human breast cancer xenografts at 4 h p.i. Each tumor-bearing mouse was administered with 0.5 mg of Cu(L2) in 100 μL PBS via the tail vein injection. In all cases, no acute toxicity was observed during and after imaging studies.
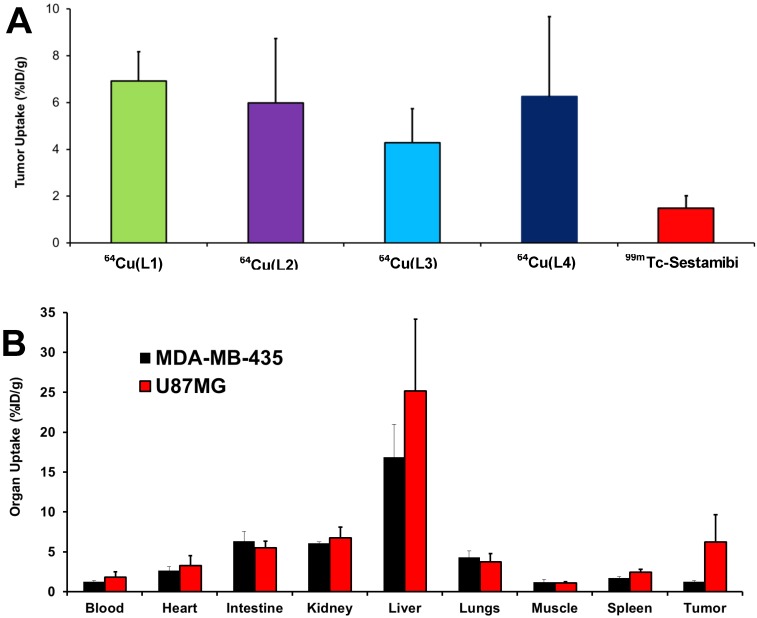
Biodistribution. Biodistribution remains the most accepted method for accurate determination of the amount of molecular probes in a specific organ. In this study, 64Cu(L) (L = L1 - L4) were evaluated in athymic nude mice bearing U87MG glioma xenografts. Their biodistribution data were listed in Table 2. Biodistribution was performed at 4 h p.i. since the tumor uptake of 64Cu(L1) peaked at this time point 30. 64Cu(L1) was re-evaluated to prove that the glioma-bearing animals were same as those used in our previous studies 30, 31. We found that the biodistribution data of 64Cu(L1) was almost identical to that reported in previous study 30. All four radiotracers had high glioma uptake (64Cu(L1): 5.71± 1.43 %ID/g; 64Cu(L2): 5.98 ± 2.75 %ID/g; 64Cu(L3): 4.28 ± 1.45 %ID/g; and 64Cu(L4): 6.25 ± 3.42 %ID/g), which was better than that of 99mTc-Sestamibi (Figure 3A). Even though 64Cu(L3) seemed to have lower tumor uptake than the other 64Cu radiotracers, this difference was not significant within experimental errors. However, all 64Cu radiotracers had significantly (p < 0.05) higher liver uptake than 64Cu(L1) (Table 2). Among 64Cu-labeled rhodamine derivatives evaluated in the U87MG glioma model, 64Cu(L1) was the best with respect to its tumor/blood, tumor/liver and tumor/muscle ratios. 64Cu(L4) was also evaluated in athymic nude mice bearing MDA-MB-435 breast cancer xenografts. As expected, 64Cu(L4) had almost identical uptake in normal organs of the U87MG and MDA-MB-435-bearing animals (Figure 3B). However, its breast tumor uptake (1.26 ± 0.10% ID/g) was significantly (p < 0.001) lower than that in glioma (6.25 ± 3.42% ID/g).
Table 2.
