Abstract
The infectious form of many parasitic nematodes, which afflict over one billion people globally, is a developmentally arrested third-stage larva (L3i). The parasitic nematode Strongyloides stercoralis differs from other nematode species that infect humans, in that its life cycle includes both parasitic and free-living forms, which can be leveraged to investigate the mechanisms of L3i arrest and activation. The free-living nematode Caenorhabditis elegans has a similar developmentally arrested larval form, the dauer, whose formation is controlled by four pathways: cyclic GMP (cGMP) signaling, insulin/IGF-1-like signaling (IIS), transforming growth factor β (TGFβ) signaling, and biosynthesis of dafachronic acid (DA) ligands that regulate a nuclear hormone receptor. We hypothesized that homologous pathways are present in S. stercoralis, have similar developmental regulation, and are involved in L3i arrest and activation. To test this, we undertook a deep-sequencing study of the polyadenylated transcriptome, generating over 2.3 billion paired-end reads from seven developmental stages. We constructed developmental expression profiles for S. stercoralis homologs of C. elegans dauer genes identified by BLAST searches of the S. stercoralis genome as well as de novo assembled transcripts. Intriguingly, genes encoding cGMP pathway components were coordinately up-regulated in L3i. In comparison to C. elegans, S. stercoralis has a paucity of genes encoding IIS ligands, several of which have abundance profiles suggesting involvement in L3i development. We also identified seven S. stercoralis genes encoding homologs of the single C. elegans dauer regulatory TGFβ ligand, three of which are only expressed in L3i. Putative DA biosynthetic genes did not appear to be coordinately regulated in L3i development. Our data suggest that while dauer pathway genes are present in S. stercoralis and may play a role in L3i development, there are significant differences between the two species. Understanding the mechanisms governing L3i development may lead to novel treatment and control strategies.
Author Summary
Parasitic nematodes infect over one billion people worldwide and cause many diseases, including strongyloidiasis, filariasis, and hookworm disease. For many of these parasites, including Strongyloides stercoralis, the infectious form is a developmentally arrested and long-lived thirdstage larva (L3i). Upon encountering a host, L3i quickly resume development and mature into parasitic adults. In the free-living nematode Caenorhabditis elegans, a similar developmentally arrested third-stage larva, known as the dauer, is regulated by four key cellular mechanisms. We hypothesized that similar cellular mechanisms control L3i arrest and activation. Therefore, we used deep-sequencing technology to characterize the S. stercoralis transcriptome (RNAseq), which allowed us to identify S. stercoralis homologs of components of these four mechanisms and examine their temporal regulation. We found similar temporal regulation between S. stercoralis and C. elegans for components of two mechanisms, but dissimilar temporal regulation for two others, suggesting conserved as well as novel modes of developmental regulation for L3i. Understanding L3i development may lead to novel control strategies as well as new treatments for strongyloidiasis and other diseases caused by parasitic nematodes.
Introduction
Parasitic nematodes infect over one billion people worldwide, resulting in vast morbidity [1], as well as causing significant agricultural losses from infections of both animals and plants [2]. The infectious form of many parasitic nematodes, including those causing hookworm disease, filariasis, and strongyloidiasis, is a developmentally arrested third-stage larva (L3i), which is both stress-resistant and long-lived [3]–[5]. Upon entering a suitable host, L3i quickly resume development (activation), eventually forming parasitic adults [4], [5]. The genes and proteins constituting the pathways that control the developmental arrest and activation of L3i represent potential targets for chemotherapy as well as environmental control strategies.
Our lab uses the parasitic nematode Strongyloides stercoralis, which infects 30–100 million people globally [1], to study mechanisms controlling L3i arrest and activation [6]. S. stercoralis has a complex life-cycle (Figure 1), which includes both an obligatory parasitic generation as well as a facultative free-living generation. Parasitic females reproduce parthenogenetically to produce post-parasitic larvae, which develop either directly to L3i (homogonic/direct development) or to free-living males and females (heterogonic/indirect development). Post-free-living larvae constitutively form L3i [7]. This life cycle allows us to investigate the mechanisms underlying different developmental fates for similar larval forms. Additionally, we have developed molecular tools in S. stercoralis, which are unavailable in other parasitic nematodes, to investigate molecular mechanisms involved in L3i regulation [8]–[10].
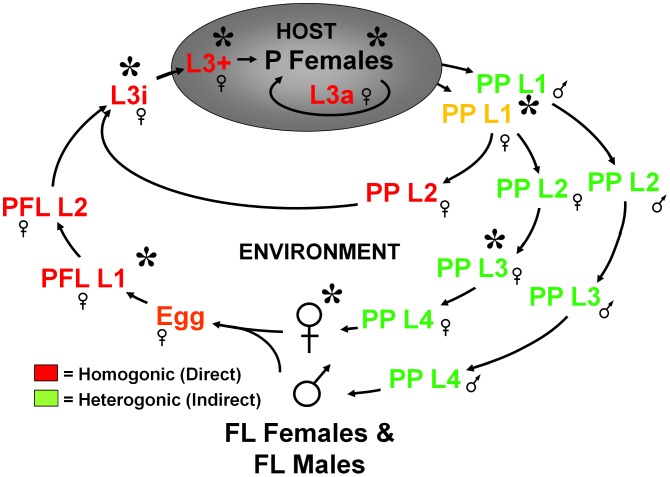
Figure 1. Diagram of the Strongyloides stercoralis life cycle.
Developmentally arrested infective third-stage larvae (L3i) can form by either a homogonic route (dark red) or a heterogonic route (light green). Female post-parasitic first-stage larvae (PP L1) passed in the feces of the infected host can develop homogonically through two larval molts directly to L3i or heterogonically through four larval molts to free-living females (FL Females). Post-parasitic L1 males invariably develop heterogonically through four molts to free-living males (FL Males). Post-free-living L1 (PFL L1), which are all female, molt twice and develop exclusively to L3i. Upon encountering and penetrating a susceptible host, activated third-stage larvae (L3+) resume feeding and development, migrate to the intestines, and molt twice into parasitic females (P Females). Post-parasitic L1 larvae can also precociously develop into auto-infective third-stage larvae (L3a) entirely within the host. Developmental stages marked with an asterisk (*) were interrogated by RNAseq. Adapted from [7].
The free-living nematode Caenorhabditis elegans has a developmentally arrested third-stage dauer larva, morphologically similar to L3i, which forms during conditions of low food abundance, high temperature, and high dauer pheromone levels reflecting high population density. Dauer larvae quickly resume development into reproductive adults once environmental conditions improve. Mutant screens in C. elegans have identified over 30 genes that are involved in dauer formation (daf), and mutations in these genes result in either dauer constitutive (daf-c) or dauer defective (daf-d) phenotypes. Extensive study has placed many of these daf genes into four dauer pathways (Figure 2): a cyclic guanosine monophosphate (cGMP) signaling pathway, an insulin/IGF-1-like signaling (IIS) pathway regulated by insulin-like peptide (ILP) ligands, a dauer transforming growth factor β (TGFβ) pathway regulated by the Ce-DAF-7 ligand, and a nuclear hormone receptor (NHR) regulated by a class of steroid ligands known as dafachronic acids (DAs) [11]. Epistatic analysis places the cGMP signaling pathway upstream of the parallel IIS and dauer TGFβ pathways, which converge on the DA biosynthetic pathway, ultimately regulating the NHR Ce-DAF-12 (Figure 2) [12]. A long-standing paradigm in the field, known as the “dauer hypothesis,” proposes that similar molecular mechanisms regulate the developmental arrest and activation of both C. elegans dauer larvae and L3i of parasitic nematodes [4], [13]–[15], despite their high degree of evolutionary divergence [16], [17].
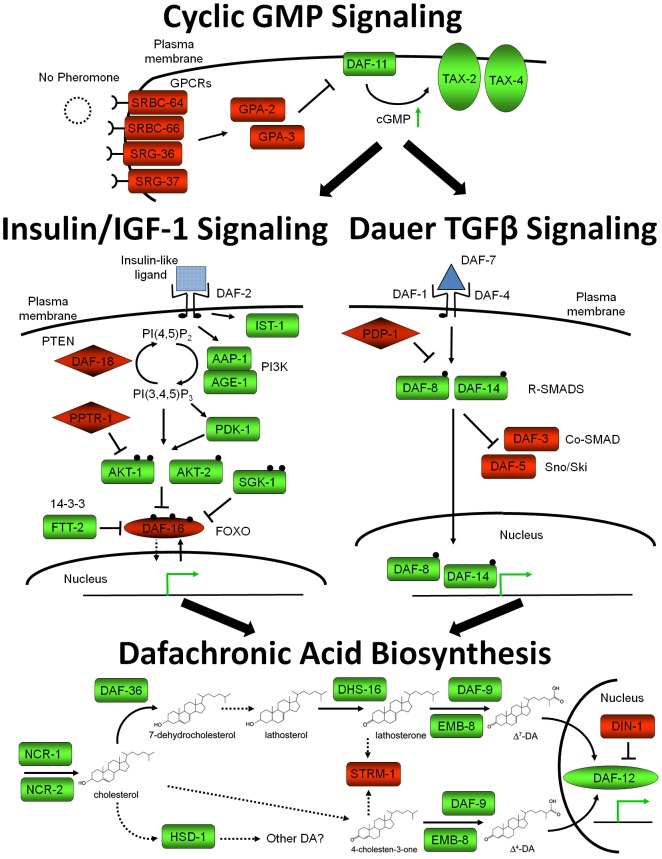
Figure 2. Caenorhabditis elegans dauer pathways during reproductive development.
Four developmental pathways regulate C. elegans dauer entry and exit: a cyclic guanosine monophosphate (GMP) signaling pathway, an insulin/insulin-like growth factor 1 (IGF-1) -like signaling pathway, a dauer transforming growth factor β (TGFβ) pathway, and a nuclear hormone receptor (DAF-12) regulated by a class of steroid ligands known as dafachronic acids (DAs). This simplified model depicts the four pathways under conditions favoring reproductive development and repression of dauer arrest. Proteins in light green are “active,” while proteins in dark red are “inactive.” Black circles represent phosphorylation sites and diamond-shaped boxes represent phosphatases. Green arrows represent either increases in metabolite concentration or increases in gene transcription. Solid black lines represent well-established pathways, while dashed lines represent putative pathways. Adapted from [12], [54], [111], [129].
Members from each of the four dauer pathways have been cloned in S. stercoralis [18]–[22]; however, it is unclear whether all members from each of the C. elegans pathways are present in this parasite, whether their anatomical and temporal regulation is similar to C. elegans, and whether they control L3i development in S. stercoralis. While we have demonstrated that S. stercoralis IIS plays a crucial role in post-free-living L3i arrest and activation [10], [22], we have also shown that an S. stercoralis TGFβ ligand encoding gene, Ss-tgh-1, is transcriptionally regulated in a manner opposite to that of the C. elegans TGFβ ligand encoding gene Ce-daf-7 [20], [23]. Studies examining the global transcriptional changes during S. stercoralis L3i development have failed to identify specific pathways regulating L3i development and have not directly shown whether pathways regulating dauer in C. elegans are similarly regulated in S. stercoralis [24], [25]. However, these studies have been hindered by a small expressed sequence tag (EST) database, which does not include homologs for many C. elegans dauer genes.
To overcome these obstacles, we used a next-generation RNA sequencing (RNAseq) approach aided by the concurrent release of draft Strongyloides ratti and S. stercoralis genome sequences. Similar to recent work in other parasitic nematode species [26]–[31], we isolated polyadenylated RNA from seven different S. stercoralis developmental stages (Figure 1), from which we constructed dsDNA libraries that were subjected to high-throughput sequencing. Using both S. ratti and S. stercoralis genomic contigs as well as de novo assembled RNAseq transcripts, we identified S. stercoralis homologs of C. elegans genes involved in dauer regulation and examined their temporal regulation throughout the S. stercoralis life cycle using a collection of over 2.3 billion paired-end reads.
While we identified S. stercoralis homologs of nearly all C. elegans dauer genes, some of which appear to have similar developmental regulation between the two species, we also identified multiple differences between C. elegans dauer genes and their S. stercoralis homologs, including protein structure, developmental regulation, and expansion of gene families. Both IIS and cGMP signaling appear to be regulated in a manner consistent with a role in L3i regulation, while genes putatively involved in DA biosynthesis were not coordinately regulated during L3i development. S. stercoralis dauer-like TGFβ signaling was regulated oppositely to that observed in C. elegans; nevertheless, this pathway may play a unique role in S. stercoralis L3i development.
Materials and Methods
Ethics statement
The S. stercoralis PV001 strain was maintained in prednisolone-treated beagles in accordance with protocols 702342, 801905, and 802593 approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). Experimental infections of S. stercoralis were conducted in Mongolian gerbils under the same IACUC-approved protocols, and animals were sacrificed by CO2 asphyxiation in accordance with standards established by the American Veterinary Medical Association. All IACUC protocols, as well as routine husbandry care of the animals, were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
S. stercoralis maintenance and RNA isolation
The S. stercoralis PV001 line, derived from a single female worm [22], was maintained and cultured as previously described [6], [32], [33]. S. stercoralis developmental stages were isolated as previously described [22]; see supplemental methods for detailed protocol (Text S1). Both L3+, which had resumed development as evidenced by changes in morphology and resumption of feeding (Figure S1), and parasitic females were derived from experimental infections of Mongolian gerbils, a permissive host [33]. All developmental stages, except for parasitic females and L3+, were rendered free of fine particle debris by migration through agarose [34] into BU buffer [35]. Worms were snap-frozen in TRIzol reagent (Life Technologies, http://www.lifetechnologies.com) in liquid nitrogen; total RNA was extracted using the manufacturer's protocol. Total RNA was quantified using the Bioanalyzer 2100 (Agilent Technologies, Inc., http://www.agilent.com), and only samples with an RNA integrity number (RIN) greater than 8.0 were used.
S. stercoralis polyadenylated RNA library construction and sequencing
Libraries were constructed using the TruSeq RNA Sample Preparation Kit (Illumina, Inc., http://www.illumina.com) according to the manufacturer's protocol. For each of the 21 libraries, 500 ng of total RNA, diluted to 10 ng/µl in de-ionized water, was used as starting material. Polyadenylated RNA enrichment was performed first using olido-dT beads and eluted polyadenylated RNA fragmented at 94°C for eight minutes to approximately 170±50 (standard deviation) bp. Subsequently, first and second strand cDNA was synthesized; unique adapters for each replicate were then ligated. dsDNA fragments with ligated adapters were enriched using 15 cycles of PCR. Libraries were assessed for fragment size distribution using the Bioanalyzer 2100.
The concentration of the dsDNA adapter-ligated libraries was then determined by quantitative PCR (qPCR) using the Kapa SYBR Fast qPCR Kit for Library Quantification (Kapa Biosystems, Inc., http://www.kapabiosystems.com) using the manufacturer's protocol. Three dilutions, at 1∶4,000, 1∶8,000, and 1∶16,000, were used to calculate the concentration of each of the 21 libraries using a calibration curve of Kapa standards. Each library was then diluted to 15 nM, and libraries from each developmental stage were pooled in equal volume quantities. The concentration of each of these pools was determined using qPCR and diluted to a final concentration of 10 nM.
The quality of the pooled libraries from each of the seven developmental stages was assessed using the High Sensitivity DNA Assay (Agilent Technologies). Pooled libraries were loaded on individual lanes of the Illumina HiSeq 2000 flow cell at 4 pM for all libraries, except for the post-free-living L1 and parasitic female libraries, which were loaded at 3 pM. Samples were then sequenced on the Illumina HiSeq 2000 with 100 bp paired-end reads, with image analysis and base calling performed with HiSeq Control Software. Raw flow-cell data was processed and demultiplexed using CASAVA version 1.8.2 (Illumina) for each of the 21 samples (ArrayExpress accession number E-MTAB-1164; http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1164).
Alignment of S. stercoralis RNAseq reads to genomic contigs
Raw reads from each of the 21 samples were independently aligned to S. stercoralis genomic contigs (6 December 2011 draft; ftp://ftp.sanger.ac.uk/pub/pathogens/HGI/) using TopHat version 1.4.1 (http://tophat.cbcb.umd.edu/), which utilized the Bowtie aligner version 0.12.7 (http://bowtie-bio.sourceforge.net/index.shtml) and SAMtools version 0.1.18 (http://samtools.sourceforge.net/). We refined the alignment parameters until TopHat accurately predicted introns and exons of several known S. stercoralis genes. Default parameters were used, but with the following options: mate inner distance of 25; mate standard deviation of 50; minimum anchor length of 6; minimum intron length of 30; maximum intron length of 20,000; micro exon search; minimum segment intron of 30; and maximum segment intron of 20,000. Aligned reads from each developmental stage were inspected using the Integrated Genome Viewer (IGV) version 2.0.34 (http://www.broadinstitute.org/igv/).
De novo assembly of developmental stage-specific S. stercoralis transcripts
RNAseq reads from the sample with the greatest number of reads for each stage were independently de novo assembled into transcripts. First, forward and reverse read pairs were merged to form a single “contig” using SeqPrep (https://github.com/jstjohn/SeqPrep), with a quality score cutoff of 35, a minimum merged read length of 100 bp, and no mismatches in the overlapping region. The two read contigs were then trimmed with the FASTX toolkit quality trimmer (http://hannonlab.cshl.edu/fastx_toolkit/) to remove bases from the ends with a quality score less than 35. These high quality contigs were then de novo assembled via Trinity release 2012-04-27 (http://trinityrnaseq.sourceforge.net/) using “jellyfish” for k-mer counting. The de novo assembled transcripts from each developmental stage (ArrayExpress accession number E-MTAB-1184; http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1184) were tagged with the name of the developmental stage from which they were derived and merged into a single FASTA file. This FASTA file was then searched using the custom BLAST feature in Geneious version 5.5.6 (http://www.geneious.com/) [36] to search for S. stercoralis homologs of C. elegans genes.
Identification and annotation of S. stercoralis genes
BLAST searches of the S. stercoralis (ftp://ftp.sanger.ac.uk/pub/pathogens/HGI/) and S. ratti (http://www.sanger.ac.uk/resources/downloads/helminths/strongyloides-ratti.html) genomic contigs using C. elegans protein sequences (http://www.wormbase.org/) were performed using Geneious set to the least restrictive parameters. Putative S. stercoralis homologs were identified through reverse BLAST searches using NCBI's pBLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [37] against C. elegans and/or phylum Nematoda sequences. Putative homologs were then manually annotated using aligned reads from all seven developmental stages by a combination of IGV and Geneious. Manually annotated S. stercoralis transcripts (Data S1, Data S2) were used to determine predicted protein sequences (Data S3).
