Abstract
Reconstruction algorithms for diffuse optical tomography (DOT) and bioluminescence tomography (BLT) have been developed based on diffusion theory. The algorithms numerically solve the diffusion equation using the finite element method. The direct measurements of the uncalibrated light fluence rates by a camera are used for the reconstructions. The DOT is self-calibrated by using all possible pairs of transmission images obtained with external sources along with the relative values of the simulated data and the calculated Jacobian. The reconstruction is done in the relative domain with the cancelation of any geometrical or optical factors. The transmission measurements for the DOT are used for calibrating the bioluminescence measurements at each wavelength and then a normalized system of equations is built up which is self-calibrated for the BLT. The algorithms have been applied to a three dimensional model of the mouse (MOBY) segmented into tissue regions which are assumed to have uniform optical properties. The DOT uses the direct method for calculating the Jacobian. The BLT uses a reduced space of eigenvectors of the Green's function with iterative shrinking of the permissible source region. The reconstruction results of the DOT and BLT algorithms show good agreement with the actual values when using either absolute or relative data. Even a small calibration error causes significant degradation of the reconstructions based on absolute data.
OCIS codes: (170.6960) Tomography, (170.3880) Medical and biological imaging
1. Introduction
Bioluminescence imaging (BLI) is a noninvasive technique that can monitor biological processes at a molecular level and visualize disease progression and response to treatment [1–4]. In its simplest form, a sensitive camera is used to image light that reaches the surface from the internal bioluminescence source. These images can provide useful information but they are not quantitative. Bioluminescence tomography (BLT) attempts to reconstruct the three dimensional distribution of the bioluminescent source (i.e. optical power per unit volume) and to provide true quantitative information about its magnitude and location [5–9]. Many different strategies have been proposed for BLT [10–18] but, in general, three components are necessary: a forward model of light propagation from the internal source to the animal surface, a calibration of the camera that links the image (e.g. counts per pixel) to the emittance (optical power per unit area), and a reconstruction algorithm, usually iterative, that estimates the source distribution that minimizes the difference between the observed images and those calculated by application of the forward model.
The second component, camera calibration, typically requires three sub-components: an assumption that the emission from the animal surface is Lambertian [19], modeling of the camera optics that transfers the light to the image plane, and measurement of the absolute camera response on a pixel-by-pixel basis. Recent work by Kienle and Foschum showed that the Lambertian approximation is a poor description of the true surface emission [20]. In addition, measurement of the camera response and modeling of the optics are challenging technical problems. In this paper we propose an alternative strategy for BLT. In addition to the bioluminescence image, a transmission image (or images) is acquired using an external fiber optic source (or sources) of known optical power. The reconstruction algorithm uses the ratio of the internal source image to the external source image as input data. Because the angular dependence of the emission at any point on the surface is very likely the same for both sources, and because the same camera is used to capture both images, the camera calibration problem is reduced to a simple mapping of the animal surface to the camera image plane. In our system [21] the surface anatomy is acquired by an integral x-ray CT scanner but other methods, such as structured light imaging, have been applied [22].
Several investigators have concluded that better BLT reconstructions can be achieved if forward calculations take into account the true three dimensional distribution of scattering and absorption coefficients [23,24]. In previous papers [25,26] we have described a strategy whereby this information can be obtained by diffuse optical tomography (DOT). Transmission images acquired with external fiber optics sources and the BLI camera can be used to reconstruct the optical properties which, in turn, are used in the BLT algorithm. Our previous algorithms assumed that a perfect camera calibration (as described in the previous paragraph) was available to link the transmission images to the actual emittance produced by each source. In this paper we propose an alternative strategy for DOT: instead of “absolute data”, ratio images or “relative data” obtained with different source positions are used. If, for example, nine external sources are employed and nine transmission images are acquired, 36 ratio images can be formed and used as input to a DOT algorithm. The relative power of each of the fiber optic sources must also be known. In the simulations presented in this paper we demonstrate that the optical property reconstructions based on these ratios are generally not as accurate as those based on absolute calibration. However, the BLT solutions based on the relative data are comparable in accuracy to those based on absolute data. In addition, we show by example that the relative method outperforms the absolute method if the calibration process is imperfect.
2. DOT algorithm
Although we will use only the continuous wave (CW) solution, for generality we begin with the frequency domain diffusion equation and the Robin-type boundary condition at a specific wavelength given by [27,28]
| (1) |
| (2) |
where is the light fluence rate at position r, wavelength λ, and light modulation frequency f given by ; Ω and are the domain and its boundary, respectively, and the source distribution is given by . The spatial distribution of the tissue optical properties at wavelength λ is given by the absorption coefficient and the diffusion coefficient , where the diffusion coefficient is defined by κ(r, λ) =, where is the reduced scattering coefficient is the scattering coefficient, and g is the anisotropy factor. is a unit vector pointed outwardly normal to , and ξ is derived from Fresnel’s law as [29] , where the critical angle and , n is the tissue refractive index, and the refractive index of the surrounding medium is assumed to be 1.