Selected biodistribution data for 64Cu radiotracers in athymic nude mice bearing U87MG human glioma xenografts at 4 h p.i. The organ uptake is expressed as %ID/g.
| Compounds |
64Cu(L1) (n = 3) |
64Cu(L2) (n = 5) |
64Cu(L3) (n = 5) |
64Cu(L4) (n = 5) |
|---|---|---|---|---|
| Blood | 0.77 ± 0.11 | 3.06 ± 1.01 | 2.26 ± 0.13 | 1.40 ± 0.38 |
| Brain | 0.18 ± 0.02 | 0.25 ± 0.05 | 0.42 ± 0.27 | 0.25 ± 0.04 |
| Heart | 1.89± 0.04 | 4.07 ± 1.42 | 3.01 ± 0.74 | 2.87 ± 0.49 |
| Intestine | 4.41 ± 0.99 | 5.83 ± 2.19 | 5.24 ± 1.00 | 6.02 ± 0.84 |
| Kidney | 4.40 ± 0.18 | 7.46 ± 2.22 | 5.25 ± 0.30 | 6.77 ± 1.33 |
| Liver | 10.98 ± 0.66 | 40.66 ± 4.98 | 30.90 ± 6.52 | 20.50 ± 4.99 |
| Lungs | 4.25± 1.01 | 4.50 ± 1.57 | 3.16 ± 0.39 | 3.93 ± 0.96 |
| Muscle | 0.15 ± 0.08 | 1.45 ± 0.51 | 2.01 ± 0.63 | 1.19 ± 0.27 |
| Spleen | 0.92 ± 0.07 | 3.23 ± 1.11 | 2.30 ± 0.27 | 2.15 ± 0.36 |
| U87MG | 5.71± 1.43 | 5.98 ± 2.75 | 4.28 ± 1.45 | 6.25 ± 3.42 |
| Tumor/Blood | 6.94 ± 0.75 | 2.00 ± 0.69 | 1.90 ± 0.66 | 4.12 ± 3.24 |
| Tumor/Liver | 0.48 ± 0.03 | 0.14 ± 0.05 | 0.15 ± 0.08 | 0.32 ± 0.17 |
| Tumor/Lungs | 1.35 ± 0.24 | 1.31 ± 0.35 | 1.35 ± 0.40 | 1.60 ± 0.52 |
| Tumor/Muscle | 35.3 ± 12.6 | 4.32 ± 1.62 | 2.25 ± 0.89 | 6.02 ± 1.88 |
Figure 3.
A: Comparison of the tumor uptake values of 64Cu radiotracers with that of 99mTc-Sestamib in the athymic nude mice bearing U87MG glioma xenografts. 99mTc-Sestamibi was used as “control” for comparison purposes, and it tumor uptake was obtained from our previous study 13. B: Direct comparison of organ uptake values of 64Cu(L4) at 4 h p.i. in athymic nude mice bearing U87MG glioma and MDA-MB-435 breast cancer xenografts.
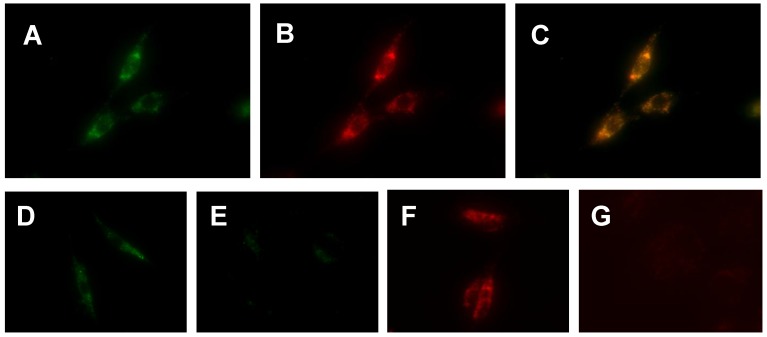
Localization Mechanism and Tumor Selectivity. We used cellular staining assays to demonstrate the localization mechanism and tumor selectivity of Cu-labeled rhodamine derivatives. Figure 4 (A - C) shows microscopic images of the U87MG glioma cells stained with 20 µM of Cu(L4). We were interested in Cu(L4) due to its structural difference with the other Cu(II) complexes. LRB-SC was used as the “control” since it is a well-known mitochondrion-targeted fluorescent dye. The orange color in overlay images indicated that Cu(L4) was able to co-localize with LRB-SC in mitochondria of U87MG cells. To demonstrate their selectivity, we obtained microscopic images (Figure 4: D - G) of U87MG cells and human fibroblast cells after incubation with Cu(L2) and Cu(L4), respectively. Fibroblast cells were used as the “negative control” due to their low mitochondrial potential as compared to that in tumor cells. We found that Cu(L2) and Cu(L4) were able to localize in the mitochondria of both human fibroblast cells and U87MG glioma cells. The fluorescent intensity in the U87MG glioma cells was much stronger than that in the human fibroblast cells using the same brightness and contrast.