Additional searches for ILP motifs in the S. stercoralis and S. ratti genomes were performed by translating the contigs in all six reading frames and searching for conserved A and B peptide motifs using Geneious. Similarly, we searched the S. stercoralis de novo assembled transcripts for ILP motifs by assembling the contigs from all developmental stages using Geneious, translating into all six reading frames, and searching for the B peptide motifs, C-11X-C and CPPG-11X-C, as well as the A peptide motifs, C-12X-CC, C-13X-CC, C-14X-CC, CC-3X-C-8X-CC, CC-4X-C-8X-CC, CC-3X-C-8X-C, and CC-3X-C-9X-C, where X represents any amino acid except for cysteine.
Protein alignments and phylogenetic analysis
Protein alignments and phylogenetic analyses were performed when several S. stercoralis or C. elegans homologs with similar e-values were identified in an attempt to resolve the homology of the S. stercoralis genes. Predicted protein sequences for S. stercoralis genes were derived from manually annotated transcripts using Geneious. Protein alignments using related S. stercoralis, C. elegans, phylum Nematoda, and other kingdom Animalia protein sequences were generated with Clustal W, using a BLOSUM matrix, or MUSCLE and neighbor-joining phylogenetic trees constructed using Geneious. Accession numbers for protein alignments referred to in the text can be found in Data S4.
A protein alignment for full-length guanylyl cyclases, similar to Ce-DAF-11, was performed with Clustal W in Geneious (Data S5). A neighbor-joining tree with 100 iterations of boot-strapping was constructed using Geneious and inspected for clear homology between Ce-DAF-11 and nematode homologs (Figure S2).
A protein alignment for the TGFβ super-family ligands (Data S6) was performed using only the ligand domain, truncated at the first conserved cysteine residue [38], with Clustal W in Geneious. A neighbor-joining tree with 100 iterations of boot-strapping was constructed using Geneious. A protein alignment for the TGFβ ligand domains that included all cysteine residues was performed using MUSCLE in Geneious and manually corrected (Figure S3).
A protein alignment for the full-length SMADs (Data S7) using every publicly available phylum Nematoda sequence was performed with Clustal W in Geneious. A neighbor-joining tree with 100 iterations of boot-strapping was constructed using Geneious and inspected for clear homology between C. elegans proteins and other nematode homologs (Figure S4). Similarly, a protein alignment for full-length short-chain dehydrogenases related to Ce-DHS-16 (Data S8) was used to construct a neighbor-joining phylogenetic tree (Figure S5) to find an S. stercoralis homolog most similar to Ce-DHS-16. A similar approach was used for cytochrome P450 proteins related to Ce-DAF-9 to generate a protein alignment (Data S9) and construct a neighbor-joining phylogenetic tree (Figure S6) to find the S. stercoralis homolog most similar to Ce-DAF-9.
Differential analysis of S. stercoralis transcripts
Transcript abundances of manually annotated S. stercoralis genes were calculated using Cufflinks version 2.0.0 (http://cufflinks.cbcb.umd.edu/) as fragments per kilobase of exon per million mapped reads (FPKM), with paired-end reads counted as single sampling events [39]. FPKM values for coding sequences (CDS) were calculated for each gene in each of the 21 samples and FPKM values for entire transcripts were calculated for each isoform in each of the 21 samples (Data S10). Log transformed values, ±95% confidence intervals, were plotted in Prism version 5.03 (GraphPad Software, Inc., http://www.graphpad.com/), and the y-axis was scaled from zero to 3.5 to aid comparisons between genes. Significant differences in FPKM values between developmental stages and p-values were determined using Cuffdiff version 1.3.0, a program with the Cufflinks package [40].
Results
RNAseq of seven S. stercoralis developmental stages
Many genes involved in C. elegans dauer regulation are transcriptionally regulated, including genes encoding ILPs [41], the dauer TGFβ ligand-encoding gene Ce-daf-7 [42], and the genes encoding biosynthetic enzymes for DA [43] that regulate the NHR Ce-DAF-12 [44]. To acquire a comprehensive transcriptomic profile of the S. stercoralis homologs of these genes, as well as other genes potentially involved in S. stercoralis L3i developmental regulation, we undertook a next-generation RNA sequencing (RNAseq) approach using Illumina HiSeq technology.
Since S. stercoralis has a unique life cycle with a single free-living generation (Figure 1), several pair-wise comparisons can be made between life stages fated for free-living versus parasitic development. For RNAseq analysis, we examined the following developmental stages: gravid free-living females (FL Females), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), gravid parasitic females (P Females), predominantly (>95%) heterogonically developing post-parasitic first-stage larvae (PP L1), and post-parasitic larvae at approximately the third-stage developing heterogonically to free-living adults and enriched for females (PP L3).
We isolated total RNA, in biological triplicate, from these seven developmental stages, using an S. stercoralis strain derived from a single free-living female (Data S11) [22] to decrease the number of nucleotide polymorphisms, which can confound alignment [30]. Using these samples, we constructed 21 polyadenylated RNA libraries, which we sequenced with 100 base-pair (bp) paired-end reads on an Illumina HiSeq 2000 instrument, generating a total of 2.36 billion reads (Figure 3). We independently aligned reads from each sample to the approximately 41 megabases of S. stercoralis genomic contigs using TopHat [40], [45], [46], a strategy used in the clade III parasitic nematode species Ascaris suum [31] and Brugia malayi [27]. Of the 2.36 billion reads initially sequenced, 1.75 billion (74%) aligned to genomic contigs (Figure 3). The roughly one quarter of reads that did not align to the genome may have come from contaminants such as gut bacteria or the gerbil host, contained sequencing errors, or originated from parts of the S. stercoralis genome that remain unsequenced.
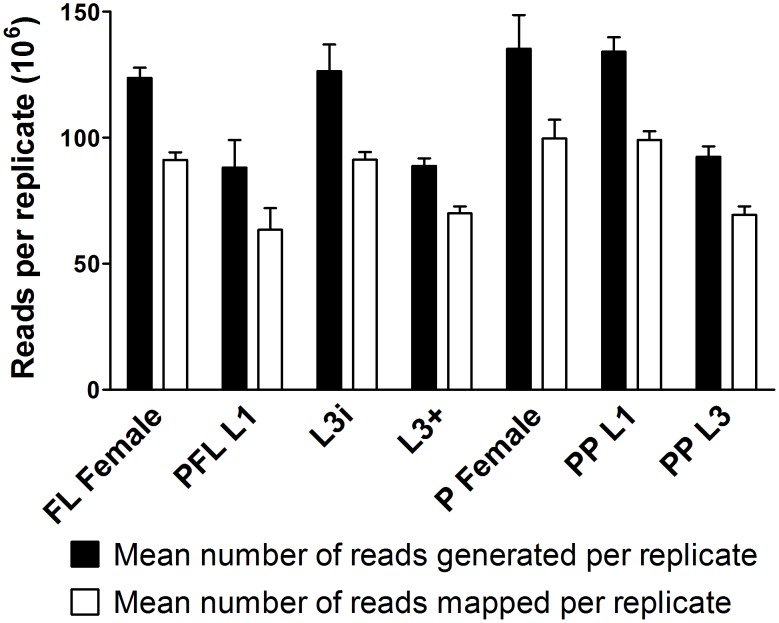
Figure 3. S. stercoralis RNAseq mean library sizes and number of reads aligning to the genome.
A total of 21 libraries were derived from polyadenylated RNA and sequenced from seven developmental stages, each in biological triplicate. Paired-end 100 base-pair (bp) reads were generated from the following developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), predominantly (>95%) heterogonically developing post-parasitic first-stage larvae (PP L1), and post-parasitic approximately third-stage larvae heterogonically developing to free-living adults and enriched for females (PP L3). The mean number of reads generated per replicate refers to the mean number of 100 bp reads sequenced (black bars) per biological replicate from each developmental stage. The mean number of mapped reads per replicate refers to the mean number of 100 bp reads aligned to S. stercoralis genomic contigs using TopHat (white bars) per biological replicate from each developmental stage. Error bars represent +1 standard deviation.
Identification of S. stercoralis genes encoding homologs of C. elegans dauer genes
To identify S. stercoralis homologs of the critical components involved in cGMP signaling, IIS, TGFβ signaling, as well as DA biosynthesis and NHR regulation, we performed BLAST searches of the S. stercoralis draft genome using C. elegans protein sequences. To confirm hits, we performed reverse BLAST searches to compare the manually annotated S. stercoralis sequences with C. elegans and phylum Nematoda databases [47]. When several homologs with similar e-values were present, we performed protein alignments and phylogenetic analysis to attempt to resolve the homology of S. stercoralis genes using related S. stercoralis, C. elegans, and phylum Nematoda protein sequences. For a few genes, we were unable to identify clear C. elegans homologs in S. stercoralis due to the lack of sequence similarity between the two species. We also noted several cases where either S. stercoralis or C. elegans had several closely related genes for which there was a single homolog in the other species, highlighting the evolutionary divergence between these two species, which are members of clade IV and clade V, respectively [16], [17].
We were unable to identify S. stercoralis homologs of several C. elegans genes within the S. stercoralis or closely related S. ratti genome sequences. To determine if these genes are absent from the genome assemblies, but present in the transcriptome, we performed de novo assembly of S. stercoralis transcripts with Trinity [48]. Using one sample from each developmental stage, we first merged each forward and reverse read pair to form a single, high quality “contig.” These merged single-read contigs were quality filtered and independently assembled to form expressed transcripts for each developmental stage. The seven expressed transcript libraries were merged to form a database on which we performed BLAST searches for C. elegans homologs not present in the draft S. stercoralis or S. ratti genomes. This S. stercoralis expressed transcript database contains a total of 210,709 developmental stage-specific transcripts; however, this includes redundant, fragmented, and un-spliced transcripts as well as contaminating sequences from gerbil and other environmental sources.
Due to the compactness of the S. stercoralis genome, we were unable to use Cufflinks [40], [49] to reliably predict transcripts because this program merged transcripts with untranslated region (UTR) overlap into single transcripts. Thus, we used aligned reads from all seven developmental stages to manually annotate exons and predict coding sequences for all isoforms of transcripts of interest. We then determined transcript abundances using Cufflinks to calculate fragments per kilobase of exon per million mapped reads (FPKM), with paired-end reads counted as single sampling events [39]. FPKM values were calculated for each gene or isoform in each developmental stage (Data S10), and significant differences between developmental stages were determined using the three biological replicates and Cuffdiff [40].
Cyclic GMP signaling components are up-regulated in S. stercoralis L3i
In C. elegans, formation of dauer larvae is regulated by dauer pheromone [50], [51], a constitutively produced complex mixture of ascarosides [52], [53], which is indicative of population density. Dauer entry is promoted by dauer pheromone, which is sensed by several GTP-binding protein (G protein)-coupled receptors (GPCRs), including Ce-SRBC-64, Ce-SRBC-66, Ce-SRG-36, and Ce-SRG-37 [54], [55]. When bound by specific ascarosides, GPCRs activate G protein alpha subunits [55], including Ce-GPA-2 and Ce-GPA-3 [56], resulting in repression of the transmembrane guanylyl cyclase Ce-DAF-11 [57] and a decrease in cGMP levels. Intracellular cGMP levels regulate cyclic nucleotide-gated ion channels [58], composed of the Ce-TAX-4 α subunits [59] and Ce-TAX-2 β subunits, which result in neuron depolarization when activated. The C. elegans cGMP signaling pathway is epistatic to the TGFβ pathway [60] (Figure 2) and may regulate the production of the Ce-DAF-7 TGFβ ligand [61] as well as the IIS agonists Ce-DAF-28 and Ce-INS-7 [62], [63]. Other daf mutants have been identified that are critical both in the localization of these cGMP signaling pathway proteins to the cilia as well as in the formation of proper ciliary structures [64]. Developmental regulation of C. elegans cGMP signaling pathway genes during dauer arrest has not been well studied, although Ce-gpa-2, Ce-gpa-3, Ce-daf-11, Ce-tax-2, and Ce-tax-4 are all down-regulated following dauer recovery in microarray analysis [43].
Outside of C. elegans, the role of ascarosides and cGMP pathway signaling in parasitic nematodes has been nearly overlooked. Muscarinic agonists and the cGMP analog 8-bromo-cGMP have been shown to activate Ancylostoma caninum L3i [65], [66], and we have previously cloned S. stercoralis homologs of Ce-gpa-2 and Ce-gpa-3 [18]. Recently, several groups have reported the presence of ascarosides in parasitic nematodes, which appear to differ in structure and composition between species and may play a role in L3i formation [67]–[69]. Thus, we sought to determine whether the components of a cGMP signaling pathway are present in S. stercoralis and whether these transcripts are developmentally regulated (Table 1).
Table 1. Comparison of cGMP signaling pathway homologs and transcript abundances in S. stercoralis and C. elegans.
| C. elegans gene(s) | S. stercoralis homolog(s) | S. stercoralis transcript abundance profile | Regulation consistent with C. elegans (+, +/−, −)1 |
| Ce-gpa-2 & -3 | Ss-gpa-2 & -3 | peak in L3i | + |
| Ce-daf-11 | Ss-gcy-11 2 | peak in L3i and L3+ | + |
| Ce-tax-2 | Ss-tax-2 | peak in L3i | + |
| Ce-tax-4 | Ss-tax-4 | peak in L3i | + |
(+) similar, (+/−) unclear, and (−) dissimilar transcript abundance patterns.
Homology is by phylogenetic similarity only.
We identified an S. stercoralis gene encoding a putative guanylyl cyclase that phylogenetically groups with Ce-DAF-11, which we termed Ss-gyc-11 (Figure S2, Data S5). We also identified genes encoding homologs of the two cGMP-gated ion channels, Ce-TAX-2 and Ce-TAX-4, which we termed Ss-tax-2 and Ss-tax-4 respectively. We were unable to identify clear homologs of the GPCR genes, as many of the seven transmembrane receptor families have undergone rapid expansion in C. elegans [55]. Examination of the transcript abundance profiles for each of the five S. stercoralis genes putatively involved in cGMP pathway signaling revealed strikingly similar temporal regulation (Figure 4), with the steady-state level of each transcript at its peak in L3i and its nadir in both free-living and parasitic females. Interestingly, this developmental transcript abundance profile was also observed for two other guanylyl cyclases similar to Ss-gyc-11 (Figure 4).
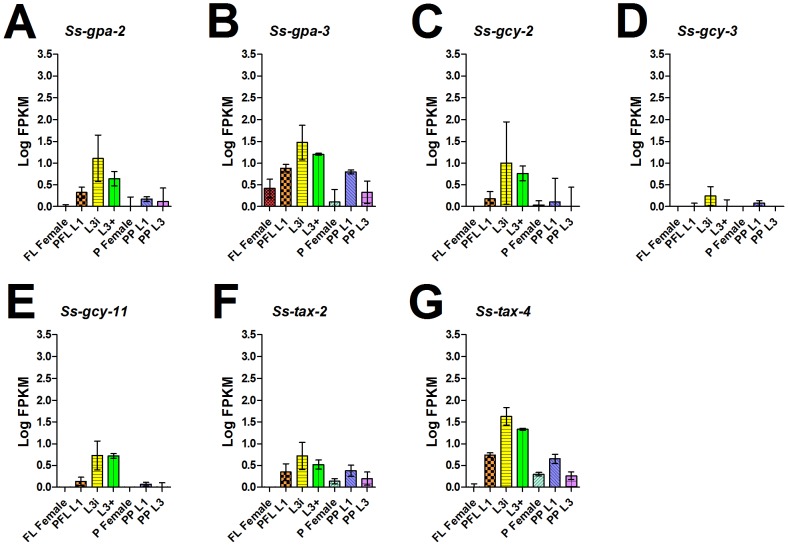
Figure 4. S. stercoralis cGMP signaling pathway homologs are coordinately up-regulated in L3i.
Transcript abundances were determined for the coding region of: (A) Ss-gpa-2 and (B) Ss-gpa-3, the genes encoding homologs of the G-protein α subunits Ce-GPA-2 and Ce-GPA-3, respectively; (C) Ss-gcy-2, (D) Ss-gcy-3, and (E) Ss-gcy-11, genes which all encode guanylyl cyclase homologs, of which Ss-gcy-11 encodes the S. stercoralis homolog most similar to the guanylyl cyclase Ce-DAF-11; (F) Ss-tax-2 and (G) Ss-tax-4, genes encoding homologs of the cyclic nucleotide-gated ion channels Ce-TAX-2 and Ce-TAX-4, respectively. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
Insulin-like peptide transcripts are regulated during S. stercoralis development
IIS plays a critical role in both dauer arrest and recovery in C. elegans. Both microarray [70] and careful transcript quantification experiments [41] have shown that regulation of C. elegans IIS transcripts during dauer development takes place at the level of the ILPs, while the intracellular signaling component transcripts are always present. We have previously shown that IIS in S. stercoralis plays a crucial role in L3i arrest [10] and activation [22]. However, neither the presence nor regulation of ILPs has been reported in S. stercoralis or any other parasitic nematodes.