Equation (1) can be solved numerically using the method of finite elements (FE) [30–32], and the discrete version of Eq. (1) is given by
| (3) |
where A is the forward matrix obtained by discretizing the diffusion operator in Eq. (1). Assuming the object can be segmented into N regions where each element within the region has the same scattering and absorption coefficients, the number of unknown optical coefficients is 2N and the optical properties vector can be written in its transpose form as where and are the scattering and absorption coefficients of region n. Differentiating both sides of Eq. (3) with respect to gives
| (4) |
where the differentiation of the source with respect to the optical properties gives zero, since the source is not a function of the optical properties. The derivative of the forward matrix A with respect to is obtained by changing by and calculating the difference in the forward matrix . The first order derivative of the light fluence rate with respect to the optical properties is obtained from Eq. (4):
| (5) |
The first order derivative of the light fluence rate with respect to the optical properties at the detectors around the object is given by
| (6) |
where d is the detector index and is a vector of length , where is the number of detectors and the matrix consists of the d rows and all columns of the matrix . If the detectors do not coincide with the FE nodes at the boundary, interpolation can be used to get the value from the surrounding nodes. It is assumed that the values of the light fluence rate at the detector locations can be obtained from the measurements of the corresponding pixels of the CCD camera. For a phasor representation of the light fluence rate where , Eq. (6) can be written as
| (7) |
Therefore
| (8) |
The light fluence rate data captured by the detectors around the object can be approximated by a first order Taylor expansion such that
| (9) |
where and are the magnitude and phase of the light fluence rate corresponding to the initial guess . In matrix form, Eq. (9) is given by
| , | (10) |
which can be concisely written as
| (11) |
For multiple external sources, measured data are obtained for each source and so Eq. (11) for sources is modified to
| (12) |
or, concisely, . This equation can be solved iteratively for such that
| (13) |
Equation (13) can be used to reconstruct the optical properties for absolute (or completely calibrated) data, where the magnitude and phase of the light fluence rate at the object boundary can be obtained by heterodyne detection. In the case of non-contact camera detection, usually only the magnitude is measured (i.e., f = 0).
If the measured light fluence rate is not calibrated, the magnitude of the light fluence rate at the detectors is multiplied by an unknown vector of calibration coefficients α and the phase is shifted by an unknown vector of phase delay β. Therefore the measured light fluence rate is given by
| (14) |
and for multiple source measurements
| (15) |
A system of equations to reconstruct the optical properties as in Eq. (12) can be obtained by using the relative uncalibrated measured data to remove the magnification factor vector α and the phase delay vector β such that
| (16) |
or, concisely, .
Therefore, the optical properties can be reconstructed iteratively from the relative measured data and the relative simulated data at the initial guess by using the following equation:
| (17) |
Equation (17) can be used to reconstruct the optical properties by using the relative measurement, and the reconstructed optical properties at iteration i are given by
| (18) |
The relative simulated data are recalculated at every iteration, using the updated optical properties. The relative Jacobian can be updated at every iteration if there is high nonlinearity or less frequently if there are no large changes in the optical properties during these iterations. The solution is obtained when the maximum relative error between the relative measured data and the relative simulated data goes below a certain limit.
3. BLT algorithm
The diffusion equation in Eq. (3) can be written as [26]
| (19) |
where is the Green’s function matrix at wavelength λ and s(λ) is the bioluminescence source distribution. Assuming that the source magnitude is not a function of wavelength or, equivalently, the source spectrum is known in the range of measurement, the light fluence rate at the detectors on the object surface is given by
| (20) |
where d is the detector index at the boundary , and R is the permissible source region index which contains the index of nodes of the finite element mesh inside the domain Ω.
The light fluence rate measured by the CCD camera is related to the light fluence rate at the “detectors” on by
| (21) |
where α is an unknown scaling vector that represents all calibrations required to obtain the right values of the light fluence rate at the detectors. Using Eq. (21), the logarithm of the measured light fluence rates corresponding to the bioluminescence source and the external sources are given by
| (22) |
where is the Green's function where all columns of the matrix are included, which means the permissible region is the whole object. The external sources used for the DOT reconstruction and the measurements resulted from these sources are also used for calibrating the bioluminescence measurements. The calibrating vector α can be removed from Eq. (21), and a calibrated equation can be obtained as
| (23) |