Figure 4.
Top: Representative microscopic images (magnification: 400×) of the cultured U87MG cells stained with 20 µM Cu(L4) (A: green color) and 10 µM LRB-SC (B: red color) for 12 h. The orange color in the overlay image (C) indicates that Cu(L4) was able to co-localize with LRB-SC in mitochondria of U87MG human glioma cells. Bottom: Representative microscopic images (magnification: 400×) of the cultured U87MG glioma cells (D and F) and human fibroblasts (E and G) stained for 12 h with 20 µM Cu(L4) (D and E) and 10 µM Cu(L2) (F and G). These images were obtained to demonstrate the tumor-selectivity of Cu-labeled rhodamine derivatives and its relationship with the higher mitochondrial potential in U87MG cells than that in human fibroblasts.
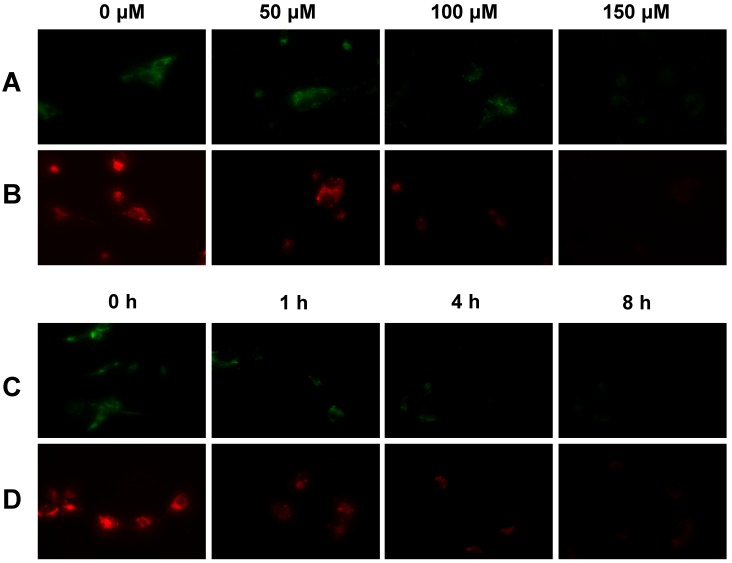
Tumor Selectivity and Mitochondrial Potential. We used the uncoupling experiments to illustrate that the tumor selectivity of Cu-labeled rhodamine derivatives is caused by the higher mitochondrial potential in U87MG glioma cells. In these experiments, CCCP was used as the mitochondrial uncoupling agent. Figure 5 illustrates representative microscopic images of the cultured U87MG cells stained with 10 µM Cu(L2) and 20 µM Cu(L4), respectively, with or without the CCCP treatment. Apparently, the intracellular fluorescent intensity decreased rapidly with the increase of CCCP concentration (Figure 5: A and B). When the concentration of CCCP is fixed at 50 µM, the intracellular fluorescent intensity decreased very fast with the incubation time (Figure 5: C and D). These data clearly showed that the intracellular fluorescent intensity was potential-dependent.
Figure 5.
Typical microscopic images (magnification: 400×) of the cultured U87MG glioma cells stained with 20 µM Cu(L4) (A) and 10 µM Cu(L2) (B) after treated with CCCP at 0, 50, 100 and 150 µM for 1 h; and stained with 20 µM Cu(L4) (C) and 10 µM Cu(L2) (D) after treated with 50 µM CCCP for 0 , 1, 4, and 8 h. These images were obtained to show that the tumor-selectivity of Cu-labeled rhodamine derivatives is indeed associated with the higher mitochondrial potential in U87MG cells as compared to that in human fibroblasts.