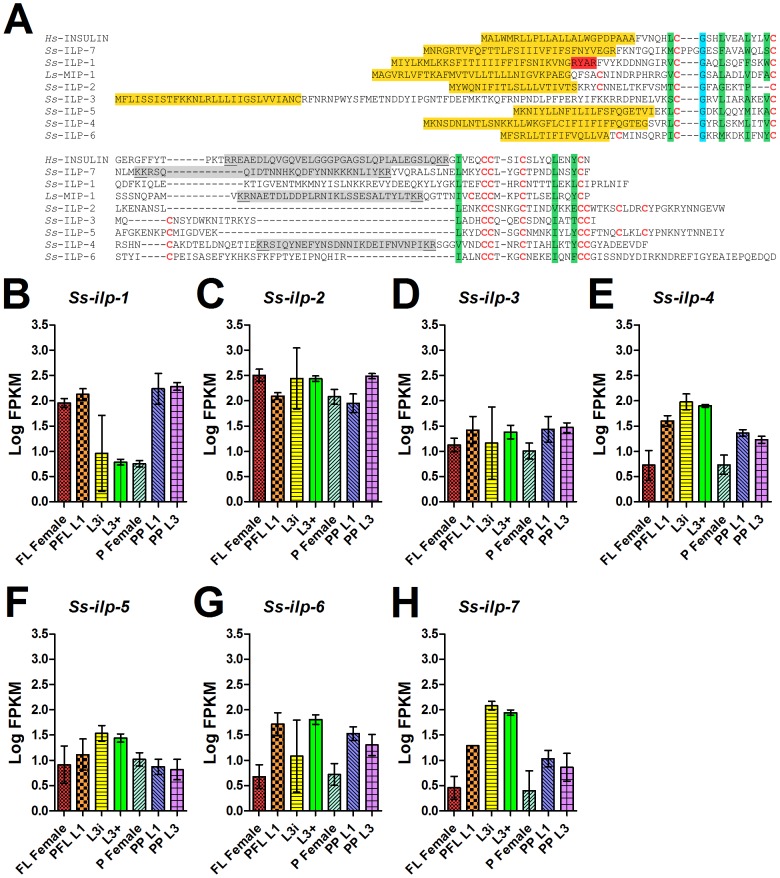
In C. elegans, 40 ILPs have been discovered and are thought to play redundant and complex roles in regulating dauer as well as other forms of development, with some ILPs agonizing and others antagonizing IIS [62], [71]. To find S. stercoralis ILPs, we performed BLAST searches of the draft genomes of S. stercoralis and S. ratti as well as our de novo assembled S. stercoralis transcripts using both C. elegans ILP protein sequences and conserved cysteine motifs in the A and B peptides [71]. In total, we identified seven S. stercoralis ILPs (Figure 5A, Table 2), which are also present in S. ratti (data not shown). The predicted protein sequences of the S. stercoralis ILPs are highly divergent from C. elegans homologs, except for several conserved cysteine residues which are predicted to form disulfide bonds. In contrast to both C. elegans and Homo sapiens, S. stercoralis ILPs lack the conserved intron located between N-terminal B peptide and C-terminal A peptide, and all but one lack a predicted furin cleavage site [71]–[73]. Furthermore, cleavable C peptides, located between the B and A peptides, are not conserved between species.
Figure 5. Protein sequence diversity and temporal regulation of S. stercoralis insulin-like peptides.
(A) A predicted protein sequence alignment of seven S. stercoralis insulin-like peptides (ILPs), Ss-ILP-1 through -7, was constructed using human insulin (Hs-INSULIN) and Lymnaea stagnalis molluscan insulin-related peptide I (Ls-MIP-1) as the references. Cysteine residues, which are predicted to form disulfide bonds, are in red letters. Predicted signal sequences are highlighted in yellow, predicted furin recognition motifs are highlighted in red, hydrophobic residues important for helix formation are highlighted in green, and a conserved glycine is highlighted in blue. Predicted C peptides are highlighted in gray with dibasic predicted cleavage sites underlined. The B peptide is N-terminal of the C peptide, while the A peptide is C-terminal of the C peptide. (B-H) Transcript abundances were determined for the coding region of seven S. stercoralis ILP-encoding genes (Ss-ilp-1 through -7) in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
Table 2. Comparison of IIS pathway homologs and transcript abundances in S. stercoralis and C. elegans.
| C. elegans gene(s) | S. stercoralis homolog(s) | S. stercoralis transcript abundance profile | Regulation consistent with C. elegans (+, +/−, −)1 |
| Insulin-like Peptides | |||
| Type α: Ce-ins -20 to -30, and -33 to -36 | None identified | ||
| Type β: Ce-daf-28, and Ce-ins -1 to -10 | Ss-ilp-3 | present in all stages examined | +/− |
| Ss-ilp-4 | decreased in FL and P Females | +/− | |
| Ss-ilp-6 | increased from L3i to L3+ | + | |
| Type γ: Ce-ins-11 to -19, -31, -32, and -37 | Ss-ilp-1 | decreased in PFL generation | + |
| Ss-ilp-7 | increased in L3i and L3+ | + | |
| Type δ: None | Ss-ilp-2 & -5 | present in all stages examined | +/− |
| Intracellular signaling components | |||
| Ce-asna-1 | Ss-asna-1 | present in all stages examined | + |
| Ce-daf-2 | Ss-daf-2 | increased in PFL generation | + |
| Ce-ist-1 | Ss-ist-1 & -2 | present in all stages examined | + |
| Ce-aap-1 | Ss-aap-1 | increased in FL and P Females | + |
| Ce-age-1 | Ss-age-1 | increased in L3+ | + |
| Ce-daf-18 | Ss-pten-1 & -2 | present in all stages examined | + |
| Ce-pdk-1 | Ss-pdk-1 | increased in L3i | + |
| Ce-sgk-1 | Ss-sgk-1 | absent in L3i and L3+ | - |
| Ce-akt-1 and -2 | Ss-akt-1 | increased in L3i and L3+ | +/− |
| Ce-pptr-1 2 | Ss-pptr-1 | present in all stages examined | + |
| Ce-ftt-2 | Ss-ftt-2 | present in all stages examined | + |
| Ce-daf-16 | Ss-daf-16 | decreased in FL and P Females | +/− |
| Ce -DAF-16 regulated genes | |||
| Ce-sod-3 3 | Ss-sod-1 | present in all stages examined | − |
| Ce-daf-15 | Ss-daf-15 | present in all stages examined | − |
| Ce-acs-19 | Ss-acs-19 | present in all stages examined | − |
| Ce-ldb-1 | Ss-limdb-1 & -2 | present in all stages examined | − |
| Ce-pitp-1 | Ss-pitp-1 | present in all stages examined | − |
| Ce-Y105E8B.9 | Ss- Y105E8B.9 | increased in developing larvae | +/− |
(+) similar, (+/−) unclear, and (−) dissimilar transcript abundance patterns.
A homolog for the closely related gene Ce-pptr-2 was identified and termed Ss-pptr-2.
The closely related gene Ce-sod-2 was accounted for; only one sod gene was identified in S. stercoralis.
The S. stercoralis putative ILPs—Ss-ILP-3, Ss-ILP-4, and Ss-ILP-6—have type β cysteine architecture [71]. In C. elegans, the type β family includes several agonistic ligands including Ce-DAF-28 [62], Ce-INS-6 [62], [74], [75], and Ce-INS-7 [76], as well as the antagonistic ligand Ce-INS-1 [71], [74], [77]–[79]. The type β family also includes the Lymnaea stagnalis molluscan insulin-related peptide I (MIP-1) [80]. In contrast, Ss-ILP-1 and Ss-ILP-7 have type γ cysteine architecture, similar to that found in human insulin [71]. In C. elegans, the type γ family includes the putative antagonist Ce-INS-18, which has a PPG motif between the conserved cysteine and glycine residues in the B peptide [71], [81]. Interestingly, Ss-ILP-7 is the only S. stercoralis ILP to share this motif (Figure 5A). Unlike the six cysteine residues found in type α and γ ILPs or the eight found in type β ILPs, Ss-ILP-2 and Ss-ILP-5 have 10 cysteine residues. We propose that Ss-ILP-2 and Ss-ILP-5 represent a novel class of nematode ILPs, which we term type δ.
To determine whether S. stercoralis ILP transcripts are developmentally regulated, we compared FPKM values for each transcript between developmental stages (Figure 5B–H). In contrast to many C. elegans ILPs which are only expressed at one or a few developmental stages [41], transcripts encoding all seven S. stercoralis ILPs were detected in all developmental stages examined. We noted that Ss-ilp-1 transcripts are decreased in L3i and significantly down-regulated in L3+ and parasitic females compared to the other developmental stages examined (p<0.001). We also noted that transcripts for both Ss-ilp-4 and Ss-ilp-7, encoding the only two S. stercoralis ILPs with predicted C peptides that are cleaved, are at their peak in L3i. Additionally, we observed high variability in the transcript abundances of several ILP-encoding genes in the L3i developmental stage, evidenced by the large 95% confidence intervals. Since we isolated L3i incubated at 21°C after 8 and 10 days of culture or 25°C after 7 of days of culture (Data S11), we plotted transcript abundance for each ilp gene by relative age for each biological replicate (Figure S7). This analysis revealed that the error was not stochastic, but rather a developmental trend dependent upon the relative age of the L3i. In this analysis, we observed a one log increase in the transcript abundance of Ss-ilp-6 from the oldest L3i to the L3+.
Intracellular IIS component transcripts are always present in S. stercoralis
While C. elegans ILPs are developmentally regulated, intracellular IIS components are always present [41], [70]. We have previously cloned and detected transcripts throughout the life cycle of S. stercoralis homologs of both the forkhead transcription factor daf-16 [82] and the age-1 catalytic subunit of the phosphatidylinositol-3 kinase (PI3K) [22]. Recently, we have also cloned and characterized the S. stercoralis genes encoding the Ss-AAP-1 PI3K accessory/regulatory subunit [22] and the Ss-DAF-2 insulin-like receptor (Massey, HC, et al., in preparation). In this study, we asked whether homologs of the remaining IIS components are present in S. stercoralis and, if so, whether their transcripts are also present throughout the life cycle (Table 2).
Downstream of the DAF-2 IIS receptor, we identified two genes encoding homologs of the insulin receptor substrate Ce-IST-1 [83], which we termed Ss-ist-1 and Ss-ist-2. Interestingly, we also found two homologs of the gene encoding the C. elegans phosphatase and tensin (PTEN) homolog Ce-DAF-18, which opposes the function of the PI3K Ce-AGE-1 when IIS is activated [84]. We termed these genes Ss-pten-1 and Ss-pten-2. We also identified Ss-pdk-1 as a homolog of the gene encoding the 3-phosphoinositide-dependent kinase Ce-PDK-1, which phosphorylates and activates Ce-AKT-1 and -2 when IIS is activated [85]. We identified Ss-akt-1 as a single homolog of the genes encoding the C. elegans serine/threonine kinases Ce-AKT-1 and Ce-AKT-2 [86], which phosphorylate Ce-DAF-16 when IIS is activated [87], [88]. In C. elegans, AKT-1 is negatively regulated by Ce-PPTR-1, a B56 regulatory subunit of the PP2A phosphatase [89]. We identified an S. stercoralis gene encoding a similar phosphatase, which we termed Ss-pptr-1. We also found a gene encoding the C. elegans homolog of the serum- and glucocorticoid-inducible kinase Ce-SGK-1 that regulates Ce-DAF-16 [90], which we termed Ss-sgk-1. We identified a homolog of the gene encoding the 14-3-3 protein Ce-FTT-2 [91] that regulates Ce-DAF-16 [92], which we termed Ss-ftt-2. Additionally, we found Ss-asna-1, a homolog of the gene encoding the ATPase Ce-ASNA-1, which regulates ILP secretion in C. elegans [93]. Together, these S. stercoralis homologs reconstruct a complete IIS pathway similar to that found in C. elegans and other metazoans [94].
Transcripts for each of the S. stercoralis genes encoding IIS cytoplasmic signaling proteins, except for Ss-sgk-1, were detected in every developmental stage examined (Figure S8), suggesting that the IIS cytoplasmic signaling proteins are present throughout the S. stercoralis life cycle. We observed varying degrees of transcript up-regulation in the post-free-living generation of genes encoding the core IIS cytoplasmic signaling proteins Ss-DAF-2, Ss-AGE-1, Ss-PDK-1, Ss-AKT-1, and Ss-DAF-16. Interestingly, the increases in Ss-akt-1 transcripts in the L3i and L3+ stages were largely due to expression of a second isoform, Ss-akt-1b, which encodes a predicted peptide with a shortened N-terminus that results in a 33 amino acid deletion from the AKT pleckstrin homology (PH) domain and which is only present in these two stages (Figure S9). Conversely, we noted an absence of Ss-sgk-1 transcripts in L3i and L3+ (Figure S8).
Homologs of Ce-DAF-16-regulated genes are not similarly regulated in S. stercoralis development
To determine whether IIS regulates similar genes in S. stercoralis and C. elegans, we then asked whether homologs of genes transcriptionally regulated by Ce-DAF-16 were similarly regulated over the course of S. stercoralis development (Table 2). In C. elegans, multiple studies have examined the genes regulated by the transcription factor Ce-DAF-16 [76], [95]–[98]. The superoxide dismutase encoding gene Ce-sod-3 is a well-characterized gene that is up-regulated by Ce-DAF-16 in the dauer stage [41], [99], [100], while the RAPTOR ortholog-encoding gene Ce-daf-15 is down-regulated by Ce-DAF-16 in low IIS conditions [101]. We identified a single superoxide dismutase-encoding gene in S. stercoralis that phylogenetically grouped with Ce-sod-2 and Ce-sod-3, which we termed Ss-sod-1, as well as a homolog of Ce-daf-15, which we termed Ss-daf-15. Additionally, we identified S. stercoralis homologs of Ce-acs-19, Ce-ldb-1, Ce-pitp-1, and Ce-Y105E8B.9, all of which were identified as Ce-DAF-16 targets by ChIPseq, are differentially regulated in Ce-daf-16(mu86) mutants, and have a phenotype associated with loss of Ce-DAF-16 function upon RNAi knock-down [98]. We termed these homologs Ss-acs-19, Ss-limdb-1 and -2, Ss-pitp-1, and Ss-Y105E8B.9, respectively.
Surprisingly, transcript abundance profiles for each of these six genes (Figure S10) revealed that neither Ss-sod-1, Ss-daf-15, nor the other five genes were up- or down-regulated in L3i. In fact, no large differences in Ss-sod-1 or Ss-daf-15 transcript levels were observed among any of the seven developmental stages examined.
The DAF-7-like TGFβ ligand family is expanded in S. stercoralis
In C. elegans, mutation of the TGFβ ligand-encoding gene daf-7 results in temperature sensitive dauer arrest and is the only TGFβ ligand in the C. elegans genome in the same family as human TGFβ1, Inhibin/Activin, and Myostatin [42], [102]. Ce-daf-7 transcripts are at their peak in L1 larvae and are up-regulated during recovery from both L1 and dauer arrested states [23], [42], [43]. In C. elegans, DAF-7 is most likely produced in response to food cues and functions in parallel with other pathways to promote continuous development.
Previous work in S. stercoralis, S. ratti, and Parastrongyloides trichosuri has identified Ce-DAF-7-like TGFβ ligand-encoding genes, named Ss-tgh-1, Sr-daf-7, and Pt-daf-7, respectively [20], [23]. In stark contrast to C. elegans, these clade IV parasitic nematode TGFβ ligands are significantly up-regulated in the developmentally arrested L3i and down-regulated in activated L3i—a pattern directly opposite to that predicted under the dauer hypothesis. Similarly, transcripts encoding a DAF-7-like TGFβ ligand, termed tgh-2, have been described in the clade V parasitic nematodes Ancylostoma caninum [103], [104], Heligmosomoides polygyrus, Nippostrongylus brasiliensis, Haemonchus contortus, and Teladorsagia circumcincta [105], as well as the clade III parasitic nematodes Brugia malayi and Brugia pahangi [106]. For many of these nematode species, the tgh-2 transcripts are up-regulated in the L3i. These observations have led some groups to question the relevance of using C. elegans dauer pathways to predict pathways regulating infectious larval development in parasitic nematodes [107].
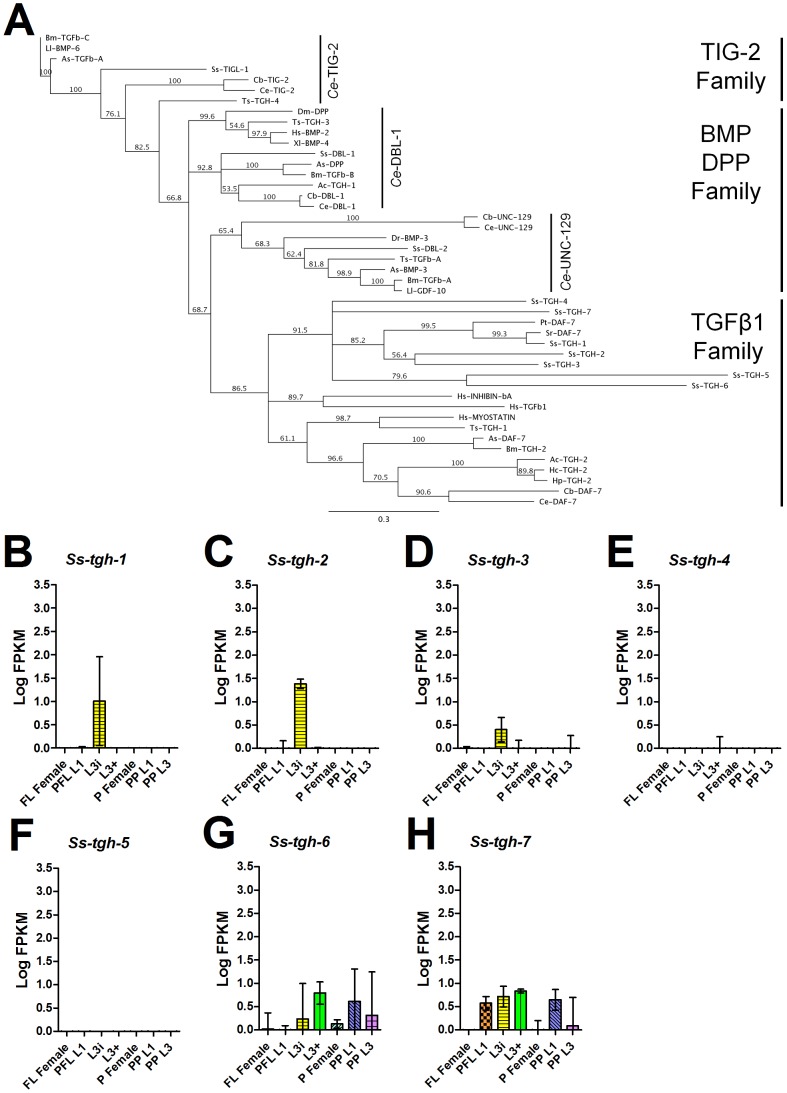
In addition to Ce-DAF-7, C. elegans also has four other TGFβ ligands that have different cysteine architecture and are not involved in dauer regulation; thus, we sought to identify homologs of all the TGFβ ligands in S. stercoralis to ensure proper classification. To our surprise, we discovered a total of 10 TGFβ ligands in both the S. stercoralis draft genome (Figure 6A) and S. ratti draft genome (data not shown). Protein alignment and phylogenetic analysis placed seven of these ligands in the same family as Ce-DAF-7, which also includes the previously described Ss-TGH-1 (Figure 6A, Figure S3, Data S6). We named these additional Ss-tgh-1-like genes Ss-tgh-2 through -7 (Table 3). Interestingly, the putative Ss-TGH-6 and Ss-TGH-7 ligands are not predicted to have propeptides, an observation previously reported in TGH-2 from N. brasiliensis [105], Schistosoma mansoni SmInAct [108], and a few TGFβ ligands from Ctenophores (marine invertebrates commonly called comb jellies) [38]. The three additional S. stercoralis TGFβ ligands grouped with homologs of Ce-DBL-1, Ce-UNC-129, and Ce-TIG-2 [109] by both phylogenetic analysis (Figure 6A) and protein alignment (Figure S3). We termed the genes encoding these ligands Ss-dbl-1, Ss-dbl-2, and Ss-tigl-1, respectively.