where are the calibrated bioluminescence measurements. For multi-spectral measurements, Eq. (23) for N wavelengths in a normalized form can be given by
| (24) |
or, concisely, . A symmetric system of equations can be obtained from Eq. (24):
| (25) |
As described in detail in a previous paper [33], Eq. (25) can be solved by assuming that the bioluminescence source is written as a superposition of the eigenvectors of the matrix corresponding to non-zero eigenvalues such that , where is a set of eigenvectors of the matrix corresponding to non-zero eigenvalues and M is the number of chosen eigenvectors. The vector consists of the coefficients of the expansion [33]. The equation described above is solved several times for different permissible source regions starting from the whole object size and ending with a few nodes. For every solution, the objective function , which gives the difference between the calculated and measured light fluence rate, is calculated. After all iterations, the solution that minimizes the objective function is considered the best solution. The shrinking of the permissible region after every iteration is done by removing source nodes from the domain R corresponding to low source magnitude as described in [33]
4. Results and discussion
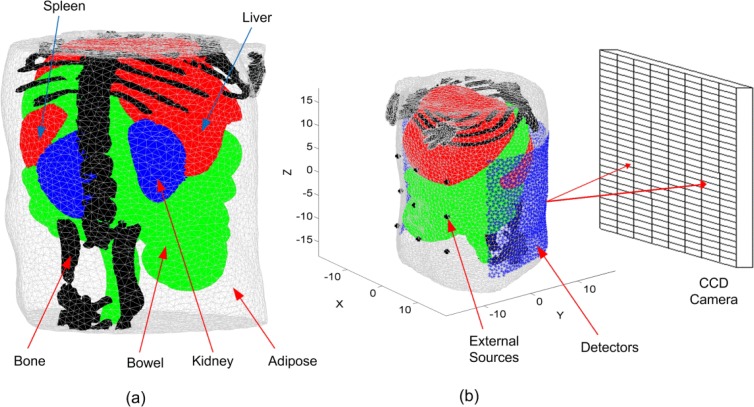
To demonstrate these algorithms, a highly detailed anatomy for a laboratory mouse (MOBY) [34] is used to create the FE mesh. The 3D object (Fig. 1(a) ) represents a finite element mesh of a section of a mouse abdomen where adipose, liver and spleen, bowel and intestine, kidney, and bone tissues are identified and assigned realistic wavelength-dependent optical properties. It is assumed that all elements within each region have the same optical properties. There are 9 optical fibers used as external sources located on the ventral surface of the mouse as shown in Fig. 1(b). It is assumed that these fibers produce Gaussian sources located one transport length inside the object (~1 mm) with a maximum value of 1 nW/mm3 and standard deviation of 1 mm. The measurements corresponding to these sources are used for the DOT reconstruction and BLT calibration at all wavelengths. The detectors shown in Fig. 1(b) are 2183 triangular surface elements and form a field of view of 225 degrees on the dorsal surface. It is assumed that the light fluence rate at these detectors can be obtained from CCD camera images by appropriate geometrical mapping [35].
Fig. 1.
(a) A 3D finite element mesh for the object used in the simulation. The mesh is a regular Delaunay tetrahedral with 17966 nodes and 99140 tetrahedrons generated by iso2mesh [36]. The object represents a section of a mouse abdomen which illustrates 5 different tissue regions: (1) adipose, (2) liver and spleen, (3) bowel and intestine, (4) kidneys, and (5) bone. (b) A schematic of the object showing the sources and detectors set up and the coordinate system.
In the first step, the optical properties of all tissue regions at the wavelengths 580, 590, 600, 610, 620, and 630 nm were reconstructed using the DOT algorithm described above. The light fluence rate at each detector is determined for excitation of each source fiber. However these measurements are not calibrated and are shifted in both magnitude and phase from the correct values as shown in Eq. (15). Equations (16) and (17) show that the optical properties can be reconstructed by using relative measured data, relative simulated data, and the relative Jacobian as shown in Eq. (16). The use of relative values in Eq. (16) cancels out the shift in magnitude and phase of the measured data from the correct calibrated data shown in Eq. (15). The DOT algorithm described above can be used for both frequency domain or continuous wave (CW) . In case of CW, the phase θ and the derivatives in Eq. (10) will be zeros, which can be removed from the equations or left, as there is no effect on the results. The initial estimates used for scattering and absorption for all tissues at all wavelengths are 1 mm−1 and 0.01 mm−1 respectively. The DOT algorithm stops when the maximum relative error of the measured and simulated light fluence rates is less than 1% or the number of iterations exceeds 60.
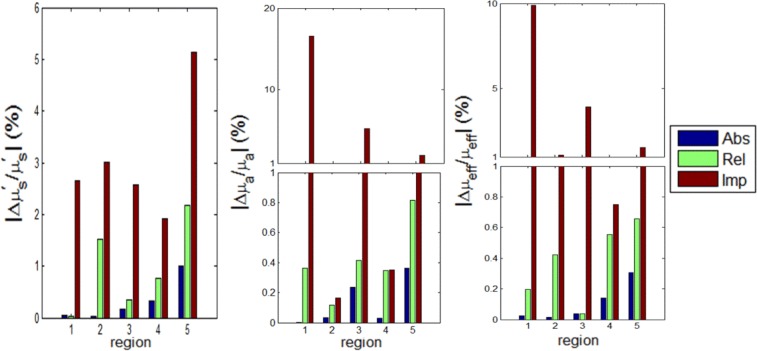
Figure 2 shows a comparison of the relative percentage error in reconstructing the optical properties for CW at wavelength 590 nm using absolute data, relative data, and imperfect calibrated data. The forward model, Eq. (3), is used to simulate the measured data using the actual optical properties and 2% Gaussian noise is added to the calculated data to simulate realistic conditions. The true scattering and absorption coefficients were calculated using the data reported in [24,37] and are shown in Tables 1 and 2 respectively. The corresponding relative errors of reconstructing scattering, absorption, and effective attenuation are shown in the figure. The errors using relative data are larger than when using absolute data. The maximum relative errors obtained in the range 580 nm to 630 nm are 2% for scattering, 3% for absorption, and 1% for effective attenuation when using absolute data while the maximum errors are 9% for scattering, 12% for absorption, and 4% for effective attenuation when using relative data. The simulation took 32 minutes for 60 iterations on a pc with Intel processor with a clock of 3.33 GHz and 24 GB RAM. The Jacobian was recalculated using the new reconstructed optical properties every 10 iterations.