DISCUSSION
In this study, we showed that Cu-labeled rhodamine derivatives were useful as fluorescent probes for tumor imaging. Both MDR-negative U87MG gliomas and MDR-positive MDA-MB-435 breast tumors could be visualized by optical imaging with Cu(L1) as the molecular probe. However, the fluorescent signals were detected only in the body parts closest to the camera due to significant tissue attenuation of the fluorescent light. The fluorescent intensity was affected significantly by the orientation of the organs to be imaged. Thus, 64Cu(L) (L = L1 - L4) were used to determine the tumor uptake and biodistribution properties of complexes Cu(L) (L = L1 - L4). It was found that the aromatic cations had a significant impact on biodistribution properties of 64Cu radiotracers. For example, the glioma uptake of 64Cu(L2) (log P = -1.5) was 5.98 ± 2.75 %ID/g while 64Cu(L3) (log P = -1.7) had a glioma uptake of 4.28 ± 1.45 %ID/g at the same time point. 64Cu(L4) (log P = -1.7) had the glioma uptake of 6.25 ± 3.42 %ID/g, which was well comparable to that (5.71± 1.43 %ID/g) of 64Cu(L1) (log P = -1.4) within experimental errors in the same tumor-bearing animal model (Figure 3: top). Apparently, there was no clear-cut optimal lipophilicity to achieve high tumor uptake.
We were very intrigued by the higher liver uptake of 64Cu(L) (L = L2 - L4) than that of 64Cu(L1). It has been suggested that metabolic instability of 64Cu-labeled DOTA-biomolecule conjugates contributes significantly to their high liver uptake 41-43. However, this can't explain their liver uptake differences since all four radiotracers share the same 64Cu(DOTA) chelate. Therefore, their liver uptake difference is most likely caused by the capability of aromatic cations to localize in the liver. At this moment, it remains unclear why 64Cu(L1) has the best tumor/liver and tumor/lung ratios among the four 64Cu radiotracers with similar structure.
The mitochondrial localization was demonstrated by cellular staining assays. For example, Cu(L4) co-localized with LRB-SC (Figure 3). Cu(L2) and Cu(L4) were able to localize in the mitochondria of U87MG glioma cells, and their mitochondrion-localization was potential-driven since the mitochondrial fluorescent intensity decreased rapidly with the increase of CCCP concentration or incubation time. These results provide strong support for our hypothesis that their tumor localization and selectivity are caused by the increased negative mitochondrial potential in U87MG glioma cells as compared to that in fibroblast cells of the surrounding normal tissues.
MDR Pgps and MRPs belong to a large family of ATP-trafficking proteins that mediate the transport of amphipathic drug molecules 44-49. The net effect of MDR Pgp and MRP overexpression is the reduced intracellular drug accumulation through energy-dependent drug efflux in the MDR-positive cancer cells. MDA-MB-435 is an estrogen-independent human breast cancer cell line (metastatic ductal adenocarcinoma). It has a low expression of MDR1 and high levels of MRP2 and MRP4 50-52. In our previous report, we showed that the xenografted MDA-MB-435 tumors had high expression levels of MDR1 Pgp, MRP2 and MRP4 (Figure 4A) 13. It is not unexpected that 64Cu(L4) had lower uptake in the xenografted MDA-MB-435 breast tumors than that in glioma (Figure 3: bottom). However, this doesn't explain why Cu(L1) was able to retain in the xenografted MDA-MB-435 breast tumors.
It is well-established that rhodamine derivatives are substrates of MDR Pgps and MRPs 53-56. Since only a small amount (<1 μg) of 64Cu(L4) was used in biodistribution studies, the MDR Pgps and MRPs were able to efficiently exclude 64Cu(L4) from the MDR-positive tumor cells. For optical imaging, however, Cu(L1) was used at the dose of 0.5 - 0.7 mg/mouse (20 - 30 mg/kg). It is possible that excess Cu(L1) is able to “overload” the MDR Pgps and MPs, which allows a small portion of Cu(L1) to localize in the tumor cells. It must be noted that this explanation remains largely speculation in the absence of more experimental evidence.
CONCLUSIONS
The key finding of this study is that Cu(II)-labeled rhodamine derivatives, such as Cu(L1), are useful as fluorescent probes for in vitro cellular staining assays and tumor imaging by optical methods, and their 64Cu complexes have the potential as radiotracers for tumor imaging by PET. This study represents a good example of dual modality imaging (PET and optical) using two agents, 64Cu(L) and Cu(L), with identical chemical composition. Future research will focus on development of fluorescent probes with longer wavelength and the reduced liver uptake.