Figure 6. Phylogenetic analysis and temporal regulation of S. stercoralis TGFβ ligands.
(A) Phylogenetic analysis of the transforming growth factor β (TGFβ) super-family ligands was performed; nematode TGFβ ligands resolved into three main families that share the same cysteine architecture. Ss-TIGL-1 groups with the Ce-TIG-2-like family; Ss-DBL-1 and Ss-DBL-2 group with the D. melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP) family; and Ss-TGH-1 through -7 group with the human TGFβ1 family that also includes Ce-DAF-7. A Clustal W alignment of the TGFβ ligands truncated at the first conserved cysteine was used to construct the neighbor-joining tree with 100 iterations of boot-strapping. Abbreviations: Ancylostoma caninum (Ac), Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Danio rerio (Dr), Drosophila melanogaster (Dm), Haemonchus contortus (Hc), Heligmosomoides polygyrus (Hp), Homo sapiens (Hs), Loa loa (Ll), Parastrongyloides trichosuri (Pt), Strongyloides ratti (Sr), Strongyloides stercoralis (Ss), Trichinella spiralis (Ts), and Xenopus laevis (Xl). The scale bar represents substitutions per position. Accession numbers are listed in Data S4. (B–H) Transcript abundances were determined for the coding region of seven S. stercoralis genes, Ss-tgh-1 through -7, encoding putative TGFβ ligands similar to Ce-DAF-7 in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
Table 3. Comparison of dauer TGFβ signaling pathway homologs and transcript abundances in S. stercoralis and C. elegans.
| C. elegans gene(s) | S. stercoralis homolog(s) | S. stercoralis transcript abundance profile | Regulation consistent with C. elegans (+, +/−, −)1 |
| Ce-daf-7 2 | Ss-tgh-1 to -3 | L3i only | − |
| Ss-tgh-4 & -5 | not present in stages examined | +/− | |
| Ss-tgh-6 | increased from PFL L1 to L3+ | − | |
| Ss-tgh-7 | not present in FL or P Females | +/− | |
| Ce-daf-1 | Ss-daf-1 | increased in L3i and L3+ | − |
| Ce-daf-4 3 | Ss-daf-4 | increased in L3i | − |
| Ce-bra-1 | Ss-bra-1 | present in all stages examined | +/− |
| Ce-daf-3, -8, & -14 4 | Ss-smad-1 | present in all stages examined | +/− |
| Ss-smad-5 | decreased in P Females | +/− | |
| Ss-smad-7 | present in all stages examined | +/− | |
| Ss-smad-8 | decreased in FL and P Females | +/− | |
| Ce-pdp-1 | Ss-pdp-1 | present in all stages examined | +/− |
| Ce-daf-5 5 | Ss-daf-5 | present in all stages examined | +/− |
(+) similar, (+/−) unclear, and (−) dissimilar transcript abundance patterns.
Homologs for the Ce-dbl-1, Ce-unc-129, and Ce-tig-2 were identified and termed Ss-dbl-1, Ss-dbl-2, and Ss-tigl-1, respectively.
A homolog for the related gene Ce-sma-6 was identified and termed Ss-sma-6.
Homologs for Ce-sma-2, Ce-sma-3, and Ce-sma-4 were identified and termed Ss-smad-2, Ss-smad-3, and Ss-smad-4, respectively.
A homolog for Ce-sma-9 was identified and termed Ss-sma-9.
We investigated whether the transcript abundance patterns of the seven genes encoding S. stercoralis TGH ligands were similar to Ss-tgh-1 (Figure 6B–H). Interestingly, Ss-tgh-1, -2, and -3 transcripts were detected exclusively in L3i, while Ss-tgh-4 and -5 were not detected in any of the life stages examined. Ss-tgh-6 and -7 had more complex transcript abundance patterns; Ss-tgh-6 was up-regulated in L3+ in comparison to L3i (p<0.001), while Ss-tgh-7 was not expressed in either the free-living or parasitic females. Similar to the ILP-encoding genes, the tgh genes also had a high degree of variability in the transcript abundances in the L3i developmental stage. As with the ilp genes, the variability of the tgh genes in L3i represented developmental trends that are dependent upon the relative age of the L3i (Figure S7). We also determined transcript abundances for Ss-dbl-1, Ss-dbl-2, and Ss-tigl-1, which are not predicted to signal through the dauer TGFβ signaling pathway (Figure S11).
Dauer TGFβ signaling pathway components are present in S. stercoralis, but have high sequence divergence
Components of the C. elegans dauer TGFβ signaling pathway all have a temperature sensitive dauer phenotype when mutated [60]. Recent studies have presented an integrated model for dauer TGFβ signaling [110], [111], where under well-fed conditions, the Ce-DAF-7 ligand is expressed [42], [112] and binds the type I receptor Ce-DAF-1 [113] and type II receptor Ce-DAF-4 [114], overcoming the inhibition of Ce-DAF-1 by Ce-BRA-1 [115]. This results in phosphorylation and activation of the cytoplasmic R-SMADs Ce-DAF-8 [110] and Ce-DAF-14 [116], which together repress the Co-SMAD Ce-DAF-3 [117] and allow for reproductive development. However, when the Ce-DAF-7 ligand is not present, Ce-DAF-3 is active [110] and, together with the Sno/Ski-like transcriptional co-factor Ce-DAF-5 [118], represses expression of Ce-daf-7 and Ce-daf-8 [110], thereby promoting dauer development (Figure 2). In C. elegans, Ce-DAF-8 and Ce-DAF-14 are also inhibited by the phosphatase Ce-PDP-1, which also appears to control components of IIS, including ILPs, suggesting cross-talk between these pathways [111].
Proteins of the C. elegans dauer TGFβ pathway have diverged from those of other metazoans in both structure and function. Ce-DAF-1 can signal to some extent without Ce-DAF-4 [119], and a truncated Ce-DAF-4 protein expressed in dauers can negatively regulate Ce-DAF-7 signaling [120]. Consensus SMADs have both an MH1 (DNA-binding) and an MH2 (protein-protein interacting) domain and are activated by TGFβ signaling [121]; however, Ce-DAF-14 does not contain a consensus MH1 domain [116] and Ce-DAF-3 is repressed by Ce-DAF-7 signaling [117]. Temporal regulation of multiple components has been observed, including an up-regulation of Ce-DAF-1 [119] and Ce-DAF-8 [110] in L1 similar to Ce-daf-7 transcriptional regulation [42], as well as a decrease in full-length Ce-daf-4 transcripts in dauer larvae [120].
Since we observed a marked increase in the number of Ce-DAF-7-like TGFβ ligands in S. stercoralis, we asked whether the dauer TGFβ cytoplasmic signaling components were conserved in both protein structure and temporal regulation (Table 3). We sought to differentiate these components from those in the C. elegans small body size and male tail abnormal (Sma/Mab) TGFβ pathway. We identified homologs of the genes encoding the Ce-DAF-1 type I receptor and the Ce-DAF-4 type II receptor, which we termed Ss-daf-1 and Ss-daf-4, respectively. We also identified a homolog of the gene encoding the Ce-DAF-1 negative regulator Ce-BRA-1, which we termed Ss-bra-1. The C. elegans Sma/Mab TGFβ pathway, which uses the Ce-DBL-1 ligand [122], [123], also utilizes the Ce-DAF-4 type II receptor but with Ce-SMA-6 as the type I receptor [124]. To ensure proper classification of the type I receptors, we identified a gene encoding a homolog of Ce-SMA-6, which we termed Ss-sma-6.
Identification of homologs for each of the SMADs proved difficult and was confounded by structurally similar SMADs involved in the dauer and Sma/Mab TGFβ signaling pathways present in C. elegans [102]. We identified a gene encoding a homolog of Ce-DAF-14 that did not include a MH1 domain, which we termed Ss-smad-1. We identified three S. stercoralis genes, termed Ss-smad-5, Ss-smad-7, and Ss-smad-8, which encode SMADs similar to Ce-DAF-3 and Ce-DAF-8; however, we were unable to resolve homology further by protein alignment or phylogenetic analysis (Figure S4, Data S7). Interestingly, we were able to clearly resolve genes encoding Sma/Mab TGFβ pathway SMADs similar to Ce-SMA-2, Ce-SMA-3, and Ce-SMA-4, which we termed Ss-smad-2, Ss-smad-3, and Ss-smad-4, respectively.
We identified a gene encoding a dauer TGFβ pathway Ce-DAF-5-like transcriptional co-factor, which we termed Ss-daf-5. The gene encoding a homolog of the Sma/Mab TGFβ pathway Ce-SMA-9-like transcriptional co-factor, which we termed Ss-sma-9, was clearly differentiable from Ss-daf-5. We also identified a gene encoding a phosphatase similar to Ce-PDP-1, which we termed Ss-pdp-1.
Examination of the transcript abundance patterns of the S. stercoralis genes encoding dauer pathway TGFβ homologs revealed several interesting trends (Figure S12). In direct contrast to the down-regulation of the type I and type II receptors observed in C. elegans dauer larvae [119], [120], Ss-daf-1 and Ss-daf-4 transcripts are at their peak in L3i and L3+. Likewise, Ss-smad-8 transcripts were also at their peak in L3i. These observations are consistent with the expression of the Ss-tgh-1, Ss-tgh-2, and Ss-tgh-3 transcripts exclusively in L3i (Figure 6B–D). We also noted a significant decrease in Ss-smad-5 transcripts in parasitic females in comparison to the other six developmental stages examined (p<0.001). We did not observe any changes greater than one log in the transcript abundance of Ss-bra-1, Ss-smad-1, Ss-smad-7, or Ss-daf-5 in the seven developmental stages examined. Additionally, we examined the transcript abundances of the components in the Sma/Mab TGFβ pathway and noted that transcript levels for the receptor-encoding genes, Ss-sma-6 and Ss-daf-4, as well as the Ss-sma-9 transcriptional co-factor, are at their peak in L3i (Figure S12).
A putative dafachronic acid biosynthetic pathway is present in S. stercoralis
In C. elegans dauer development, epistatic analysis has placed both the IIS and dauer TGFβ pathways upstream of the NHR Ce-DAF-12 [125] (Figure 2). Ce-DAF-12 is broadly expressed [126] and is regulated by at least two steroid-like ligands, known as Δ4- and Δ7-dafachronic acid (DA) [44]. These DAs are synthesized from cholesterol, which is trafficked intracellularly by Ce-NCR-1 and -2 [127]. For Δ7-DA synthesis, cholesterol is first modified by the Rieske-like oxygenase Ce-DAF-36 [128], followed by the short-chain dehydrogenase Ce-DHS-16 [129]. In the final step, the cholesterol side chain is oxidized by the cytochrome P450 Ce-DAF-9 [130], [131], with likely assistance from the cytochrome P450 reductase Ce-EMB-8 [129]. The enzymes that synthesize the precursors of Δ4-DA are unknown, although the final oxidation step(s) are carried out by Ce-DAF-9 and Ce-EMB-8, similarly to Δ7-DA [129]. The 3β-hydroxysteriod dehydrogenase/Δ5-Δ4 isomerase Ce-HSD-1 has previously been reported to play a role in Δ4-DA biosynthesis [132]; however, a recent study has shown that this is not the case and that Ce-HSD-1 may be involved in synthesizing other DAs [129]. Additionally, the Ce-STRM-1 methyltransferase modifies DA precursors and can influence dauer development [133].
In favorable environmental conditions and when dauer larvae resume development, DAs are synthesized and bind Ce-DAF-12 [44] to promote reproductive development. However, in unfavorable environmental conditions, DAs are not synthesized and Ce-DAF-12, along with its co-repressor Ce-DIN-1 [134], promotes dauer development. Expression of GFP reporter constructs from Ce-daf-36 [128] and Ce-daf-12 [126] promoters is down-regulated in dauers, while microarray evidence has shown that Ce-daf-9 and Ce-daf-36 transcripts are up-regulated during dauer recovery [43]. Somewhat contradictorily, Ce-daf-12 transcripts have been shown to be up-regulated during dauer formation [135].
The S. stercoralis homolog of DAF-12 has been cloned [21], and recent evidence from our lab has demonstrated that exogenous application of Δ7-DA to S. stercoralis L3i results in potent activation, as measured by resumption of feeding, in the absence of all host-like cues [136]. Furthermore, Δ7-DA applied to S. stercoralis post-free-living larvae results in failure to arrest as L3i and development to free-living L4, which we have termed an “L3i bypass” phenotype [136]. In the closely related parasite Strongyloides papillosus, which has a life cycle outside the host very similar to that of S. stercoralis, application of Δ7-DA to post-free-living larvae results in a second free-living generation of reproductively competent females [137]. In both S. stercoralis and S. papillosus, Δ7-DA results in stronger L3i activation or L3i bypass phenotypes than does Δ4-DA [136], [137].
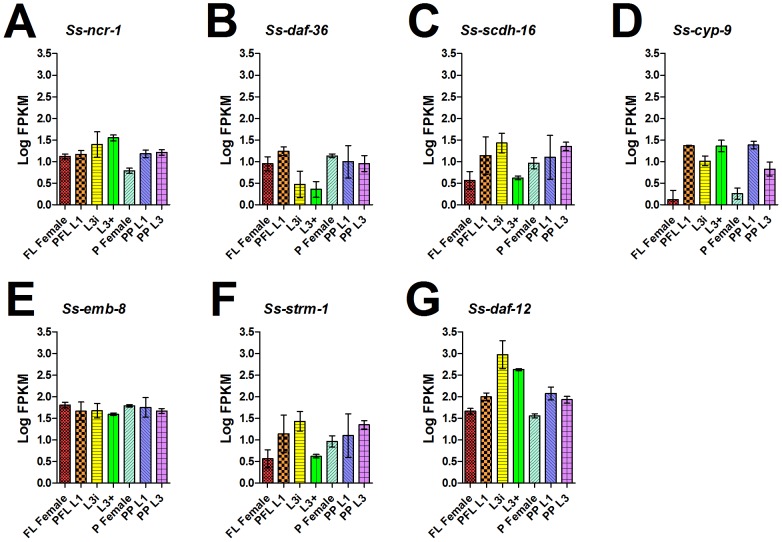
Therefore, we asked whether a biosynthetic pathway for NHR DA ligand(s) similar to that found in C. elegans was present in S. stercoralis and had similar developmental regulation (Table 4). We identified a single S. stercoralis gene encoding a homolog of Ce-NCR-1 and -2, which we termed Ss-ncr-1, as well as a gene encoding a homolog of Ce-DAF-36, which we termed Ss-daf-36. We identified several S. stercoralis genes encoding putative short-chain dehydrogenases similar to Ce-DHS-16; one of these genes, which we termed Ss-scdh-16, encoded a predicted protein that phylogenetically grouped closely with Ce-DHS-16 (Figure S5, Data S8). Similarly, we identified several S. stercoralis genes putatively encoding cytochrome P450s similar to Ce-DAF-9; one of these, which we termed Ss-cyp-9, encoded a putative peptide that grouped with Ce-DAF-9 by phylogenetic analysis (Figure S6, Data S9). We also identified a gene encoding a homolog of Ce-EMB-8, which we termed Ss-emb-8, as well as a gene encoding a homolog of Ce-STRM-1, which we termed Ss-strm-1. Curiously, we were unable to identify genes encoding S. stercoralis homologs of Ce-HSD-1 or Ce-DIN-1 in the S. stercoralis draft genome, the S. ratti draft genome, or our de novo assemblies of S. stercoralis transcripts. We also found that the Ss-daf-12 locus encoded a total of seven transcripts encoding three different proteins, with the variability confined to the N-terminus of the predicted protein before the DNA-binding domain, similar to that found in Ce-daf-12 [126], [135].
Table 4. Comparison of NHR pathway homologs and transcript abundances in S. stercoralis and C. elegans.
| C. elegans gene(s) | S. stercoralis homolog(s) | S. stercoralis transcript abundance profile | Regulation consistent with C. elegans (+, +/−, −)1 |
| Dafachronic acid biosynthesis | |||
| Ce-ncr-1 & -2 | Ss-ncr-1 | decreased in P Females | +/− |
| Ce-daf-36 | Ss-daf-36 | decreased in L3i and L3+ | +/− |
| Ce-dhs-16 | Ss-scdh-16 2 | decreased from L3i to L3+ | − |
| Ce-daf-9 | Ss-cyp-9 2 | decreased in FL and P Females | − |
| Ce-emb-8 | Ss-emb-8 | present in all stages examined | +/− |
| Ce-strm-1 | Ss-strm-1 | decreased from L3i to L3+ | + |
| Nuclear hormone receptor | |||
| Ce-daf-12 | Ss-daf-12 | peak in L3i | + |
| Ce-din-1 | Not identified | ||
| Ce -DAF-12 regulated genes | |||
| Ce-gck-2 | Ss-gck-2 | present in all stages examined | − |
| Ce-lev-9 | Ss-lev-9 | decreased in FL and P Females | − |
| Ce-lit-1 | Ss-lint-1 | decreased in FL and P Females | − |
| Ss-lint-2 | present in all stages examined | − | |
| Ce-ugt-65 | Ss-udpgt-1 | not present in L3i or L3+ | + |
| Ss-udpgt-2 | low expression, peak in L3i | − |
(+) similar, (+/−) unclear, and (−) dissimilar transcript abundance patterns.
Homology is by phylogenetic similarity only.