Fig. 2.
The relative percent error in the reconstructions of scattering , absorption ,and effective attenuation , for the 5 tissue regions (1) adipose, (2) liver, (3) bowel, (4) kidney, and (5) bone at 590 nm using absolute, relative, and imperfect calibrations. Note the split vertical scale.
Table 1. True [24,37] reduced scattering coefficients (mm−1) for each region in the heterogeneous object at six different wavelengths.
| 580 nm | 590 nm | 600 nm | 610 nm | 620 nm | 630 nm | |
|---|---|---|---|---|---|---|
| Adipose | 1.30 | 1.29 | 1.28 | 1.27 | 1.26 | 1.25 |
| Liver | 0.79 | 0.77 | 0.76 | 0.75 | 0.74 | 0.72 |
| Bowel | 1.37 | 1.35 | 1.32 | 1.29 | 1.27 | 1.24 |
| Kidney | 2.80 | 2.73 | 2.66 | 2.60 | 2.53 | 2.47 |
| Bone | 3.08 | 3.01 | 2.93 | 2.86 | 2.80 | 2.73 |
Table 2. True [24,37] absorption coefficients (mm−1) for each region in the heterogeneous object at six different wavelengths.
| 580 nm | 590 nm | 600 nm | 610 nm | 620 nm | 630 nm | |
|---|---|---|---|---|---|---|
| Adipose | 0.0073 | 0.0043 | 0.0021 | 0.0014 | 0.0010 | 0.0008 |
| Liver | 0.6474 | 0.3993 | 0.1899 | 0.1200 | 0.0824 | 0.0647 |
| Bowel | 0.0198 | 0.0128 | 0.0063 | 0.0040 | 0.0028 | 0.0023 |
| Kidney | 0.1209 | 0.0746 | 0.0356 | 0.0226 | 0.0156 | 0.0123 |
| Bone | 0.1040 | 0.0670 | 0.0325 | 0.0207 | 0.0142 | 0.0112 |
In Fig. 2 we also show an example of the results obtained when the calibration process is imperfect. There are many possible sources of error in this multi-step process but here we use a simple illustration. One way to calibrate the camera is to use a reference object with known optical properties [38]. We have assumed that this reference object has the same external shape as the Moby phantom and uniform internal optical properties. We further assume that the reduced scattering coefficient is nominally 1.00 mm−1 but actually 1.05 mm−1 and the absorption coefficient is nominally 0.0100 mm−1 but actually 0.0105 mm−1. We performed forward calculations of the transmission for each source-detector pair under nominal and actual conditions. The ratio is the imperfect calibration factor used in all subsequent simulations including the BLT calculations. This 5% error in scattering and absorption coefficients is relatively small and likely underestimates the cumulative error in a real calibration process. With this imperfect calibration the maximum relative errors are 8% for scattering, 55% for absorption, and 35% for effective attenuation.
For BLT, to simulate the measured data using Eq. (23), the true source distribution and the Green's function calculated using the true optical properties at each wavelength given in Tables 1 and 2 were used. 2% Gaussian noise was added to simulate realistic data: in Eq. (23). Two cases are simulated for in Eq. (23): the first case is to set each element in α equal to 1 in Eq. (22) when the data are calibrated and match the correct values at the detectors around the object. In this case there is no need to use external sources for calibrations and in Eq. (23). The second case is to set each element in α equal to 1000 in Eq. (22) to simulate uncalibrated data and normalization with external sources is needed as described in Eq. (23). To simulate the measured data due to external sources, the Green's functions calculated using the true optical properties at each wavelength were used in Eq. (22). The external sources are simulated by Gaussian sources as described before and the elements of α equal 1000. In Eq. (23), the Green’s functions and were calculated using the reconstructed optical properties, by the DOT algorithm described above, using absolute data, relative data, and imperfect calibrated data. The permissible region R was initially chosen to be a cylinder of 20 mm diameter and height 20 mm with its center located at (0, 3, 3). At each iteration, the permissible region is slightly shrunk by an arbitrary factor γ The permissible region initially containing 5010 mesh nodes is finally shrunk to 10 nodes in 60 iterations, γ = (5010/10)(1/60) = 1.1092. The 60 iterations took approximately 30 minutes on a PC with Intel processor with a clock of 3.33 GHz and 24 GB RAM. The execution time varies greatly according to the choice of mesh size, the volume of the initial permissible region, and number of iterations. It is useful to start with a small size mesh and fewer iterations to get a quick initial solution before solving the complete problem. This can provide a more focused permissible region and hence greatly reduce the simulation time and give more accurate results.
Figure 3 shows transverse and coronal cross sections for the actual and reconstructed source distributions. The transverse section passes through the two kidneys at and the coronal section passes through the two kidneys at . The actual sources are spheres of 4 mm diameters located in the two kidneys at (−5, 3, 3) and (6, 3, 3) with magnitudes of 2 nW/ mm3 and 1 nW/ mm3 respectively and total spatially integrated powers of 67.2 nW and 39 nW respectively.
Fig. 3.