SUPPLEMENTARY MATERIAL
Fig.SI1-SI3.
Acknowledgments
This work was supported by Purdue University, and research grants: R01 CA115883 (S.L.) from the National Cancer Institute (NCI) and KG111333 (Y.Z. and S.L.) from the Susan G. Komen Breast Cancer Foundation.
Abbreviations
- DOTA
1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid
- DOTA-NHS
1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid mono (N-hydroxysuccinimide)
- DOTA(OBu-t)3-NHS
1,4,7,10-tetraazacyclododecane-1,4,7-tris(t-butyl acetate)-10-acetate mono (N-hydroxysuccinimide)
- H3L1
2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)-5-(N-(2-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecan-1-yl)acetamido)ethyl)sulfamoyl)benzenesulfonate
- H3L2
5-(N-(1-carboxy-5-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecan-1-yl)acetamido)pentyl)sulfamoyl)-2-(6-(diethylamino)-3-(diethyliminio)-3H-xanthen-9-yl)benzenesulfonate
- H3L3
N-(9-(2-carboxy-4-((1-carboxy-5-(2-(4,7,10-tris(carboxymethyl)-1,4,7,10-tetraazacyclododecan-1-yl)acetamido)pentyl)carbamoyl)phenyl)-6-(dimethylamino)-3H-xanthen-3-ylidene)-N-methylmethanaminium
- H3L4
2,2',2''-(10-(2-((5-carboxy-5-(3-carboxy-4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzamido)pentyl)amino)-2-oxoethyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid
- MDR Pgps
multidrug resistance P-glycoproteins
- MRPs
multidrug resistance-associated proteins
- PET
positron emission tomography.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:1–24. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–42. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- 3.Modica-Napolitano JS, Aprille JR. Delocalized lipophilic cations selectively target the mitochondria of carcinoma Cells. Adv Drug Deliv Rev. 2001;49:63–70. doi: 10.1016/s0169-409x(01)00125-9. [DOI] [PubMed] [Google Scholar]
- 4.Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Mannella CA. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta. 2006;1762:140–7. doi: 10.1016/j.bbadis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Summerhayes IC, Lampidis TJ, Bernal SD, Nadakavukaren JJ, Nadakavukaren KK, Shepard EL, Chen LB. Unusual retention of Rhodamine 123 by mitochondria in muscle and carcinoma cells. Proc Natl Acad Sci USA. 1982;77:990–4. doi: 10.1073/pnas.79.17.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modica-Napolitano JS, Aprille JR. Basis for the selective cytotoxicity of Rhodamine 123. Cancer Res. 1987;47:4361–5. [PubMed] [Google Scholar]
- 8.Davis S, Weiss MJ, Wong JR, Lampidis TJ, Chen LB. Mitochondrial and plasma membrane potentials cause unusual accumulation and retention of Rhodamine 123 by human breast adenocarcinoma-derived MCF-7 cells. J Biol Chem. 1985;260:13844–50. [PubMed] [Google Scholar]
- 9.Dairkee SH, Hackett AJ. Differential retention of Rhodamine 123 by breast carcinoma and normal human mammary tissue. Breast Cancer Res Treat. 1991;18:57–61. doi: 10.1007/BF01975444. [DOI] [PubMed] [Google Scholar]
- 10.Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of Cancer. Expert Rev Mol Med. 2002;4:1–19. doi: 10.1017/S1462399402004453. [DOI] [PubMed] [Google Scholar]
- 11.Gottlieb E, Thompson CB. Targeting the mitochondria to enhance tumor suppression. Methods Mol Biol. 2003;223:543–54. doi: 10.1385/1-59259-329-1:543. [DOI] [PubMed] [Google Scholar]