We then examined the developmental regulation of the S. stercoralis genes potentially involved in a DA biosynthetic pathway (Figure 7). We found that Ss-ncr-1 transcripts peak in L3+ and then significantly decrease in parasitic females (p<0.001), while Ss-daf-36 transcripts are at their nadir in L3i and L3+ developmental stages. Counterintuitively, we also found that Ss-cyp-9 transcripts are down-regulated in both free-living and parasitic females compared to the other developmental stages examined. Perhaps our most interesting observation was that Ss-daf-12 transcript levels peak in L3i and that the differences in expression also reflected significant changes in the promoter usage and coding forms (Figure S9).
Figure 7. Temporal regulation of S. stercoralis DAF-12 and genes putatively involved in dafachronic acid synthesis.
Transcript abundances were determined for the coding region of: (A) Ss-ncr-1, a gene encoding a homolog of the intracellular cholesterol transporters Ce-NCR-1 and Ce-NCR-2; (B) Ss-daf-36, a gene encoding a homolog of the Rieske-like oxygenase Ce-DAF-36; (C) Ss-scdh-16, a gene encoding a short-chain dehydrogenase homolog most similar to Ce-DHS-16; (D) Ss-cyp-9, a gene encoding a cytochrome P450 homolog most similar to Ce-DAF-9; (E) Ss-emb-8, a gene encoding a homolog of the cytochrome P450 reductase Ce-EMB-8; (F) Ss-strm-1, a gene encoding a homolog of the methyltransferase Ce-STRM-1; and (G) Ss-daf-12, the homolog of the nuclear hormone receptor Ce-DAF-12. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
Homologs of Ce-DAF-12-regulated genes are not similarly regulated in S. stercoralis development
We asked whether homologs of genes transcriptionally regulated by Ce-DAF-12 during dauer development were similarly regulated during S. stercoralis L3i development. We selected C. elegans genes that are directly linked to DAF-12 response elements, are differentially regulated during dauer development [138], and for which we could identify clear homologs in S. stercoralis (Table 4). We identified S. stercoralis homologs of Ce-lev-9 and Ce-gck-2, which are up-regulated during both dauer induction [138] and following dauer recovery [43], that we termed Ss-lev-9 and Ss-gck-2, respectively. We also identified two S. stercoralis homologs of Ce-lit-1, which is up-regulated in dauers [138], that we termed Ss-lint-1 and Ss-lint-2. Additionally, we identified two S. stercoralis homologs of Ce-ugt-65, a gene down-regulated during dauer formation by Ce-DAF-12 [138], which we termed Ss-udpgt-1 and Ss-udpgt-2. Intriguingly, we were unable to identify S. stercoralis homologs of the Ce-let-7 microRNA family [139] in the S. stercoralis or S. ratti draft genomes or in our de novo assembled S. stercoralis transcripts. Members of this microRNA family are directly regulated by DAF-12 in C. elegans and control several dauer developmental programs [140].
We did not observe any consistent regulation of the S. stercoralis homologs during L3i formation (Figure S13). In the seven developmental stages examined, Ss-gck-2 and Ss-lint-2 had no differences in transcript abundance greater than one log, Ss-udpgt-1 and Ss-udpgt-2 were expressed at very low levels in all stages examined, and Ss-lev-9 and Ss-lint-1 appeared to have decreased transcripts in parasitic and free-living females in comparison to the other developmental stages. This lack of consistent regulation of target genes between the two species appeared similar to that observed in S. stercoralis homologs of genes regulated by Ce-DAF-16 in C. elegans.
Discussion
In this study, we determined which homologs of C. elegans genes involved in dauer arrest and/or activation (Figure 2) are present in S. stercoralis and whether these S. stercoralis genes are developmentally regulated in a manner consistent with the regulation of their C. elegans counterparts. Our results have provided important insights into which developmental pathways are conserved between the morphologically similar dauer and L3i stages, thereby illuminating potential mechanisms governing L3i development. In our searches of the S. stercoralis and S. ratti draft genomes as well as our de novo assembled S. stercoralis transcript database, we were able to identify S. stercoralis homologs for nearly every C. elegans gene directly involved in the four canonical dauer pathways. While these pathways are well conserved in metazoans, they regulate a wide variety of functions; thus, we were specifically interested in whether they regulate S. stercoralis L3i arrest and/or activation.
In previous work, we demonstrated that both IIS and DAs play a role in S. stercoralis L3i arrest and activation [10], [22], [136]. However, we have also found that an S. stercoralis TGFβ ligand similar to Ce-DAF-7 is transcriptionally regulated in opposition to its C. elegans homolog [20]. A recent study found that genes involved in dauer recovery differ considerably between the clade V nematodes Pristionchus pacificus and C. elegans [141], further suggesting potential developmental differences between S. stercoralis and C. elegans, which are far more evolutionarily divergent [16]. Together, these studies, along with others in multiple parasitic nematode species, have demonstrated that while some C. elegans dauer pathway genes and metabolites appear to play a role in L3i development, others appear to be uninvolved [4]. Therefore, in this study, we used an RNAseq approach to globally examine the developmental regulation of S. stercoralis homologs in each of the four canonical dauer pathways and to gain key insights into their potential role in regulating S. stercoralis L3i development.
The role of cGMP signaling in S. stercoralis development
The pronounced up-regulation in L3i and the striking similarity of the transcriptional profiles of the S. stercoralis genes putatively involved in a cGMP signaling pathway (Figure 4) suggest a role in transducing host cues during the infective process. The similar up-regulation of putative guanylyl cyclases that do not phylogenetically group with Ce-DAF-11 suggests broad up-regulation of cGMP pathway components in S. stercoralis L3i and is reminiscent of studies in C. elegans showing that genes with similar temporal regulatory patterns often have similar genetic functions [142]. Since S. stercoralis L3i are attracted to chemical and thermal host cues [143]–[145] and are activated in host-like conditions [136], “priming” L3i for infection by up-regulating signaling components that relay these host cues would impart a selective advantage. Therefore, we hypothesize that cGMP signaling plays an important role in transducing signals of host recognition, consistent with studies in A. caninum, which demonstrated that a cGMP analog can stimulate L3i activation [66], [146].
This proposed role for cGMP signaling in S. stercoralis is somewhat at odds with the role of cGMP pathway signaling in C. elegans, where dauer pheromone, composed of a complex mixture of ascarosides, utilizes the cGMP signaling pathway to control dauer arrest [50], [52], [53]. This would suggest that an as yet undiscovered ascaroside helps to control S. stercoralis L3i formation. Recent reports suggest that ascarosides play a role in L3i formation in the closely related nematode P. trichosuri [68] as well as the entomopathogenic nematode Heterorhabditis bacteriophora [67]. Both of these species have multiple free-living generations, allowing ascaroside concentration to build up over time. In contrast, S. stercoralis has only one free-living generation, the progeny of which constitutively form L3i regardless of the population density. This makes it difficult to envisage a role for an environmentally secreted ascaroside by either the parasitic or free-living female, although an ascaroside that acts in utero on the developing embryo remains a possibility.
The role of IIS in S. stercoralis development
In previous studies, we have demonstrated that S. stercoralis IIS is crucial to both L3i arrest [10] and activation [22]. Down-regulation of IIS is necessary for L3i formation, since a Ss-DAF-16 dominant interfering construct designed to block the function of native Ss-DAF-16 results in L3i bypass phenotypes [10]. Furthermore, up-regulation of IIS is important during L3i activation, since pharmacological inhibition of PI3Ks, which include Ss-AGE-1, results in a dramatic decrease in L3i activation [22].
In this study, we demonstrate that the transcripts of intracellular IIS signaling components are always present in all developmental stages examined, with the exception of Ss-sgk-1 (Figure S8). These results are consistent with findings in C. elegans, where IIS signaling is thought to be regulated at the level of the ILPs, while intracellular signaling components are always present [41]. However, we did observe developmental regulation of several IIS signaling component transcripts in the post-free living generation, including increases in the transcript abundances for Ss-daf-2, Ss-age-1, Ss-pdk-1, Ss-akt-1, and Ss-daf-16 (Figure S8). We also noted an absence of Ss-sgk-1 transcripts in L3i and L3+ (Figure S8). In C. elegans, loss of Ce-sgk-1 results in increased stress resistance and lifespan extension [90]. These two attributes are key features of S. stercoralis L3i and we postulate that Ss-sgk-1 plays a role in these processes. Perhaps our most interesting observation is that Ss-akt-1b transcripts, encoding an isoform that is predicted to have a truncated PH domain and may not be subject to regulation by phosphatidylinositol lipids, are found almost exclusively in L3i and L3+ (Figure S9). We hypothesize that Ss-AKT-1B modulates S. stercoralis IIS during L3i development, potentially by interfering with Ss-AKT-1A or its substrates. Together, these data suggest that S. stercoralis IIS may be modulated at the level of the intracellular signaling proteins; however, the developmental transcript abundance profiles suggest that the core components are always present.
Upstream regulation of IIS by ILPs has never been demonstrated in parasitic nematodes, and it has generally been assumed that such regulation would be highly complex and redundant, similar to that of C. elegans, which has 40 known ILPs [41], [71]. In this study, extensive searches of the S. stercoralis and S. ratti draft genomes as well as de novo assembled S. stercoralis transcripts identified only seven ILPs (Figure 5A). These are conserved between these two parasite species but are highly divergent from the ILPs in C. elegans. We do not discount the possibility that other ILPs may be present in S. stercoralis; however, they would almost certainly have non-canonical cysteine architecture, given our search algorithm. Although we have no direct evidence to support their role in L3i development, we hypothesize that Ss-ilp-1, Ss-ilp-6, and Ss-ilp-7 encode ligands that regulate S. stercoralis IIS during L3i development. Determining whether an ILP acts as an agonist or antagonist is complicated by the fact that IIS regulates functions other than dauer development in C. elegans, including life-span [76].
We hypothesize that Ss-ilp-7 encodes a type γ antagonistic IIS ligand that promotes developmental arrest, due to the conservation of a unique PPG motif found in Ce-INS-18, which acts as an IIS antagonist in C. elegans [71], [81]. This hypothesis is supported by our observation that Ss-ilp-7 transcripts are significantly up-regulated in the post-free-living generation and peak in L3i, which are developmentally arrested (Figure 5H). However, Ss-ilp-7 transcripts remain at an elevated level in L3+, which are developmentally activated. The similar levels of Ss-ilp-7 transcripts in L3i and L3+ may reflect the fact that both forms are third-stage larvae and that the L3+ has yet to complete all the developmental programs associated with activation, which may only commence after molting and establishment in the intestine.
We also hypothesize that Ss-ilp-1 encodes an agonistic ligand that, when down-regulated, allows parasitic development, as it is the only other gene we identified to encode a type γ ILP, a family that also includes the human agonists insulin and IGF-1 [71]. The fact that Ss-ilp-1 transcripts are significantly down-regulated in L3i, L3+, and parasitic females supports this characterization (Figure 5B). Continued down-regulation of Ss-ilp-1 transcripts in L3+ and parasitic females is difficult to reconcile with a strictly developmental regulatory role in L3i. However, it should be noted that parasitic females retain characteristics of L3i, including an extended lifespan. Parasitic females live for many months in contrast to a lifespan of a few days for their free-living counterparts [147]. We hypothesize that negative regulation of lifespan by IIS is reversed in the long-lived parasitic forms and that Ss-ILP-1 participates in this effect.
We also speculate that Ss-ilp-6 encodes an agonistic ligand that promotes larval growth and development in both homogonic and heterogonic phases of the life cycle. This is based on our observation that Ss-ilp-6 transcript levels decrease during post-free-living development, reaching a minimum in physiologically older L3i, and then increase by one log in L3+, which have resumed feeding (Figure S7). Additionally, Ss-ilp-6 transcripts appear to increase in rapidly developing post-parasitic larvae (Figure 5G).
In future studies, we will test whether these three S. stercoralis ILPs, and possibly others, directly regulate L3i development. Due to the significantly smaller number of ILPs in S. stercoralis compared to C. elegans, as well as their highly divergent amino acid sequences and even novel cysteine architecture, we believe that these two species differ in the diversity and complexity of ligands regulating the DAF-2 receptor. Nevertheless, the data we report here are consistent with roles for S. stercoralis ILPs in the regulation of larval development and lifespan via the IIS pathway.
Differences in IIS regulated genes between S. stercoralis and C. elegans are suggested by our observation that none of the S. stercoralis homologs of Ce-DAF-16-regulated genes we examined, including Ss-sod-1 and Ss-daf-15, had transcript abundance differences greater than one log between any of the S. stercoralis developmental stages examined (Figure S10). We found this observation interesting because Ss-DAF-16 can heterologously complement C. elegans daf-16 mutants [19], suggesting similar biochemical capabilities. These results illustrate the important caveat that heterologous rescue does not prove that homologous genes fulfill similar genetic functions. However, no Ce-DAF-16-regulated genes have been shown to be “master regulators” of dauer development, and thus it is difficult to determine which target genes are most important. Ce-DAF-16 regulates several biological processes in addition to dauer development, including longevity, stress responses, and metabolism, and this has complicated the identification of target genes for specific processes [98]. The S. stercoralis genes in this study were selected because in C. elegans, the homologs are transcriptionally regulated directly by Ce-DAF-16 during dauer development and have dauer-associated phenotypes upon RNAi knock-down of their transcripts [98]. Our previous work points to S. stercoralis IIS regulating L3i arrest and activation, but the genes regulated by Ss-DAF-16 to carry out this process are unknown. In future studies, we hope to determine which S. stercoralis genes are regulated by Ss-DAF-16 using a chromatin immunoprecipitation and deep sequencing (ChIPseq) approach by constructing a stable transgenic S. stercoralis line that expresses a tagged version of Ss-DAF-16 [148].
The role of dauer-like TGFβ signaling in S. stercoralis development
Previously, we identified an S. stercoralis TGFβ ligand similar to Ce-DAF-7 [20], a finding repeated in two closely related parasitic nematodes [23]. In contrast to C. elegans, these putative TGFβ ligand-encoding genes are up-regulated in L3i, while Ce-daf-7 is down-regulated in the dauer stage [23]. In this study, we identified six additional S. stercoralis genes encoding TGFβ1 family ligands similar to Ce-DAF-7 (Figure 6A), which is the only dauer pathway TGFβ ligand in C. elegans [42], [102]. We were not only surprised by the increase in the number of genes encoding Ce-DAF-7-like ligands in S. stercoralis, but also by their temporal regulation.
We noted that Ss-tgh-1, Ss-tgh-2, and Ss-tgh-3 transcripts are found exclusively in L3i (Figure 6B–D), suggesting a similar function. As previously proposed, the S. stercoralis TGFβ-like ligands encoded by Ss-tgh-1, Ss-tgh-2, and Ss-tgh-3 may play a role in L3i arrest [23] or may be stored in L3i and secreted into the host following activation for purposes of immunomodulation [106]. Recent work has shown that H. polygyrus excretory-secretory antigen binds and activates the host TGFβ receptor, potentially supporting an immunomodulatory role for nematode TGFβ ligands [149]. Additionally, we noted highly variable transcript abundances for these three genes, as well as several others, in the L3i developmental stage, evidenced by the large 95% confidence intervals. Since we isolated L3i incubated at 21°C or 25°C and after 7, 8, or 10 days of culture (Data S11), we plotted the transcript abundances for the genes encoding both the ILP and TGH ligands over the course of post-free-living larval development, with each L3i biological replicate plotted by relative age (Figure S7). We observed that the large 95% confidence intervals were not stochastic, but rather represented underlying developmental trends dependent upon the relative age of the L3i. Therefore, we concluded that L3i may not be the static population originally assumed. Instead, physiologic age of developmentally arrested L3i, which is a function of temperature and time, may influence the transcriptomic profile of a synchronous population of these infectious larvae. These observations lead us to favor the hypothesis that the up-regulation of Ss-tgh-1, Ss-tgh-2, and Ss-tgh-3 during L3i development may play a role in L3i arrest; however, this role is not mutually exclusive with immunomodulation, given the plurality of TGH ligands in S. stercoralis.
In this study, we also identified four other genes encoding Ce-DAF-7-like ligands (Figure 6A). Both Ss-tgh-4 and Ss-tgh-5 transcripts were not detected in any developmental stage examined (Figure 6E–F), while Ss-tgh-6 and Ss-tgh-7 transcripts encoded putative peptides without a pro-peptide domain. We do not know whether Ss-tgh-4 or Ss-tgh-5 are ever robustly expressed during S. stercoralis development or are pseudo-genes; however, two important developmental stages, free-living males and auto-infective L3, were absent from this study. Additionally, the function of the putative ligands encoded by Ss-tgh-6 and Ss-tgh-7 are altogether unknown. Advances in the technologies to knock down genes by RNAi in S. stercoralis, which to date has been intractable to this approach, would facilitate our ability to address these questions [150].
Examination of intracellular signaling components of the dauer TGFβ pathway in S. stercoralis also led to some perplexing observations. While we were able to identify clear S. stercoralis homologs of genes encoding the Type I and Type II dauer pathway TGFβ receptors, Ss-daf-1 and Ss-daf-4, as well as the homolog of the Ce-DAF-5 transcriptional co-factor, Ss-DAF-5, we were unable to clearly identify homologs of the SMADs Ce-DAF-8, Ce-DAF-14, and Ce-DAF-3 (Figure S4). The large protein sequence divergence of dauer pathway TGFβ SMADs in S. stercoralis and all other sequenced nematodes, evidenced by our inability to resolve them phylogenetically, indicates a high degree of evolutionary divergence and suggests the potential for rapid evolution of these genes. This is in stark contrast to the SMADs in the Sma/Mab TGFβ pathway, for which clear sequence-level relationships exist for all nematode species examined (Figure S4). However, future testing of the functional consequences of this sequence-level divergence of dauer pathway TGFβ SMADs will be challenging due to the current limitations of functional genomic methods in S. stercoralis.
The regulation of dafachronic acid biosynthesis and DAF-12 in S. stercoralis development
We previously demonstrated that Δ7-DA is a potent activator of L3i and can act as a ligand for the nuclear hormone-receptor Ss-DAF-12 [136]. Furthermore, Δ7-DA can promote L3i bypass phenotypes in the post-free-living generation [136]. From these observations, we hypothesized that S. stercoralis synthesizes DAs in vivo and that the homologs of the enzymes responsible for DA biosynthesis would be up-regulated during reproductive development and down-regulated in L3i. This would also be consistent with observations in C. elegans, where Ce-daf-9 and Ce-daf-36 are up-regulated during dauer recovery [43].