BLT reconstruction of two uniform bioluminescence sources localized in the left and right kidneys (4 mm diameters). The “anatomy” is shown in Fig. 1. The actual sources in transverse section (a) and coronal section (b) have magnitudes 2 nW/ mm3 and 1 nW/ mm3 for the left and right source respectively. The reconstructed sources are shown in (c) and (d) using absolute data, (e) and (f) using relative data, and (g) and (h) using imperfect calibrated data. The total powers of the actual and reconstructed sources and the normalized magnitude errors of the reconstructed sources using different reconstruction methods are shown in the figure.
Two objective metrics have been calculated to assess the accuracy of the BLT reconstruction of each source. The first is simply the total power integrated over three dimensions, , where is the volume of the element i in the FE mesh and the summation includes all elements that include the source. The second we define as the “normalized magnitude error” calculated according to
This element-by-element summation is sensitive to the shape and position of the reconstructed source and is a more stringent metric than the total power.
The reconstructed sources in Figs. 3(c) and 3(d) using absolute data and in 3(e) and 3(f) using relative data show good agreement with the actual sources in Figs. 3(a) and 3(b) in terms of location, total power reconstructed (within 0.75%) and magnitudes with normalized magnitude errors of 0.27. There are no significant differences in the reconstruction results when using absolute or relative data. The reconstructed sources using imperfect calibrated data are shown in Figs. 3(g) and 3(h) with total power reconstructed (within 6%) and normalized magnitude error of 0.54.
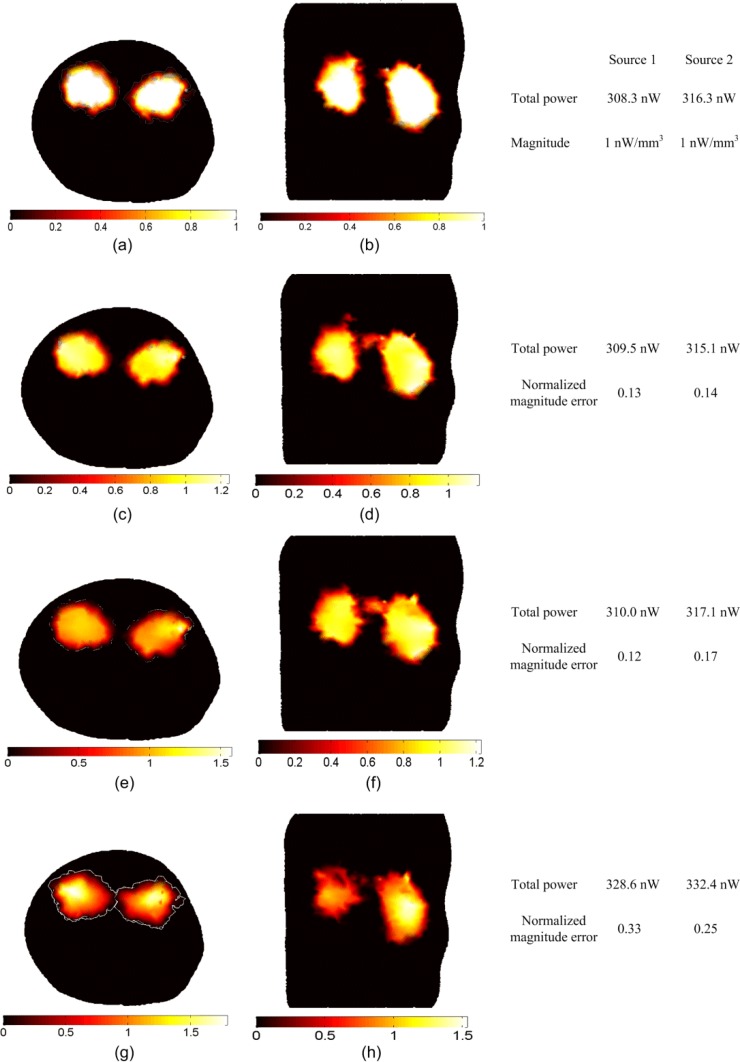
Figure 4 shows the result of the reconstruction of large uniform sources that fill the entire two kidneys; the actual sources in Figs. 4(a) and 4(b) are of uniform magnitude 1 nW/mm3. The total powers of the sources are 308.3 nW in the left kidney and 316.3 nW in the right kidney. The reconstructed sources in Figs. 4(c) and 4(d) using absolute data and in 4(e) and 4(f) using relative data show comparable results in terms of the total power reconstructed which is within 0.63% of the actual values and the normalized magnitude errors which are 0.14 and 0.17 respectively. The reconstructed sources using imperfect calibration in Figs. 4(g) and 4(h) have total power reconstructed within 6.3% and normalized magnitude error of 0.54.
Fig. 4.
BLT reconstruction of large distributed sources that fill the entire two kidneys. The magnitude of the actual sources in (a) and (b) is 1 nW/mm3, and the reconstructed sources are shown in (c) and (d) using absolute data, (e) and (f) using relative data, and (g) and (h) using imperfect calibrated data. The total powers of the actual and reconstructed sources and the normalized magnitude errors of the reconstructed sources using different reconstruction methods are shown in the figure.