- 12.Ross MF, Kelso GF, Blaikie FH, James AM, Cocheme HM, Filipovska A, Da Ros T, Hurd TR, Smith RA, Murphy MP. Lipophilic triphenylphosphonium cations as tools in mitochondrial bioenergetics and free radical biology. Biochemistry (Mosc) 2005;70:222–30. doi: 10.1007/s10541-005-0104-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Kim YS, Zhai S, Shi J, Hou G. Evaluation of 64Cu(DO3A-xy-TPEP) as a potential PET radiotracer for monitoring tumor multidrug resistance. Bioconj Chem. 2009;20:790–8. doi: 10.1021/bc800545e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Yang CT, Kim YS, Sreerama SG, Cao Q, Li ZB, He Z, Chen X, Liu S. 64Cu-Labeled triphenylphosphonium and triphenylarsonium cations as highly tumor-selective imaging agents. J Med Chem. 2007;50:5057–69. doi: 10.1021/jm0704088. [DOI] [PubMed] [Google Scholar]
- 15.Kim YS, Yang C, Wang J, Sreerama SG, Cao Q, Li Z, He Z, Chen X, Liu S. Effects of targeting moiety, linker, bifunctional chelator, and molecular charge on biological properties of 64Cu-labeled triphenylphosphonium cations. J Med Chem. 2008;51:2971–84. doi: 10.1021/jm7015045. [DOI] [PubMed] [Google Scholar]
- 16.Yang CT, Kim YS, Wang J, Wang L, Shi J, Li ZB, Chen X, Fan M, Li JJ, Liu S. 64Cu-Labeled 2-(diphenylphosphoryl)ethyldiphenylphosphonium cations as highly selective tumor imaging agents: effects of linkers and chelates on radiotracer biodistribution characteristics. Bioconj Chem. 2008;19:2008–22. doi: 10.1021/bc8002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichtshtein D, Kaback HR, Blume AJ. Use of a lipophilic cation for determination of membrane potential in neuroblastoma-glioma hybrid cell suspensions. Proc Natl Acad Sci USA. 1979;76:650–4. doi: 10.1073/pnas.76.2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hockings PD, Rogers PJ. The Measurement of transmembrane electrical potential with lipophilic cations. Biochim Biophys Acta. 1996;1282:101–6. doi: 10.1016/0005-2736(96)00045-4. [DOI] [PubMed] [Google Scholar]
- 19.Huang SG. Development of a high throughput screening assay for mitochondrial membrane potential in living cells. J Biomol Screening. 2002;7:383–9. doi: 10.1177/108705710200700411. [DOI] [PubMed] [Google Scholar]
- 20.Lefevre C, Kang HC, Haugland RP, Malekzadeh N, Arttamangkul S, Haugland RP. Texas Red-X and Rhodamine Red-X, new derivatives of sulforhodamine 101 and Lissamine Rhodamine B with improved labeling and fluorescence properties. Bioconj Chem. 1996;7:482–9. doi: 10.1021/bc960034p. [DOI] [PubMed] [Google Scholar]
- 21.Mao C, Kisaalita WS. Determination of resting membrane potential of individual neuroblastoma cells (IMR-32) using a potentiometric dye (TMRM) and confocal microscopy. J Fluoresc. 2004;14:739–43. doi: 10.1023/b:jofl.0000047224.41328.f8. [DOI] [PubMed] [Google Scholar]
- 22.Scaduto R. Jr, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76:469–77. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licha K, Olbrich C. Optical imaging in drug discovery and diagnostic applications. Adv Drug Deliv Rev. 2005;57:1087–1108. doi: 10.1016/j.addr.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 24.Jakobs S. High resolution imaging of live mitochondria. Biochim Biophys Acta. 2006;1763:561–75. doi: 10.1016/j.bbamcr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Lavis LD, Chao TY, Raines RT. Fluorogenic label for biomolecular imaging. ACS Chem Biol. 2006;1:252–60. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hama Y, Urano Y, Koyama Y, Kamiya M, Bernardo M, Paik RS, Shin IS, Paik CH, Choyke PL, Kobayashi H. A target cell-specific activatable fluorescence probe for in vivo molecular imaging of cancer based on a self-quenched Avidin-Rhodamine conjugate. Cancer Res. 2007;67:2791–9. doi: 10.1158/0008-5472.CAN-06-3315. [DOI] [PubMed] [Google Scholar]