Contrary to our hypothesis, we did not observe consistent developmental regulation of S. stercoralis homologs putatively involved in DA biosynthesis (Figure 7). Most puzzling was the significant decrease in Ss-cyp-9 transcripts, which encode a putative cytochrome P450 most similar to Ce-DAF-9, in both free-living and parasitic females (Figure 7D). Although Ss-daf-36 transcripts, which encode a putative Rieske-like oxygenase, appeared to be decreased in L3i and L3+ in comparison to other developmental stages, there was no significant difference between L3i and L3+ (Figure 7B). We expected both Ss-cyp-9 and Ss-daf-36 transcripts to be down-regulated in the developmentally arrested L3i and up-regulated in activated L3+; however, the L3+ may not have fully initiated all programs associated with resumption of development, as discussed for the ILPs. Only Ss-strm-1, the homolog of which in C. elegans encodes a methyltransferase that decreases DA levels when active, was down-regulated from L3i to L3+ and consistent with our hypothesis (Figure 7F). This inconsistent regulation of putative DA biosynthetic enzymes may be a result of our misidentification of several enzymes, such as Ss-cyp-9 and Ss-scdh-16, of which several closely related homologs are present in S. stercoralis (Figures S5 and S6). Additionally, these inconsistent results may be a result of additional layers of regulation which await discovery. In future studies, we hope to verify the role of these enzymes in a DA biosynthetic pathway.
Interestingly, we noted that unlike Ce-DAF-12, which is down-regulated in the dauer stage [126], Ss-daf-12 transcripts were at their peak in L3i (Figure 7G) and that this up-regulation was isoform specific (Figure S9). The differences in promoter usage as well as the predicted differences in the N-terminus of Ss-DAF-12 may represent additional layers of regulation. Until the native Ss-DAF-12 ligand(s) are identified and quantified for each developmental stage in future studies, the endogenous role of DAs in S. stercoralis development will remain difficult to assess.
Transcriptional regulation of S. stercoralis homologs of Ce-DAF-12-regulated genes was more difficult to interpret than for the S. stercoralis homologs of Ce-DAF-16-regulated genes. While some genes, including Ss-lev-9 and Ss-lint-1, appeared to be regulated similarly to Ss-cyp-9, others, including Ss-gck-2 and Ss-lint-2, did not display substantial changes in their transcript levels in the developmental stages examined (Figure S13). Our inability to identify S. stercoralis homologs of the Ce-let-7 miRNA family, which in C. elegans are directly regulated by Ce-DAF-12 to control development [140], further confounded our ability to interpret these results. In future studies, a ChIPseq approach using a tagged Ss-DAF-12 construct expressed in a stable transgenic S. stercoralis line would allow for the identification of native Ss-DAF-12-regulated genes in different developmental stages [148].
Putative mechanisms controlling S. stercoralis L3i development
Our data, as well as that from previous studies in other parasitic nematodes, point to several key regulatory pathways that may govern S. stercoralis L3i arrest and activation. The potent up-regulation of many cGMP signaling pathway components in L3i (Figure 4) is striking. We hypothesize that this pathway is directly involved in sensing/transducing host cues when L3i encounter a favorable host.
Perhaps the most interesting observation in this study is the paucity of S. stercoralis ILPs in comparison to C. elegans and the fact that several of these ILP encoding genes are dramatically up-/down-regulated during the course of L3i development (Figure 5). These data support a role for ILPs in regulating L3i arrest by modulating IIS, in agreement with our previous findings that Ss-DAF-16 regulates L3i arrest [10] and that S. stercoralis PI3Ks play a role in L3i activation [22]. We hypothesize that both L3i arrest in the environment and activation in the host are functions of the balance between agonistic and antagonistic ILPs. Under this hypothesis, down-regulation of agonistic ILPs and up-regulation of antagonistic ILPs would drive L3i arrest, while the reciprocal balance of ILPs would stimulate L3i activation and resumption of development in the host.
We were surprised by the increased number of genes encoding S. stercoralis TGFβ ligands similar to the single C. elegans dauer TGFβ ligand and the fact that three of these are expressed solely in L3i (Figure 6). As previously proposed [4], we hypothesize that these Ce-DAF-7-like TGFβ ligands play an important role in regulating L3i arrest. It is also possible that these TGFβ ligands may modulate host immunity.
Our lab and others have demonstrated that DAs are potent activators of L3i as well as stimulators of heterogonic development [136], [137]. However, the transcriptional profiles of the S. stercoralis DA biosynthetic enzyme homologs identified in this study do not demonstrate any coordinated regulation (Figure 7). Careful dissection of this pathway in future studies will be needed to determine the in vivo role of DA biosynthesis and Ss-DAF-12 regulation during L3i activation and heterogonic development. Together, these four pathways present several exciting avenues of future research in understanding the mechanisms controlling S. stercoralis L3i arrest and activation. Additionally, the transcriptomic data that formed the basis of this study provide a rich source of information for future unbiased global surveys of genes differentially regulated during L3i development and for many other aspects of parasitic nematode biology.
Supporting Information
Morphological comparison of S. stercoralis L3i and L3+. S. stercoralis third-stage larvae were photographed as (A) developmentally arrested and non-feeding third-stage larvae (L3i) or (B) as feeding third-stage larvae activated in vivo for three days in a permissive host (L3+). Overall, the body of the (A) L3i is more radially constricted than that of the (B) L3+. Similarly, the (A) L3i has a filariform pharynx (f) that is radially constricted forming a long thin tube extending almost to the midpoint of the worm, while the (B) L3+ has a pharynx that is pumping, is not radially constricted, and has differentiated into both muscular (m) and glandular(g) segments. The fork-shaped tail (arrow) in both the (A) L3i and (B) L3+ is characteristic of third-stage larvae in Strongyloides species and is lost in subsequent larval stages. Scale bar represents 100 µM.
(TIF)
Phylogenetic analysis of phylum Nematoda guanylyl cyclases similar to Ce -DAF-11. A protein alignment, generated with Clustal W, of several predicted guanylyl cyclases similar to Ce-DAF-11 from S. stercoralis and other parasitic nematodes was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted Ss-GCY-11 protein grouped with Ce-DAF-11. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Heterodera glycines (Hg), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Protein alignment of TGFβ ligand domains by cysteine architecture. Protein sequence alignment of TGFβ ligand domains including all cysteine residues in S. stercoralis and other metazoans revealed three distinct sets of cysteine architecture: Hs-TGFβ1-like ligands, including Ce-DAF-7, which have nine conserved cysteine residues; Drosophila melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP) -like ligands, including Ce-DBL-1 and Ce-UNC-129 subfamilies, which have seven conserved cysteine residues; and Ce-TIG-2-like ligands, which have six conserved cysteine residues. The conserved cysteine residues, critical for disulfide bond formation and “cysteine knot” folding, are in red. Abbreviations: Ancylostoma caninum (Ac), Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Danio rerio (Dr), Drosophila melanogaster (Dm), Haemonchus contortus (Hc), Heligmosomoides polygyrus (Hp), Homo sapiens (Hs), Loa loa (Ll), Parastrongyloides trichosuri (Pt), Strongyloides ratti (Sr), Strongyloides stercoralis (Ss), Trichinella spiralis (Ts), and Xenopus laevis (Xl). Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda SMAD homologs. A protein alignment, generated with Clustal W, of all publicly available phylum Nematoda SMAD homolog predicted proteins, was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The small body size and male tail abnormal (Sma/Mab) TGFβ pathway SMADs, including Ce-SMA-2, Ce-SMA-3, and Ce-SMA-4, resolved into distinct clades while the dauer TGFβ pathway SMADs, including Ce-DAF-3, Ce-DAF-8, and Ce-DAF-14, did not. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Heterodera glycines (Hg), Homo sapiens (Hs), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda short-chain dehydrogenase homologs similar to Ce -DHS-16. A protein alignment, generated with Clustal W, of phylum Nematoda short-chain dehydrogenase homologs similar to Ce-DHS-16 was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted protein for Ss-SCDH-16 grouped closest to Ce-DHS-16. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda cytochrome P450 homologs similar to Ce -DAF-9. A protein alignment, generated with Clustal W, of phylum Nematoda cytochrome P450 homologs similar to Ce-DAF-9 was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted protein for Ss-CYP-9 grouped closest to Ce-DAF-9. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Bursaphelenchus xylophilus (Bx), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Regulation of ILP and TGFβ ligand genes during post-free-living development. Transcript abundances were determined for the coding region of (A–G) seven S. stercoralis insulin-like peptide (ILP) -encoding genes (Ss-ilp-1 through -7) and (H–N) seven S. stercoralis DAF-7-like TGFβ ligand encoding genes (Ss-tgh-1 through -7) over the course of post-free living larval development. Transcript abundances were examined for post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), and in vivo activated third-stage larvae (L3+). The mean of both PFL L1 and L3+, from the three biological replicates, was plotted along with the 95% confidence intervals. Each biological sample of L3i was plotted individually according to relative age: L3i incubated at 21°C for 8 days (young L3i), L3i incubated at 25°C for 7 days (medium L3i), and L3i incubated at 21°C for 10 days (old L3i). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of S. stercoralis homologs of intracellular IIS genes. Transcript abundances were determined for the coding region of S. stercoralis homologs of genes encoding insulin/IGF-1-like signaling (IIS) pathway proteins, including: (A) Ss-asna-1, a homolog of Ce-asna-1, which encodes a putative membrane transporter involved in insulin-like peptide secretion; (B) Ss-daf-2, a homolog of Ce-daf-2, which encodes an insulin-like receptor; (C) Ss-ist-1 and (D) Ss-ist-2, homologs of the insulin receptor substrate encoding gene Ce-ist-1; (E) Ss-aap-1, a homolog of Ce-aap-1, which encodes a phosphatidylinositol-3 (PI3) kinase accessory/regulatory subunit; (F) Ss-age-1, a homolog of Ce-age-1, which encodes a PI3 kinase catalytic subunit; (G) Ss-pten-1 and (H) Ss-pten-2, homologs of Ce-daf-18, which encodes a phosphatase opposing Ce-age-1 function; (I) Ss-pdk-1, a homolog of Ce-pdk-1, which encodes a PTEN-like kinase; (J) Ss-sgk-1, a homolog of the serum- and glucocorticoid-inducible kinase Ce-sgk-1; (K) Ss-akt-1, a homolog of Ce-akt-1 and -2, which encode AKT kinases; (L) Ss-pptr-1, a homolog of Ce-pptr-1, which encodes a B56 regulatory subunit of the PP2A phosphatase opposing Ce-AKT-1 function; (M) Ss-ftt-2, a homolog of the 14-3-3 encoding gene Ce-ftt-2; and (N) Ss-daf-16, a homolog of Ce-daf-16, which encodes a forkhead transcription factor. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental stage-specific transcripts of S. stercoralis akt-1 , daf-16 , and daf-12 isoforms. (A, B) Two transcripts, each from different promoters, are generated from the Ss-akt-1 locus: (A) Ss-akt-1a and (B) Ss-akt-1b. The Ss-akt-1b transcript encodes a putative 525 amino acid Ss-AKT-1B peptide that has 33 amino acids truncated from the conserved AKT pleckstrin homology (PH) domain at the N-terminus. The Ss-akt-1a transcript encodes a putative 580 amino acid Ss-AKT-1A protein with a conserved full-length PH domain. (C, D) The locus from which the S. stercoralis forkhead transcription factor Ss-daf-16 is transcribed also produces two transcripts, (C) Ss-daf-16a and (D) Ss-daf-16b, each from a different promoter. These transcripts encode a 741 amino acid Ss-DAF-16A predicted protein and a 566 amino acid Ss-DAF-16B predicted protein, which differ at the N-terminus. (E–G) The genomic locus encoding the S. stercoralis nuclear hormone receptor homolog Ss-DAF-12 expresses at least seven different transcripts, which we termed Ss-daf-12a-g, from several promoters. These seven transcripts encode a total of three predicted proteins, each differing at the N-terminus before the DNA-binding domain, with transcripts (E) Ss-daf-12a,d,f encoding a putative 752 amino acid Ss-DAF-12A protein, (F) Ss-daf-12b encoding a putative 947 amino acid Ss-DAF-12B protein, and (G) Ss-daf-12c,e,g encoding a putative 722 amino acid Ss-DAF-12C protein. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of transcript exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Lack of developmental regulation of S. stercoralis homologs of Ce -DAF-16-regulated genes. Transcript abundances were examined for S. stercoralis homologs of C. elegans genes that are transcriptionally regulated directly by Ce-DAF-16 and have a phenotype associated with RNAi knock-down. (A) Ss-sod-1 is the sole S. stercoralis superoxide dismutase and is a homolog of Ce-sod-3. (B) Ss-daf-15 and (C) Ss-acs-19 are homologs of Ce-daf-15 and Ce-acs-19, respectively. Both (D) Ss-limdb-1 and (E) Ss-limdb-2 are homologs of Ce-lim-1, while (F) Ss-pitp-1 and (G) Ss-Y105E8B.9 are homologs of Ce-pitp-1 and Ce-Y105E8B.9, respectively. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of additional S. stercoralis TGFβ ligand transcripts. The S. stercoralis genome contains three TGFβ super-family ligands that do not group with the human TGFβ1 and Ce-DAF-7 family: (A) Ss-tigl-1, a gene encoding a homolog of Ce-TIG-2, a TGFβ ligand that does not have any known biological function or receptor; (B) Ss-dbl-1, a gene encoding a homolog of Ce-DBL-1, a TGFβ ligand homolog of D. melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP), which signals through the small body size and male tail abnormal (Sma/Mab) pathway; and (C) Ss-dbl-2, a gene encoding a homolog of the Ce-UNC-129 TGFβ ligand that regulates cell migration and axon guidance and acts independently of type I or type II receptors. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of TGFβ signaling homologs in S. stercoralis . Developmental expression patterns were assessed for members of both the dauer and small body size and male tail abnormal (Sma/Mab) TGFβ pathways: (A) Ss-daf-4, a gene encoding a homolog of the type II receptor Ce-DAF-4; (B) Ss-sma-6, a gene encoding a homolog of the Sma/Mab type I receptor Ce-SMA-6; (C) Ss-daf-1, a gene encoding a homolog of the dauer type I receptor Ce-DAF-1; (D) Ss-bra-1, a gene encoding a homolog of the Ce-DAF-1 negative regulator Ce-BRA-1; (E) Ss-smad-1, a gene encoding a SMAD similar to Ce-DAF-14; (F) Ss-smad-2, (G) Ss-smad-3, and (H) Ss-smad-4, genes encoding SMAD homologs similar to Ce-SMA-2, Ce-SMA-3, and Ce-SMA-4, respectively; (I) Ss-smad-5, (J) Ss-smad-7, and (K) Ss-smad-8, genes encoding SMAD homologs similar to Ce-DAF-3 and Ce-DAF-8; (L) Ss-pdp-1, a gene encoding a homolog of the phosphatase Ce-PDP-1; (M) Ss-sma-9, a gene encoding a homolog of the Sma/Mab transcriptional co-factor Ce-SMA-9; and (N) Ss-daf-5, a gene encoding a Sno/Ski transcription factor homolog similar to Ce-DAF-5. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of S. stercoralis homologs of Ce -DAF-12-regulated genes. Transcript abundances of S. stercoralis homologs of C. elegans genes transcriptionally regulated directly by Ce-DAF-12 during dauer development were examined: (A) Ss-gck-2, a gene encoding a homolog of Ce-GCK-2; (B) Ss-lev-9, a gene encoding a homolog of Ce-LEV-9; (C) Ss-lint-1 and (D) Ss-lint-2, genes encoding homologs of Ce-LIT-1; and (E) Ss-udpgt-1 and (F) Ss-udpgt-2, genes encoding homologs of Ce-UGT-65. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Strongyloides stercoralis dauer homolog genome annotations.
(GFF3)
Strongyloides stercoralis dauer homolog transcript sequences.
(FASTA)
Strongyloides stercoralis dauer homolog predicted protein sequences.
(FASTA)
Accession numbers for protein alignments.
(XLS)
Protein alignment for guanylyl cyclases similar to Ce -DAF-11.
(NEX)
Protein alignment for truncated TGFβ ligand domains.
(NEX)
Protein alignment for phylum Nematoda SMADs.
(NEX)
Protein alignment for short-chain dehydrogenases similar to Ce -DHS-16.
(NEX)
Protein alignment for cytochrome P450s similar to Ce -DAF-9.
(NEX)
FPKM values for Strongyloides stercoralis genes and isoforms.
(XLS)
RNAseq library preparation conditions and alignment statistics.
(XLS)
Supplemental methods for isolation of Strongyloides stercoralis developmental stages.
(DOC)
Acknowledgments
A special thanks to Nancy Holroyd and the Parasite Genomics group at the Wellcome Trust Sanger Institute for sharing pre-release S. stercoralis genome sequences. Also, thanks to members of the Frederick Bushman laboratory for assistance with protocols and troubleshooting scripts. Thanks to Brianna Bradford for assistance with the RNA isolation protocol and to Xinshe Lee, Najju Ranjit, and Beth Maguire for assistance with isolation of parasitic females. A special thanks to Robert Greenberg and Kristina Lewis for critical reading of the manuscript.
Funding Statement
This work was supported by U.S. National Institutes of Health (NIH) research grants AI50688 and AI22662 to JBL and RR02512 to Mark Haskins. MB is funded by the Wellcome Trust grant WT 098051. NIH training grant AI007532 helped to support JDS and AI060516 helped to support SM. This work was supported in part by the Penn Genome Frontiers Institute and a grant with the Pennsylvania Department of Health. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, et al. (2006) Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367: 1521–1532. [DOI] [PubMed] [Google Scholar]
- 2. Martin J, Abubucker S, Heizer E, Taylor CM, Mitreva M (2011) Nematode.net update 2011: addition of data sets and tools featuring next-generation sequencing data. Nucleic Acids Res 40: D720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schad GA, Chowdhury AB, Dean CG, Kochar VK, Nawalinski TA, et al. (1973) Arrested development in human hookworm infections: an adaptation to a seasonally unfavorable external environment. Science 180: 502–504. [DOI] [PubMed] [Google Scholar]
- 4. Viney ME (2009) How did parasitic worms evolve? Bioessays 31: 496–499. [DOI] [PubMed] [Google Scholar]
- 5. Dieterich C, Sommer RJ (2009) How to become a parasite - lessons from the genomes of nematodes. Trends Genet 25: 203–209. [DOI] [PubMed] [Google Scholar]
- 6.Lok JB (2007) Strongyloides stercoralis: a model for translational research on parasitic nematode biology, WormBook, ed. The C. elegans Research Community, WormBook, Available: http://www.wormbook.org/chapters/www_strongyloides/strongyloides.html. Accessed 2012 Sep 27. [DOI] [PMC free article] [PubMed]
- 7.Schad GA (1989) Morphology and life history of Strongyloides stercoralis. In: Grove DI, editor. Strongyloidiasis a major roundworm infection of man. London: Taylor and Francis. pp. 85–104.