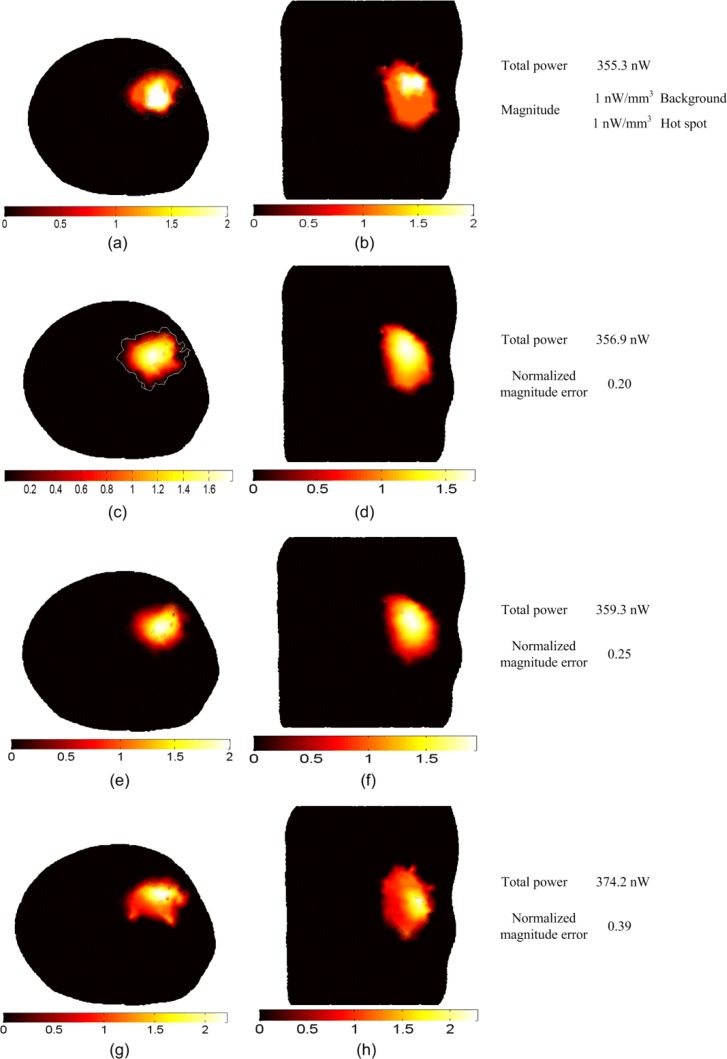
Figure 5 shows the BLT reconstruction for a large non-uniform source that fills the entire right kidney. There is a 4 mm diameter “hot spot” of 2 nW/mm3 located at (6, 3, 3) inside the kidney while the rest of the organ “background” of 1 nW/mm3 as shown in Figs. 5(a) and 5(b). The total power of the actual source is 355.3 nW which is reconstructed within 1% when using absolute data in Figs. 5(c) and 5(d) or relative data in 5(e) and 5(f). The hot spot can be recognized in both cases but the location is slightly more accurate when absolute data are used. The normalized magnitude error is 0.2 using absolute data and 0.25 while using relative data. The total power is reconstructed within 5.3% using imperfect calibration and the normalized magnitude error is 0.39. The hot spot location is less accurate than that using absolute or relative data.
Fig. 5.
BLT reconstruction of a large non-uniform source distribution that fills the entire right kidney. The actual source in (a) and (b) has a hot spot of 4 mm diameter with magnitude 2 nW/mm3 which is twice the magnitude of the background. The reconstructed source in (c) and (d) using the absolute data, (e) and (f) using the relative data, and (g) and (g) using the imperfect calibrated data. The total powers of the actual and reconstructed sources and the normalized magnitude errors of the reconstructed sources using different reconstruction methods are shown in the figure.
5. Conclusion
Self-calibrated algorithms for three dimensional DOT and BLT based on the finite element solution of the diffusion equation have been developed. Both algorithms avoid absolute calibration of the detectors as by using ratios, camera images can be used for reconstructing the optical properties and bioluminescence sources. The algorithms have been applied to simulations based on a highly detailed anatomy for a laboratory mouse (MOBY) and show good agreement between the actual values of optical properties and bioluminescence sources and the reconstructions. The DOT algorithm used the direct method to calculate the Jacobian. Using relative values for the measured data, simulated data, and Jacobian showed good reconstruction results compared to using absolute values. The same measurements due to external sources used for DOT at each wavelength were also used to calibrate the bioluminescence measurements. The reconstruction results for BLT using relative calibration show good agreements with the results using absolute data. The BLT algorithms used the eigenvectors expansion of the Green's function as a reduced space for the reconstruction, and iterative shrinking of the permissible source region. Small multiple sources and large homogeneous and inhomogeneous source distributions were reconstructed using both absolute and relative data. The results showed good agreement between the reconstructed and actual sources in terms of location and total power and magnitude. The BLT reconstructions based on relative data are slightly inferior to those based on absolute data obtained with perfect calibration. However, any calibration procedure will be imperfect, and we have demonstrated that even small errors in the calibration can lead to performance that is worse than that based on relative data.
Acknowledgment
This work was supported in part by National Institutes of Health grant RO1 CA158100.