- 27.Glunde K, Foss CA, Takagi T, Wildes F, Bhujwalla ZM. Synthesis of 6'-O-Lissamine-rhodamine B-glucosamine as a novel probe for fluorescence imaging of lysosomes in breast tumors. Bioconj Chem. 2005;16:843–51. doi: 10.1021/bc050046n. [DOI] [PubMed] [Google Scholar]
- 28.Longmire MR, Ogawa M, Hama Y, Kosaka N, Regino CA, Choyke PL, Kobayashi H. Determination of optimal rhodamine fluorophore for in vivo optical imaging. Bioconj Chem. 2008;19:1735–42. doi: 10.1021/bc800140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yova D, Atlamazoglou V, Kavantzas N, Loukas S. Development of a fluorescence-based imaging system for colon cancer diagnosis using two novel rhodamine derivatives. Lasers Med Sci. 2000;15:140–7. [Google Scholar]
- 30.Zhou Y, Kim YS, Yan X, Jacobson O, Chen X, Liu S. 64Cu-labeled Lissamine Rhodamine B: a promising PET radiotracer targeting tumor mitochondria. Mol Pharm. 2011;8:1198–1208. doi: 10.1021/mp200025m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Liu S. 64Cu-Labeled phosphonium cation as PET radiotracers for tumor imaging. Bioconj Chem. 2011;22:1459–72. doi: 10.1021/bc200106p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Z, Levi J, Xiong Z, Gheysens O, Keren S, Chen X, Gambhir SS. Near-infrared fluorescent deoxyglucose analogue for tumor optical imaging in cell culture and living mice. Bioconj Chem. 2006;17:662–9. doi: 10.1021/bc050345c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ntziachristos V, Bremer C, Weissleder R. Fluorescence imaging with near-infrared light: new technological advances that enable in vivo molecular imaging. Eur Radiol. 2003;13:195–208. doi: 10.1007/s00330-002-1524-x. [DOI] [PubMed] [Google Scholar]
- 35.Mahmood U, Weissleder R. Near-infrared optical imaging of proteases in cancer. Mol Cancer Ther. 2003;2:489–96. [PubMed] [Google Scholar]
- 36.Achilefu S. Lighting up tumors with receptor-specific optical molecular probes. Technol Cancer Res Treat. 2004;3:393–409. doi: 10.1177/153303460400300410. [DOI] [PubMed] [Google Scholar]
- 37.Sevick-Muraca EM, Houston JP, Gurfinkel M. Fluorescence-enhanced, near-infrared diagnostic imaging with contrast agents. Curr Opin Chem Biol. 2002;6:642–50. doi: 10.1016/s1367-5931(02)00356-3. [DOI] [PubMed] [Google Scholar]
- 38.Hawrysz DJ, Sevick-Muraca EM. Developments toward diagnostic breast cancer imaging using near-infrared optical measurements and fluorescent contrast agents. Neoplasia. 2000;2:388–417. doi: 10.1038/sj.neo.7900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Graaf AO, van den Heuvel LP, Dijkman HB, de Abreu RA, Birkenkamp KU, de Witte T, van der Reijden BA, Smeitink JA, Jansen JH. Bcl-2 prevents loss of mitochondria in CCCP-induced apoptosis. Exp Cell Res. 2004;299:533–40. doi: 10.1016/j.yexcr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 40.Horton KL, Stewart KM, Fonseca SB, Guo Q, Kelley SO. Mitochondria-penetrating peptides. Chem Biol. 2008;15:375–82. doi: 10.1016/j.chembiol.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Boswell CA, Sun X, Niu W, Weisman GR, Wong EH, Rheingold AL, Anderson CJ. Comparative in vivo stability of copper-64-labeled cross-bridged and conventional tetraazamacrocyclic complexes. J Med Chem. 2004;47:1465–74. doi: 10.1021/jm030383m. [DOI] [PubMed] [Google Scholar]