- 8. Li X, Massey HC Jr, Nolan TJ, Schad GA, Kraus K, et al. (2006) Successful transgenesis of the parasitic nematode Strongyloides stercoralis requires endogenous non-coding control elements. Int J Parasitol 36: 671–679. [DOI] [PubMed] [Google Scholar]
- 9. Junio AB, Li X, Massey HC Jr, Nolan TJ, Todd Lamitina S, et al. (2008) Strongyloides stercoralis: cell- and tissue-specific transgene expression and co-transformation with vector constructs incorporating a common multifunctional 3′ UTR. Exp Parasitol 118: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castelletto ML, Massey HC Jr, Lok JB (2009) Morphogenesis of Strongyloides stercoralis infective larvae requires the DAF-16 ortholog FKTF-1. PLoS Pathog 5: e1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu PJ (2007) Dauer, WormBook, ed. The C. elegans Research Community, WormBook, Available: http://www.wormbook.org/chapters/www_dauer/dauer.html. Accessed 2012 Sep 27.
- 12. Fielenbach N, Antebi A (2008) C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22: 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hotez P, Hawdon J, Schad GA (1993) Hookworm larval infectivity, arrest and amphiparatenesis: the Caenorhabditis elegans Daf-c paradigm. Parasitol Today 9: 23–26. [DOI] [PubMed] [Google Scholar]
- 14. Burglin TR, Lobos E, Blaxter ML (1998) Caenorhabditis elegans as a model for parasitic nematodes. Int J Parasitol 28: 395–411. [DOI] [PubMed] [Google Scholar]
- 15.Riddle DL (1988) The dauer larva. In: Wood WB, editor. The nematode Caenorhabditis elegans. New York: Cold Spring Harbour Laboratory Press. pp. 393–412.
- 16. Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, et al. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- 17. Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, et al. (2006) Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol Biol Evol 23: 1792–1800. [DOI] [PubMed] [Google Scholar]
- 18. Massey HC Jr, Ball CC, Lok JB (2001) PCR amplification of putative gpa-2 and gpa-3 orthologs from the (A+T)-rich genome of Strongyloides stercoralis . Int J Parasitol 31: 377–383. [DOI] [PubMed] [Google Scholar]
- 19. Massey HC Jr, Bhopale MK, Li X, Castelletto M, Lok JB (2006) The fork head transcription factor FKTF-1b from Strongyloides stercoralis restores DAF-16 developmental function to mutant Caenorhabditis elegans . Int J Parasitol 36: 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Massey HC, Castelletto ML, Bhopale VM, Schad GA, Lok JB (2005) Sst-tgh-1 from Strongyloides stercoralis encodes a proposed ortholog of daf-7 in Caenorhabditis elegans . Mol Biochem Parasitol 142: 116–120. [DOI] [PubMed] [Google Scholar]
- 21. Siddiqui AA, Stanley CS, Skelly PJ, Berk SL (2000) A cDNA encoding a nuclear hormone receptor of the steroid/thyroid hormone-receptor superfamily from the human parasitic nematode Strongyloides stercoralis . Parasitol Res 86: 24–29. [DOI] [PubMed] [Google Scholar]
- 22. Stoltzfus JD, Massey HC Jr, Nolan TJ, Griffith SD, Lok JB (2012) Strongyloides stercoralis age-1: a potential regulator of infective larval development in a parasitic nematode. PLoS ONE 7: e38587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crook M, Thompson FJ, Grant WN, Viney ME (2005) daf-7 and the development of Strongyloides ratti and Parastrongyloides trichosuri . Mol Biochem Parasitol 139: 213–223. [DOI] [PubMed] [Google Scholar]
- 24. Ramanathan R, Varma S, Ribeiro JM, Myers TG, Nolan TJ, et al. (2011) Microarray-based analysis of differential gene expression between infective and noninfective larvae of Strongyloides stercoralis . PLoS Negl Trop Dis 5: e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mitreva M, McCarter JP, Martin J, Dante M, Wylie T, et al. (2004) Comparative genomics of gene expression in the parasitic and free-living nematodes Strongyloides stercoralis and Caenorhabditis elegans . Genome Res 14: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marcilla A, Garg G, Bernal D, Ranganathan S, Forment J, et al. (2012) The transcriptome analysis of Strongyloides stercoralis L3i larvae reveals targets for intervention in a neglected disease. PLoS Negl Trop Dis 6: e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi YJ, Ghedin E, Berriman M, McQuillan J, Holroyd N, et al. (2011) A deep sequencing approach to comparatively analyze the transcriptome of lifecycle stages of the filarial worm, Brugia malayi . PLoS Negl Trop Dis 5: e1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreno Y, Gros PP, Tam M, Segura M, Valanparambil R, et al. (2011) Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl Trop Dis 5: e1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cantacessi C, Young ND, Nejsum P, Jex AR, Campbell BE, et al. (2011) The transcriptome of Trichuris suis–first molecular insights into a parasite with curative properties for key immune diseases of humans. PLoS One 6: e23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laing R, Hunt M, Protasio AV, Saunders G, Mungall K, et al. (2011) Annotation of two large contiguous regions from the Haemonchus contortus genome using RNA-seq and comparative analysis with Caenorhabditis elegans . PLoS One 6: e23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jex AR, Liu S, Li B, Young ND, Hall RS, et al. (2011) Ascaris suum draft genome. Nature 479: 529–533. [DOI] [PubMed] [Google Scholar]
- 32. Schad GA, Hellman ME, Muncey DW (1984) Strongyloides stercoralis: hyperinfection in immunosuppressed dogs. Exp Parasitol 57: 287–296. [DOI] [PubMed] [Google Scholar]
- 33. Nolan TJ, Megyeri Z, Bhopale VM, Schad GA (1993) Strongyloides stercoralis: the first rodent model for uncomplicated and hyperinfective strongyloidiasis, the Mongolian gerbil (Meriones unguiculatus). J Infect Dis 168: 1479–1484. [DOI] [PubMed] [Google Scholar]
- 34. Herbert DR, Nolan TJ, Schad GA, Lustigman S, Abraham D (2002) Immunoaffinity-isolated antigens induce protective immunity against larval Strongyloides stercoralis in mice. Exp Parasitol 100: 112–120. [DOI] [PubMed] [Google Scholar]
- 35.Stiernagle T (2006) Maintenance of C. elegans, WormBook, ed. The C. elegans Research Community, Available: http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html. Accessed 2012 Sep 27.
- 36.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2011) Geneious v5.5, Available: http://www.geneious.com. Accessed 2012 Sep 27.
- 37. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pang K, Ryan JF, Baxevanis AD, Martindale MQ (2011) Evolution of the TGF-beta signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi . PLoS One 6: e24152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 40. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, et al. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baugh LR, Kurhanewicz N, Sternberg PW (2011) Sensitive and precise quantification of insulin-like mRNA expression in Caenorhabditis elegans . PLoS One 6: e18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, et al. (1996) Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274: 1389–1391. [DOI] [PubMed] [Google Scholar]
- 43. Wang J, Kim SK (2003) Global analysis of dauer gene expression in Caenorhabditis elegans . Development 130: 1621–1634. [DOI] [PubMed] [Google Scholar]
- 44. Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, et al. (2006) Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans . Cell 124: 1209–1223. [DOI] [PubMed] [Google Scholar]
- 45. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, et al. (2011) RNAi effector diversity in nematodes. PLoS Negl Trop Dis 5: e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Golden JW, Riddle DL (1982) A pheromone influences larval development in the nematode Caenorhabditis elegans . Science 218: 578–580. [DOI] [PubMed] [Google Scholar]
- 51. Golden JW, Riddle DL (1984) The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102: 368–378. [DOI] [PubMed] [Google Scholar]
- 52. Jeong PY, Jung M, Yim YH, Kim H, Park M, et al. (2005) Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433: 541–545. [DOI] [PubMed] [Google Scholar]
- 53. Butcher RA, Fujita M, Schroeder FC, Clardy J (2007) Small-molecule pheromones that control dauer development in Caenorhabditis elegans . Nat Chem Biol 3: 420–422. [DOI] [PubMed] [Google Scholar]
- 54. McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, et al. (2011) Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, et al. (2009) Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans . Science 326: 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH (1997) Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans dauer-inducing pheromone. Genetics 145: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, et al. (2000) A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans . Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Coburn CM, Mori I, Ohshima Y, Bargmann CI (1998) A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult Caenorhabditis elegans: a distinct pathway for maintenance of sensory axon structure. Development 125: 249–258. [DOI] [PubMed] [Google Scholar]
- 59. Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y (1996) Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans . Neuron 17: 707–718. [DOI] [PubMed] [Google Scholar]
- 60. Thomas JH, Birnby DA, Vowels JJ (1993) Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans . Genetics 134: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Murakami M, Koga M, Ohshima Y (2001) DAF-7/TGF-beta expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mech Dev 109: 27–35. [DOI] [PubMed] [Google Scholar]
- 62. Li W, Kennedy SG, Ruvkun G (2003) daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev 17: 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hahm JH, Kim S, Paik YK (2009) Endogenous cGMP regulates adult longevity via the insulin signaling pathway in Caenorhabditis elegans . Aging Cell 8: 473–483. [DOI] [PubMed] [Google Scholar]
- 64. Jensen VL, Bialas NJ, Bishop-Hurley SL, Molday LL, Kida K, et al. (2010) Localization of a guanylyl cyclase to chemosensory cilia requires the novel ciliary MYND domain protein DAF-25. PLoS Genet 6: e1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tissenbaum HA, Hawdon J, Perregaux M, Hotez P, Guarente L, et al. (2000) A common muscarinic pathway for diapause recovery in the distantly related nematode species Caenorhabditis elegans and Ancylostoma caninum . Proc Natl Acad Sci USA 97: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hawdon JM, Datu B (2003) The second messenger cyclic GMP mediates activation in Ancylostoma caninum infective larvae. Int J Parasitol 33: 787–793. [DOI] [PubMed] [Google Scholar]
- 67. Noguez JH, Conner ES, Zhou Y, Ciche TA, Ragains JR, et al. (2012) A novel ascaroside controls the parasitic life cycle of the entomopathogenic nematode Heterorhabditis bacteriophora . ACS Chem Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stasiuk SJ, Scott MJ, Grant WN (2012) Developmental plasticity and the evolution of parasitism in an unusual nematode, Parastrongyloides trichosuri . Evodevo 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, et al. (2012) Ascaroside signaling is widely conserved among nematodes. Curr Biol 22: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, et al. (2003) Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans . Cold Spring Harb Symp Quant Biol 68: 159–169. [DOI] [PubMed] [Google Scholar]
- 71. Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, et al. (2001) Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev 15: 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Duckert P, Brunak S, Blom N (2004) Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 17: 107–112. [DOI] [PubMed] [Google Scholar]
- 73. Tian S, Huajun W, Wu J (2012) Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci Rep 2: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J (2011) Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138: 1183–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hua QX, Nakagawa SH, Wilken J, Ramos RR, Jia W, et al. (2003) A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor. Genes Dev 17: 826–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans . Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- 77. Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, et al. (2006) The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans . Neuron 51: 613–625. [DOI] [PubMed] [Google Scholar]
- 78. Kodama E, Kuhara A, Mohri-Shiomi A, Kimura KD, Okumura M, et al. (2006) Insulin-like signaling and the neural circuit for integrative behavior in C. elegans . Genes Dev 20: 2955–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gregoire FM, Chomiki N, Kachinskas D, Warden CH (1998) Cloning and developmental regulation of a novel member of the insulin-like gene family in Caenorhabditis elegans . Biochem Biophys Res Commun 249: 385–390. [DOI] [PubMed] [Google Scholar]
- 80. Smit AB, Vreugdenhil E, Ebberink RH, Geraerts WP, Klootwijk J, et al. (1988) Growth-controlling molluscan neurons produce the precursor of an insulin-related peptide. Nature 331: 535–538. [DOI] [PubMed] [Google Scholar]
- 81. Kawano T, Ito Y, Ishiguro M, Takuwa K, Nakajima T, et al. (2000) Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans . Biochem Biophys Res Commun 273: 431–436. [DOI] [PubMed] [Google Scholar]
- 82. Massey HC Jr, Nishi M, Chaudhary K, Pakpour N, Lok JB (2003) Structure and developmental expression of Strongyloides stercoralis fktf-1, a proposed ortholog of daf-16 in Caenorhabditis elegans . Int J Parasitol 33: 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G (2002) Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem 277: 49591–49597. [DOI] [PubMed] [Google Scholar]
- 84. Rouault JP, Kuwabara PE, Sinilnikova OM, Duret L, Thierry-Mieg D, et al. (1999) Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol 9: 329–332. [DOI] [PubMed] [Google Scholar]
- 85. Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G (1999) A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans . Genes Dev 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Paradis S, Ruvkun G (1998) Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev 12: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, et al. (2001) Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem 276: 13402–13410. [DOI] [PubMed] [Google Scholar]
- 88. Lin K, Hsin H, Libina N, Kenyon C (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145. [DOI] [PubMed] [Google Scholar]
- 89. Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, et al. (2009) A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell 136: 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hertweck M, Gobel C, Baumeister R (2004) C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell 6: 577–588. [DOI] [PubMed] [Google Scholar]
- 91. Wang W, Shakes DC (1997) Expression patterns and transcript processing of ftt-1 and ftt-2, two C. elegans 14-3-3 homologues. J Mol Biol 268: 619–630. [DOI] [PubMed] [Google Scholar]
- 92. Li J, Tewari M, Vidal M, Lee SS (2007) The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans . Dev Biol 301: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kao G, Nordenson C, Still M, Ronnlund A, Tuck S, et al. (2007) ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell 128: 577–587. [DOI] [PubMed] [Google Scholar]
- 94. Barbieri M, Bonafe M, Franceschi C, Paolisso G (2003) Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab 285: E1064–1071. [DOI] [PubMed] [Google Scholar]
- 95. Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, et al. (2001) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans . Genome Res 11: 1346–1352. [DOI] [PubMed] [Google Scholar]
- 96. Lee SS, Kennedy S, Tolonen AC, Ruvkun G (2003) DAF-16 target genes that control C. elegans life-span and metabolism. Science 300: 644–647. [DOI] [PubMed] [Google Scholar]
- 97. McElwee J, Bubb K, Thomas JH (2003) Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121. [DOI] [PubMed] [Google Scholar]
- 98. Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, et al. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257. [DOI] [PubMed] [Google Scholar]
- 99. Hunter T, Bannister WH, Hunter GJ (1997) Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans . J Biol Chem 272: 28652–28659. [DOI] [PubMed] [Google Scholar]
- 100. Honda Y, Honda S (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans . Faseb J 13: 1385–1393. [PubMed] [Google Scholar]
- 101. Jia K, Chen D, Riddle DL (2004) The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897–3906. [DOI] [PubMed] [Google Scholar]
- 102.Savage-Dunn C (2005) TGF-beta signaling, WormBook, ed. The C. elegans Research Community, WormBook. Available: http://www.wormbook.org/chapters/www_tgfbsignal/tgfbsignal.html. Accessed 2012 Sep 27.