References and links
- 1.Ntziachristos V., Ripoll J., Wang L. V., Weissleder R., “Looking and listening to light: the evolution of whole-body photonic imaging,” Nat. Biotechnol. 23(3), 313–320 (2005). 10.1038/nbt1074 [DOI] [PubMed] [Google Scholar]
- 2.Weissleder R., Mahmood U., “Molecular imaging,” Radiology 219(2), 316–333 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Willmann J. K., van Bruggen N., Dinkelborg L. M., Gambhir S. S., “Molecular imaging in drug development,” Nat. Rev. Drug Discov. 7(7), 591–607 (2008). 10.1038/nrd2290 [DOI] [PubMed] [Google Scholar]
- 4.Tian J., Bai J., Yan X. P., Bao S., Li Y., Liang W., Yang X., “Multimodality molecular imaging,” IEEE Eng. Med. Biol. Mag. 27(5), 48–57 (2008). 10.1109/MEMB.2008.923962 [DOI] [PubMed] [Google Scholar]
- 5.Chaudhari A. J., Darvas F., Bading J. R., Moats R. A., Conti P. S., Smith D. J., Cherry S. R., Leahy R. M., “Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging,” Phys. Med. Biol. 50(23), 5421–5441 (2005). 10.1088/0031-9155/50/23/001 [DOI] [PubMed] [Google Scholar]
- 6.Ahn S., Chaudhari A. J., Darvas F., Bouman C. A., Leahy R. M., “Fast iterative image reconstruction methods for fully 3D multispectral bioluminescence tomography,” Phys. Med. Biol. 53(14), 3921–3942 (2008). 10.1088/0031-9155/53/14/013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comsa D. C., Farrell T. J., Patterson M. S., “Quantification of bioluminescence images of point source objects using diffusion theory models,” Phys. Med. Biol. 51(15), 3733–3746 (2006). 10.1088/0031-9155/51/15/009 [DOI] [PubMed] [Google Scholar]
- 8.Kuo C., Coquoz O., Troy T. L., Xu H., Rice B. W., “Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging,” J. Biomed. Opt. 12(2), 024007 (2007). 10.1117/1.2717898 [DOI] [PubMed] [Google Scholar]
- 9.Dehghani H., Davis S. C., Jiang S., Pogue B. W., Paulsen K. D., Patterson M. S., “Spectrally resolved bioluminescence optical tomography,” Opt. Lett. 31(3), 365–367 (2006). 10.1364/OL.31.000365 [DOI] [PubMed] [Google Scholar]
- 10.Cong W., Wang G., Kumar D., Liu Y., Jiang M., Wang L. V., Hoffman E. A., McLennan G., McCray P. B., Zabner J., Cong A., “Practical reconstruction method for bioluminescence tomography,” Opt. Express 13(18), 6756–6771 (2005). 10.1364/OPEX.13.006756 [DOI] [PubMed] [Google Scholar]
- 11.He X., Liang J., Qu X., Huang H., Hou Y., Tian J., “Truncated total least squares method with a practical truncation parameter choice scheme for bioluminescence tomography inverse problem,” Int. J. Biomed. Imaging 2010, 12 (2010). 10.1155/2010/291874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H., Qu X., Liang J., He X., Chen X., Yang D., Tian J., “A multi-phase level set framework for source reconstruction in bioluminescence tomography,” J. Comput. Phys. 229(13), 5246–5256 (2010). 10.1016/j.jcp.2010.03.041 [DOI] [Google Scholar]
- 13.Feng J., Jia K., Yan G., Zhu S., Qin C., Lv Y., Tian J., “An optimal permissible source region strategy for multispectral bioluminescence tomography,” Opt. Express 16(20), 15640–15654 (2008). 10.1364/OE.16.015640 [DOI] [PubMed] [Google Scholar]
- 14.Lu Y., Zhang X., Douraghy A., Stout D., Tian J., Chan T. F., Chatziioannou A. F., “Source reconstruction for spectrally-resolved bioluminescence tomography with sparse a priori information,” Opt. Express 17(10), 8062–8080 (2009). 10.1364/OE.17.008062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohajerani P., Eftekhar A. A., Huang J., Adibi A., “Optimal sparse solution for fluorescent diffuse optical tomography: theory and phantom experimental results,” Appl. Opt. 46(10), 1679–1685 (2007). 10.1364/AO.46.001679 [DOI] [PubMed] [Google Scholar]
- 16.Cao N., Nehorai A., Jacobs M., “Image reconstruction for diffuse optical tomography using sparsity regularization and expectation-maximization algorithm,” Opt. Express 15(21), 13695–13708 (2007). 10.1364/OE.15.013695 [DOI] [PubMed] [Google Scholar]
- 17.Gu X., Zhang Q., Larcom L., Jiang H., “Three-dimensional bioluminescence tomography with model-based reconstruction,” Opt. Express 12(17), 3996–4000 (2004). 10.1364/OPEX.12.003996 [DOI] [PubMed] [Google Scholar]
- 18.Slavine N. V., Lewis M. A., Richer E., Antich P. P., “Iterative reconstruction method for light emitting sources based on the diffusion equation,” Med. Phys. 33(1), 61–68 (2006). 10.1118/1.2138007 [DOI] [PubMed] [Google Scholar]
- 19.Chen X., Gao X., Chen D., Ma X., Zhao X., Shen M., Li X., Qu X., Liang J., Ripoll J., Tian J., “3D reconstruction of light flux distribution on arbitrary surfaces from 2D multi-photographic images,” Opt. Express 18(19), 19876–19893 (2010). 10.1364/OE.18.019876 [DOI] [PubMed] [Google Scholar]