- 42.Boswell CA, McQuade P, Weisman GR, Wong EH, Anderson CJ. Optimization of labeling and metabolite analysis of copper-64-labeled azamacrocyclic chelators by radio-LC-MS. Nucl Med Biol. 2005;32:29–38. doi: 10.1016/j.nucmedbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Sprague JE, Peng Y, Fiamengo AL, Woodin KS, Southwick EA, Wiseman GR, Wong EH, Golden JA, Rheingold AL, Anderson CJ. Synthesis, characterization and in vivo studies of Cu(II)-64-labeled cross-bridged tetraazamacrocycle-amide complexes as models of peptide conjugate imaging agents. J Med Chem. 2007;50:2527–35. doi: 10.1021/jm070204r. [DOI] [PubMed] [Google Scholar]
- 44.Ling V. Multidrug resistance: molecular mechanisms and clinical relevance. Cancer Chemother Pharmacol. 1997;40:S3–S8. doi: 10.1007/s002800051053. [DOI] [PubMed] [Google Scholar]
- 45.Gros P, Ben Neriah YB, Croop JM, Housman DE. Isolation and expression of a complementary DNA that confers multidrug resistance. Nature. 1986;323:728–31. doi: 10.1038/323728a0. [DOI] [PubMed] [Google Scholar]
- 46.Zaman GJ, Flens MJ, Van Leusden MR, de Haas M, Mulder HS, Lankelma J, Pinedo HM, Scheper RJ, Baas F, Broxterman HJ, Borst P. The human multidrug resistance-associated-protein MRP is a plasma membrane drug-efflux pump. Proc Natl Acad Sci USA. 1994;91:8822–6. doi: 10.1073/pnas.91.19.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein I, Sarkadi B, Varadi A. An inventory of the human ABC proteins. Biochim Biophys Acta. 1999;1461:237–62. doi: 10.1016/s0005-2736(99)00161-3. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro AB, Ling V. Reconstitution of drug transport by purified P-glycoprotein. J Biol Chem. 1995;270:16167–75. doi: 10.1074/jbc.270.27.16167. [DOI] [PubMed] [Google Scholar]
- 49.Mao Q, Deeley RG, Cole SP. Functional reconstitution of substrate transport by purified multidrug resistance protein MRP1 (ABCC1) in phospholipid vesicles. J Biol Chem. 2000;275:34166–72. doi: 10.1074/jbc.M004584200. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, Sun D, Xia W, Hung MC, Yu D. Cross-reactivity of C219 anti-p170mdr-1 antibody with p185c-erbB2 in breast cancer cells: cautions on evaluating p170mdr-1. J Natl Cancer Inst. 1997;89:1524–9. doi: 10.1093/jnci/89.20.1524. [DOI] [PubMed] [Google Scholar]
- 51.Engel JB, Schally AV, Halmos G, Baker B, Nagy A, Keller G. Targeted cytotoxicity bombesin analog AN-215 effectively inhibits experimental human breast cancers with low induction of multi-drug resistance proteins. Endocrine-Related Cancer. 2005;12:999–1009. doi: 10.1677/erc.1.01022. [DOI] [PubMed] [Google Scholar]
- 52.Savas B, Kerr PE, Pross HF. Lymphokine-activated killer cell susceptibility and adhesion molecule expression of multidrug resistant breast carcinoma. Cancer Cell Int. 2006;6:24–9. doi: 10.1186/1475-2867-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seelig A. A general pattern for substrate recognition by P-glycoprotein. Eur J Biochem. 1998;251:252–61. doi: 10.1046/j.1432-1327.1998.2510252.x. [DOI] [PubMed] [Google Scholar]
- 54.Altenberg GA, Vanoye CG, Horton JK, Reuss L. Unidirectional fluxes of rhodamine 123 in multidrug-resistant cells: evidence against direct drug extrusion from the plasma membrane. Proc Natl Acad Sci USA. 1994;91:4654–7. doi: 10.1073/pnas.91.11.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkadi B, Müller M. Search for specific inhibitors of multidrug resistance in cancer. Semin Cancer Biol. 1997;8:171–82. doi: 10.1006/scbi.1997.0069. [DOI] [PubMed] [Google Scholar]
- 56.Eytan GD, Regev R, Oren G, Hurwitz CD, Assaraf YG. Efficiency of P-glycoprotein-mediated exclusion of rhodamine dyes from multidrug-resistant cells is determined by their passive transmembrane movement rate. Eur J Biochem. 1997;248:104–12. doi: 10.1111/j.1432-1033.1997.00104.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.SI1-SI3.