- 103. Freitas TC, Arasu P (2005) Cloning and characterisation of genes encoding two transforming growth factor-beta-like ligands from the hookworm, Ancylostoma caninum . Int J Parasitol 35: 1477–1487. [DOI] [PubMed] [Google Scholar]
- 104. Brand AM, Varghese G, Majewski W, Hawdon JM (2005) Identification of a DAF-7 ortholog from the hookworm Ancylostoma caninum . Int J Parasitol 35: 1489–1498. [DOI] [PubMed] [Google Scholar]
- 105. McSorley HJ, Grainger JR, Harcus Y, Murray J, Nisbet AJ, et al. (2010) daf-7-related TGF-beta homologues from Trichostrongyloid nematodes show contrasting life-cycle expression patterns. Parasitology 137: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gomez-Escobar N, Gregory WF, Maizels RM (2000) Identification of tgh-2, a filarial nematode homolog of Caenorhabditis elegans daf-7 and human transforming growth factor beta, expressed in microfilarial and adult stages of Brugia malayi . Infect Immun 68: 6402–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Viney ME, Thompson FJ, Crook M (2005) TGF-beta and the evolution of nematode parasitism. Int J Parasitol 35: 1473–1475. [DOI] [PubMed] [Google Scholar]
- 108. Freitas TC, Jung E, Pearce EJ (2007) TGF-beta signaling controls embryo development in the parasitic flatworm Schistosoma mansoni . PLoS Pathog 3: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Patterson GI, Padgett RW (2000) TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet 16: 27–33. [DOI] [PubMed] [Google Scholar]
- 110. Park D, Estevez A, Riddle DL (2010) Antagonistic Smad transcription factors control the dauer/non-dauer switch in C. elegans . Development 137: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Narasimhan SD, Yen K, Bansal A, Kwon ES, Padmanabhan S, et al. (2011) PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet 7: e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Schackwitz WS, Inoue T, Thomas JH (1996) Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans . Neuron 17: 719–728. [DOI] [PubMed] [Google Scholar]
- 113. Georgi LL, Albert PS, Riddle DL (1990) daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell 61: 635–645. [DOI] [PubMed] [Google Scholar]
- 114. Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, et al. (1993) The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- 115. Morita K, Shimizu M, Shibuya H, Ueno N (2001) A DAF-1-binding protein BRA-1 is a negative regulator of DAF-7 TGF-beta signaling. Proc Natl Acad Sci U S A 98: 6284–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Inoue T, Thomas JH (2000) Targets of TGF-beta signaling in Caenorhabditis elegans dauer formation. Dev Biol 217: 192–204. [DOI] [PubMed] [Google Scholar]
- 117. Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G (1997) The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev 11: 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. da Graca LS, Zimmerman KK, Mitchell MC, Kozhan-Gorodetska M, Sekiewicz K, et al. (2004) DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131: 435–446. [DOI] [PubMed] [Google Scholar]
- 119. Gunther CV, Georgi LL, Riddle DL (2000) A Caenorhabditis elegans type I TGF beta receptor can function in the absence of type II kinase to promote larval development. Development 127: 3337–3347. [DOI] [PubMed] [Google Scholar]
- 120. Gunther CV, Riddle DL (2004) Alternative polyadenylation results in a truncated daf-4 BMP receptor that antagonizes DAF-7-mediated development in Caenorhabditis elegans . J Biol Chem 279: 39555–39564. [DOI] [PubMed] [Google Scholar]
- 121. Herpin A, Lelong C, Favrel P (2004) Transforming growth factor-beta-related proteins: an ancestral and widespread superfamily of cytokines in metazoans. Dev Comp Immunol 28: 461–485. [DOI] [PubMed] [Google Scholar]
- 122. Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, et al. (1999) A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans . Development 126: 241–250. [DOI] [PubMed] [Google Scholar]
- 123. Morita K, Chow KL, Ueno N (1999) Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 126: 1337–1347. [DOI] [PubMed] [Google Scholar]
- 124. Krishna S, Maduzia LL, Padgett RW (1999) Specificity of TGFbeta signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans . Development 126: 251–260. [DOI] [PubMed] [Google Scholar]
- 125. Dumas KJ, Guo C, Wang X, Burkhart KB, Adams EJ, et al. (2010) Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev Biol 340: 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL (2000) daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans . Genes Dev 14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- 127. Li J, Brown G, Ailion M, Lee S, Thomas JH (2004) NCR-1 and NCR-2, the C. elegans homologs of the human Niemann-Pick type C1 disease protein, function upstream of DAF-9 in the dauer formation pathways. Development 131: 5741–5752. [DOI] [PubMed] [Google Scholar]
- 128. Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, et al. (2006) Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell 10: 473–482. [DOI] [PubMed] [Google Scholar]
- 129. Wollam J, Magner DB, Magomedova L, Rass E, Shen Y, et al. (2012) A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol 10: e1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A (2001) A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell 1: 841–851. [DOI] [PubMed] [Google Scholar]
- 131. Jia K, Albert PS, Riddle DL (2002) DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129: 221–231. [DOI] [PubMed] [Google Scholar]
- 132. Patel DS, Fang LL, Svy DK, Ruvkun G, Li W (2008) Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans . Development 135: 2239–2249. [DOI] [PubMed] [Google Scholar]
- 133. Hannich JT, Entchev EV, Mende F, Boytchev H, Martin R, et al. (2009) Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans . Dev Cell 16: 833–843. [DOI] [PubMed] [Google Scholar]
- 134. Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, et al. (2004) A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev 18: 2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Snow MI, Larsen PL (2000) Structure and expression of daf-12: a nuclear hormone receptor with three isoforms that are involved in development and aging in Caenorhabditis elegans . Biochim Biophys Acta 1494: 104–116. [DOI] [PubMed] [Google Scholar]
- 136. Wang Z, Zhou XE, Motola DL, Gao X, Suino-Powell K, et al. (2009) Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes. Proc Natl Acad Sci U S A 106: 9138–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ogawa A, Streit A, Antebi A, Sommer RJ (2009) A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol 19: 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Shostak Y, Van Gilst MR, Antebi A, Yamamoto KR (2004) Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes Dev 18: 2529–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, et al. (2003) The microRNAs of Caenorhabditis elegans . Genes Dev 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A (2009) Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324: 95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sinha A, Sommer RJ, Dieterich C (2012) Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans . BMC Genomics 13: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kim SK, Lund J, Kiraly M, Duke K, Jiang M, et al. (2001) A gene expression map for Caenorhabditis elegans . Science 293: 2087–2092. [DOI] [PubMed] [Google Scholar]
- 143. Lopez PM, Boston R, Ashton FT, Schad GA (2000) The neurons of class ALD mediate thermotaxis in the parasitic nematode, Strongyloides stercoralis . Int J Parasitol 30: 1115–1121. [DOI] [PubMed] [Google Scholar]
- 144. Forbes WM, Ashton FT, Boston R, Zhu X, Schad GA (2004) Chemoattraction and chemorepulsion of Strongyloides stercoralis infective larvae on a sodium chloride gradient is mediated by amphidial neuron pairs ASE and ASH, respectively. Vet Parasitol 120: 189–198. [DOI] [PubMed] [Google Scholar]
- 145. Safer D, Brenes M, Dunipace S, Schad G (2007) Urocanic acid is a major chemoattractant for the skin-penetrating parasitic nematode Strongyloides stercoralis . Proc Natl Acad Sci U S A 104: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Datu BJ, Loukas A, Cantacessi C, O'Donoghue P, Gasser RB (2009) Investigation of the regulation of transcriptional changes in Ancylostoma caninum larvae following serum activation, with a focus on the insulin-like signalling pathway. Vet Parasitol 159: 139–148. [DOI] [PubMed] [Google Scholar]
- 147.Viney ME, Lok JB (2007) Strongyloides spp., WormBook, ed. The C. elegans Research Community, WormBook, Available: http://www.wormbook.org/chapters/www_genomesStrongyloides/genomesStrongyloides.html. Accessed 2012 Sep 27.
- 148. Shao H, Li X, Nolan TJ, Massey HC Jr, Pearce EJ, et al. (2012) Transposon-mediated Chromosomal Integration of Transgenes in the Parasitic Nematode Strongyloides ratti and Establishment of Stable Transgenic Lines. PLoS Pathogens 8: e1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Grainger JR, Smith KA, Hewitson JP, McSorley HJ, Harcus Y, et al. (2010) Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. J Exp Med 207: 2331–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lok JB (2012) Nucleic acid transfection and transgenesis in parasitic nematodes. Parasitology 139: 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological comparison of S. stercoralis L3i and L3+. S. stercoralis third-stage larvae were photographed as (A) developmentally arrested and non-feeding third-stage larvae (L3i) or (B) as feeding third-stage larvae activated in vivo for three days in a permissive host (L3+). Overall, the body of the (A) L3i is more radially constricted than that of the (B) L3+. Similarly, the (A) L3i has a filariform pharynx (f) that is radially constricted forming a long thin tube extending almost to the midpoint of the worm, while the (B) L3+ has a pharynx that is pumping, is not radially constricted, and has differentiated into both muscular (m) and glandular(g) segments. The fork-shaped tail (arrow) in both the (A) L3i and (B) L3+ is characteristic of third-stage larvae in Strongyloides species and is lost in subsequent larval stages. Scale bar represents 100 µM.
(TIF)
Phylogenetic analysis of phylum Nematoda guanylyl cyclases similar to Ce -DAF-11. A protein alignment, generated with Clustal W, of several predicted guanylyl cyclases similar to Ce-DAF-11 from S. stercoralis and other parasitic nematodes was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted Ss-GCY-11 protein grouped with Ce-DAF-11. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Heterodera glycines (Hg), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Protein alignment of TGFβ ligand domains by cysteine architecture. Protein sequence alignment of TGFβ ligand domains including all cysteine residues in S. stercoralis and other metazoans revealed three distinct sets of cysteine architecture: Hs-TGFβ1-like ligands, including Ce-DAF-7, which have nine conserved cysteine residues; Drosophila melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP) -like ligands, including Ce-DBL-1 and Ce-UNC-129 subfamilies, which have seven conserved cysteine residues; and Ce-TIG-2-like ligands, which have six conserved cysteine residues. The conserved cysteine residues, critical for disulfide bond formation and “cysteine knot” folding, are in red. Abbreviations: Ancylostoma caninum (Ac), Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Danio rerio (Dr), Drosophila melanogaster (Dm), Haemonchus contortus (Hc), Heligmosomoides polygyrus (Hp), Homo sapiens (Hs), Loa loa (Ll), Parastrongyloides trichosuri (Pt), Strongyloides ratti (Sr), Strongyloides stercoralis (Ss), Trichinella spiralis (Ts), and Xenopus laevis (Xl). Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda SMAD homologs. A protein alignment, generated with Clustal W, of all publicly available phylum Nematoda SMAD homolog predicted proteins, was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The small body size and male tail abnormal (Sma/Mab) TGFβ pathway SMADs, including Ce-SMA-2, Ce-SMA-3, and Ce-SMA-4, resolved into distinct clades while the dauer TGFβ pathway SMADs, including Ce-DAF-3, Ce-DAF-8, and Ce-DAF-14, did not. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Heterodera glycines (Hg), Homo sapiens (Hs), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda short-chain dehydrogenase homologs similar to Ce -DHS-16. A protein alignment, generated with Clustal W, of phylum Nematoda short-chain dehydrogenase homologs similar to Ce-DHS-16 was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted protein for Ss-SCDH-16 grouped closest to Ce-DHS-16. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Phylogenetic analysis of phylum Nematoda cytochrome P450 homologs similar to Ce -DAF-9. A protein alignment, generated with Clustal W, of phylum Nematoda cytochrome P450 homologs similar to Ce-DAF-9 was used to construct a neighbor-joining tree with 100 iterations of boot-strapping. The predicted protein for Ss-CYP-9 grouped closest to Ce-DAF-9. Abbreviations: Ascaris suum (As), Brugia malayi (Bm), Bursaphelenchus xylophilus (Bx), Caenorhabditis briggsae (Cb), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Loa loa (Ll), Strongyloides stercoralis (Ss), and Trichinella spiralis (Ts). The scale bar represents substitutions per position. Accession numbers are listed in Data S4.
(TIF)
Regulation of ILP and TGFβ ligand genes during post-free-living development. Transcript abundances were determined for the coding region of (A–G) seven S. stercoralis insulin-like peptide (ILP) -encoding genes (Ss-ilp-1 through -7) and (H–N) seven S. stercoralis DAF-7-like TGFβ ligand encoding genes (Ss-tgh-1 through -7) over the course of post-free living larval development. Transcript abundances were examined for post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), and in vivo activated third-stage larvae (L3+). The mean of both PFL L1 and L3+, from the three biological replicates, was plotted along with the 95% confidence intervals. Each biological sample of L3i was plotted individually according to relative age: L3i incubated at 21°C for 8 days (young L3i), L3i incubated at 25°C for 7 days (medium L3i), and L3i incubated at 21°C for 10 days (old L3i). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of S. stercoralis homologs of intracellular IIS genes. Transcript abundances were determined for the coding region of S. stercoralis homologs of genes encoding insulin/IGF-1-like signaling (IIS) pathway proteins, including: (A) Ss-asna-1, a homolog of Ce-asna-1, which encodes a putative membrane transporter involved in insulin-like peptide secretion; (B) Ss-daf-2, a homolog of Ce-daf-2, which encodes an insulin-like receptor; (C) Ss-ist-1 and (D) Ss-ist-2, homologs of the insulin receptor substrate encoding gene Ce-ist-1; (E) Ss-aap-1, a homolog of Ce-aap-1, which encodes a phosphatidylinositol-3 (PI3) kinase accessory/regulatory subunit; (F) Ss-age-1, a homolog of Ce-age-1, which encodes a PI3 kinase catalytic subunit; (G) Ss-pten-1 and (H) Ss-pten-2, homologs of Ce-daf-18, which encodes a phosphatase opposing Ce-age-1 function; (I) Ss-pdk-1, a homolog of Ce-pdk-1, which encodes a PTEN-like kinase; (J) Ss-sgk-1, a homolog of the serum- and glucocorticoid-inducible kinase Ce-sgk-1; (K) Ss-akt-1, a homolog of Ce-akt-1 and -2, which encode AKT kinases; (L) Ss-pptr-1, a homolog of Ce-pptr-1, which encodes a B56 regulatory subunit of the PP2A phosphatase opposing Ce-AKT-1 function; (M) Ss-ftt-2, a homolog of the 14-3-3 encoding gene Ce-ftt-2; and (N) Ss-daf-16, a homolog of Ce-daf-16, which encodes a forkhead transcription factor. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental stage-specific transcripts of S. stercoralis akt-1 , daf-16 , and daf-12 isoforms. (A, B) Two transcripts, each from different promoters, are generated from the Ss-akt-1 locus: (A) Ss-akt-1a and (B) Ss-akt-1b. The Ss-akt-1b transcript encodes a putative 525 amino acid Ss-AKT-1B peptide that has 33 amino acids truncated from the conserved AKT pleckstrin homology (PH) domain at the N-terminus. The Ss-akt-1a transcript encodes a putative 580 amino acid Ss-AKT-1A protein with a conserved full-length PH domain. (C, D) The locus from which the S. stercoralis forkhead transcription factor Ss-daf-16 is transcribed also produces two transcripts, (C) Ss-daf-16a and (D) Ss-daf-16b, each from a different promoter. These transcripts encode a 741 amino acid Ss-DAF-16A predicted protein and a 566 amino acid Ss-DAF-16B predicted protein, which differ at the N-terminus. (E–G) The genomic locus encoding the S. stercoralis nuclear hormone receptor homolog Ss-DAF-12 expresses at least seven different transcripts, which we termed Ss-daf-12a-g, from several promoters. These seven transcripts encode a total of three predicted proteins, each differing at the N-terminus before the DNA-binding domain, with transcripts (E) Ss-daf-12a,d,f encoding a putative 752 amino acid Ss-DAF-12A protein, (F) Ss-daf-12b encoding a putative 947 amino acid Ss-DAF-12B protein, and (G) Ss-daf-12c,e,g encoding a putative 722 amino acid Ss-DAF-12C protein. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of transcript exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Lack of developmental regulation of S. stercoralis homologs of Ce -DAF-16-regulated genes. Transcript abundances were examined for S. stercoralis homologs of C. elegans genes that are transcriptionally regulated directly by Ce-DAF-16 and have a phenotype associated with RNAi knock-down. (A) Ss-sod-1 is the sole S. stercoralis superoxide dismutase and is a homolog of Ce-sod-3. (B) Ss-daf-15 and (C) Ss-acs-19 are homologs of Ce-daf-15 and Ce-acs-19, respectively. Both (D) Ss-limdb-1 and (E) Ss-limdb-2 are homologs of Ce-lim-1, while (F) Ss-pitp-1 and (G) Ss-Y105E8B.9 are homologs of Ce-pitp-1 and Ce-Y105E8B.9, respectively. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of additional S. stercoralis TGFβ ligand transcripts. The S. stercoralis genome contains three TGFβ super-family ligands that do not group with the human TGFβ1 and Ce-DAF-7 family: (A) Ss-tigl-1, a gene encoding a homolog of Ce-TIG-2, a TGFβ ligand that does not have any known biological function or receptor; (B) Ss-dbl-1, a gene encoding a homolog of Ce-DBL-1, a TGFβ ligand homolog of D. melanogaster decapentaplegic (DPP) and vertebrate bone morphogenetic protein (BMP), which signals through the small body size and male tail abnormal (Sma/Mab) pathway; and (C) Ss-dbl-2, a gene encoding a homolog of the Ce-UNC-129 TGFβ ligand that regulates cell migration and axon guidance and acts independently of type I or type II receptors. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of TGFβ signaling homologs in S. stercoralis . Developmental expression patterns were assessed for members of both the dauer and small body size and male tail abnormal (Sma/Mab) TGFβ pathways: (A) Ss-daf-4, a gene encoding a homolog of the type II receptor Ce-DAF-4; (B) Ss-sma-6, a gene encoding a homolog of the Sma/Mab type I receptor Ce-SMA-6; (C) Ss-daf-1, a gene encoding a homolog of the dauer type I receptor Ce-DAF-1; (D) Ss-bra-1, a gene encoding a homolog of the Ce-DAF-1 negative regulator Ce-BRA-1; (E) Ss-smad-1, a gene encoding a SMAD similar to Ce-DAF-14; (F) Ss-smad-2, (G) Ss-smad-3, and (H) Ss-smad-4, genes encoding SMAD homologs similar to Ce-SMA-2, Ce-SMA-3, and Ce-SMA-4, respectively; (I) Ss-smad-5, (J) Ss-smad-7, and (K) Ss-smad-8, genes encoding SMAD homologs similar to Ce-DAF-3 and Ce-DAF-8; (L) Ss-pdp-1, a gene encoding a homolog of the phosphatase Ce-PDP-1; (M) Ss-sma-9, a gene encoding a homolog of the Sma/Mab transcriptional co-factor Ce-SMA-9; and (N) Ss-daf-5, a gene encoding a Sno/Ski transcription factor homolog similar to Ce-DAF-5. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Developmental regulation of S. stercoralis homologs of Ce -DAF-12-regulated genes. Transcript abundances of S. stercoralis homologs of C. elegans genes transcriptionally regulated directly by Ce-DAF-12 during dauer development were examined: (A) Ss-gck-2, a gene encoding a homolog of Ce-GCK-2; (B) Ss-lev-9, a gene encoding a homolog of Ce-LEV-9; (C) Ss-lint-1 and (D) Ss-lint-2, genes encoding homologs of Ce-LIT-1; and (E) Ss-udpgt-1 and (F) Ss-udpgt-2, genes encoding homologs of Ce-UGT-65. Transcript abundances were quantified in seven developmental stages: free-living females (FL Female), post-free-living first-stage larvae (PFL L1), infectious third-stage larvae (L3i), in vivo activated third-stage larvae (L3+), parasitic females (P Female), post-parasitic first-stage larvae (PP L1), and post-parasitic third-stage larvae (PP L3). Transcript abundances were calculated as fragments per kilobase of coding exon per million mapped reads (FPKM) and log transformed. Error bars represent 95% confidence intervals. The y-axes were scaled from 0 to 3.5 to aid comparison between genes.
(TIF)
Strongyloides stercoralis dauer homolog genome annotations.
(GFF3)
Strongyloides stercoralis dauer homolog transcript sequences.
(FASTA)
Strongyloides stercoralis dauer homolog predicted protein sequences.
(FASTA)
Accession numbers for protein alignments.
(XLS)
Protein alignment for guanylyl cyclases similar to Ce -DAF-11.
(NEX)
Protein alignment for truncated TGFβ ligand domains.
(NEX)
Protein alignment for phylum Nematoda SMADs.
(NEX)
Protein alignment for short-chain dehydrogenases similar to Ce -DHS-16.
(NEX)
Protein alignment for cytochrome P450s similar to Ce -DAF-9.
(NEX)
FPKM values for Strongyloides stercoralis genes and isoforms.
(XLS)
RNAseq library preparation conditions and alignment statistics.
(XLS)
Supplemental methods for isolation of Strongyloides stercoralis developmental stages.
(DOC)