- 20.Kienle A., Foschum F., “250 years Lambert surface: does it really exist?” Opt. Express 19(5), 3881–3889 (2011). 10.1364/OE.19.003881 [DOI] [PubMed] [Google Scholar]
- 21.J. Pekar, “Multispectral bioluminescence tomography with x-ray CT spatial priors” Ph.D. thesis (McMaster University, Hamilton, 2011), Open Access Dissertations and Theses, Paper 4329, http://digitalcommons.mcmaster.ca/opendissertations/4329
- 22.Chaudhari M. A. J., Joshi A. A., Darvas F., Leahy R. M., “A method for atlas-based volumetric registration with surface constraints for optical bioluminescence tomography in small animal imaging,” Proc. SPIE 6510, 651024–, 651024–10. (2007). 10.1117/12.710197 [DOI] [Google Scholar]
- 23.Dehghani H., Pogue B. W., Davis S. C., Patterson M. S., “Modeling and image reconstruction in spectrally resolved bioluminescence tomography,” Proc. SPIE 6434, 64340V–, 64340V–9. (2007). 10.1117/12.698931 [DOI] [Google Scholar]
- 24.Alexandrakis G., Rannou F. R., Chatziioannou A. F., “Tomographic bioluminescence imaging by use of a combined optical-PET (OPET) system: a computer simulation feasibility study,” Phys. Med. Biol. 50(17), 4225–4241 (2005). 10.1088/0031-9155/50/17/021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naser M. A., Patterson M. S., “Algorithms for bioluminescence tomography incorporating anatomical information and reconstruction of tissue optical properties,” Biomed. Opt. Express 1(2), 512–526 (2010). 10.1364/BOE.1.000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naser M. A., Patterson M. S., “Improved bioluminescence and fluorescence reconstruction algorithms using diffuse optical tomography, normalized data, and optimized selection of the permissible source region,” Biomed. Opt. Express 2(1), 169–184 (2011). 10.1364/BOE.2.000169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arridge S. R., “Optical tomography in medical imaging,” Inverse Probl. 15(2), R41–R93 (1999). 10.1088/0266-5611/15/2/022 [DOI] [Google Scholar]
- 28.Klose A. D., “Transport-theory-based stochastic image reconstruction of bioluminescence sources,” J. Opt. Soc. Am. A 24(6), 1601–1608 (2007). 10.1364/JOSAA.24.001601 [DOI] [PubMed] [Google Scholar]
- 29.Dehghani H., Eames M. E., Yalavarthy P. K., Davis S. C., Srinivasan S., Carpenter C. M., Pogue B. W., Paulsen K. D., “Near infrared optical tomography using NIRFAST: Algorithm for numerical model and image reconstruction,” Commun. Numer. Methods Eng. 25(6), 711–732 (2009). 10.1002/cnm.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arridge S. R., Schweiger M., Hiraoka M., Delpy D. T., “A finite element approach for modeling photon transport in tissue,” Med. Phys. 20(2), 299–309 (1993). 10.1118/1.597069 [DOI] [PubMed] [Google Scholar]
- 31.Schweiger M., Arridge S. R., Hiraoka M., Delpy D. T., “The finite element method for the propagation of light in scattering media: boundary and source conditions,” Med. Phys. 22(11), 1779–1792 (1995). 10.1118/1.597634 [DOI] [PubMed] [Google Scholar]
- 32.Jiang H., “Frequency-domain fluorescent diffusion tomography: a finite-element-based algorithm and simulations,” Appl. Opt. 37(22), 5337–5343 (1998). 10.1364/AO.37.005337 [DOI] [PubMed] [Google Scholar]
- 33.Naser M. A., Patterson M. S., “Bioluminescence tomography using eigenvectors expansion and iterative solution for the optimized permissible source region,” Biomed. Opt. Express 2(11), 3179–3193 (2011). 10.1364/BOE.2.003179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segars W. P., Tsui B. M., Frey E. C., Johnson G. A., Berr S. S., “Development of a 4-D digital mouse phantom for molecular imaging research,” Mol. Imaging Biol. 6(3), 149–159 (2004). 10.1016/j.mibio.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 35.Da Silva A., Leabad M., Driol C., Bordy T., Debourdeau M., Dinten J. M., Peltié P., Rizo P., “Optical calibration protocol for an x-ray and optical multimodality tomography system dedicated to small-animal examination,” Appl. Opt. 48(10), D151–D162 (2009). 10.1364/AO.48.00D151 [DOI] [PubMed] [Google Scholar]
- 36.Q. Fang and D. Boas, “Tetrahedral mesh generation from volumetric binary and gray-scale images,” in IEEE International Symposium on Biomedical Imaging: from Nano to Macro,2009. ISBI '09 (IEEE, 2009), pp. 1142–1145. [Google Scholar]
- 37.S. A. Prahl, “Optical properties spectra,” (Oregon Medical Laser Clinic, 2001), http://omlc.ogi.edu/spectra/index.html
- 38.Hervé L., Koenig A., Da Silva A., Berger M., Boutet J., Dinten J. M., Peltié P., Rizo P., “Noncontact fluorescence diffuse optical tomography of heterogeneous media,” Appl. Opt. 46(22), 4896–4906 (2007). 10.1364/AO.46.004896 [DOI] [PubMed] [Google Scholar]