Abstract
Three light-regulated genes, chlorophyll a/b-binding protein (CAB), ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, and chalcone synthase (CHS), are demonstrated to be up-regulated in the high-pigment-1 (hp-1) mutant of tomato (Lycopersicon esculentum Mill.) compared with wild type (WT). However, the pattern of up-regulation of the three genes depends on the light conditions, stage of development, and tissue studied. Compared with WT, the hp-1 mutant showed higher CAB gene expression in the dark after a single red-light pulse and in the pericarp of immature fruits. However, in vegetative tissues of light-grown seedlings and adult plants, CAB mRNA accumulation did not differ between WT and the hp-1 mutant. The ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit mRNA accumulated to a higher level in the hp-1 mutant than WT under all light conditions and tissues studied, whereas CHS gene expression was up-regulated in de-etiolated vegetative hp-1-mutant tissues only. The CAB and CHS genes were shown to be phytochrome regulated and both phytochrome A and B1 play a role in CAB gene expression. These observations support the hypothesis that the HP-1 protein plays a general repressive role in phytochrome signal transduction.
Light controls many aspects of plant morphogenesis and provides energy for photosynthesis. Different regions of the spectrum are perceived by different photoreceptor molecules: the B photoreceptors, the UV photoreceptors, and the R-/FR-sensitive phytochromes. Phytochromes were physiologically identified 50 years ago, and in the last two decades different phytochrome types have been purified and cloned from several plant species. Mutants deficient in specific phytochrome family members have been isolated from several species: e.g. Arabidopsis (for review, see Smith, 1995), tomato (Lycopersicon esculentum Mill.) (van Tuinen et al., 1995a, 1995b), and pea (Weller et al., 1995). These mutants are excellent tools for studying the functions of the different members of the phytochrome family.
Although there is information about photoperception by phytochromes, little is known about the signal transduction pathways linking these receptors with gene expression. Several approaches have been used to study phytochrome signal transduction pathways. First, using microinjection, Neuhaus et al. (1993) identified several molecules that participate in phytochrome-signal transduction. The existence of two separate pathways was proposed: the cGMP-mediated pathway that leads to the regulation of CHS genes, and the Ca2+-calmodulin mediated pathway that regulates the expression of CAB and RBCS genes. In both pathways the signal is transduced from phytochrome via a heterotrimeric G protein, and subsequently the existence of a reciprocal control mechanism between the pathways has been demonstrated (Bowler et al., 1994a, 1994b).
A second approach to identify and characterize components and regulators of phytochrome-signal transduction pathways is the isolation and characterization of mutants with altered light responses. Constitutive-response mutants such as constitutive photomorphogenesis (cop), de-etiolated (det), and fusca (fus) in Arabidopsis (for review, see Wei and Deng, 1996), light-independent (lip) in pea (Frances et al., 1992), and hyp2 in tobacco (Traas et al., 1995) are excellent tools for these studies. These mutants fail to exhibit the characteristics of dark-grown seedlings and show reduced elongation and expanded leaves. Some also accumulate anthocyanin in the dark. The cloning of the corresponding genes from Arabidopsis allowed the biochemical characterization of the affected gene products and provided information about the possible role and function of these components in phytochrome-signal transduction (Wei and Deng, 1996). In addition, mutants in genes affecting phyA and B signaling have been reported (Whitelam et al., 1993; Ahmad and Cashmore, 1996; Wagner et al., 1997; Hoecker et al., 1998).
A third approach to identify the components of phytochrome-signal transduction is to screen directly for mutants altered in the regulation of particular light-regulated genes. Li et al. (1994, 1995) isolated Arabidopsis mutants altered in the regulation of CAB gene expression. These mutants were named doc (for dark overexpression of CAB) and cue1 (for CAB underexpressed). The doc mutation affects the expression of CAB genes and the cue1 mutation affects the expression of both CAB and RBCS genes. The expression of CHS genes was neither modified in doc nor in cue1 mutant plants. In contrast, the increased CHS expression (icx1) mutant of Arabidopsis shows enhanced induction of CHS gene expression by light, but no alteration in the level of CAB transcript accumulation (Jackson et al., 1995).
In this paper we examine a putative phytochrome signal transduction mutant of tomato, the hp-1 (high-pigment-1) mutant. This monogenic recessive hp-1 mutant was first identified in 1917 (Reynard, 1956) and exhibits higher anthocyanin content, shorter hypocotyl (Kerr, 1965; Mochizuki and Kamimura, 1985; Peters et al., 1989), and darker green foliage (Jarret et al., 1984) and fruits (Thompson, 1962) when compared with WT. The HP-1 gene has been recently mapped to chromosome 2 (Yen et al., 1997). Soressi (1975) identified a recessive hp-2 mutant, which is phenotypically similar but nonallelic to hp-1 and maps to chromosome 1 (van Tuinen et al., 1997). Attempts to isolate Arabidopsis counterparts of the tomato hp mutants have been reported (Ichikawa et al., 1996), but await detailed analysis.
Although the nature of the hp mutations is still unclear, detailed physiological characterization of the hp-1 mutant provided a valuable insight into phytochrome signal transduction processes. The hp-1 mutant has high levels of anthocyanin and reduced height of light-grown seedlings (Peters et al., 1992a; Kerckhoffs et al., 1997a). Furthermore, the photoinduction of several enzymes in biochemical pathways: Phe ammonia lyase (Goud et al., 1991), nitrate reductase, nitrite reductase, and amylase (Goud and Sharma, 1994), are amplified in the hp-1 mutant compared with WT. All of these features have been shown previously to be phytochrome regulated, and therefore, it was concluded that the hp-1 mutant shows exaggerated phytochrome responses (Kerckhoffs and Kendrick, 1997). The apparent phenocopying of the hp-1 mutant's phenotype and immature fruit color as a result of phyA overexpression in tomato (Boylan and Quail, 1989) is consistent with this idea. However, in vivo spectrophotometric and immunochemical analysis failed to provide evidence that the hp-1 mutant is a photoreceptor mutant (Peters et al., 1992b; Kerckhoffs et al., 1997a). Therefore, it was proposed that the hp-1 mutation is associated with an amplification step in the phytochrome-transduction chain (Peters et al., 1992b; Kerckhoffs et al., 1997a; Kerckhoffs and Kendrick, 1997). This conclusion is supported by the recent observation using specific phytochrome family-member-deficient mutants, that it is phyA and phyB1 that play a dominant role in the seedling anthocyanin response (Kerckhoffs et al., 1997b). In the phytochrome-amplification model, phytochrome responses are envisaged to be under the constraint of the HP-1 gene product. Both B and the hp-1 mutation appear to be able to relieve this constraint (Peters et al., 1989, 1992b).
The dark-green immature fruit color of the hp-1 mutant compared with WT is due to higher chlorophyll levels (Sanders et al., 1975; Kerckhoffs, 1996) and the mature hp-1-mutant fruits have a higher lycopene and carotene content and increased levels of ascorbic acid than those of WT (Thompson, 1962). Recently, the plastid copy number in the hypocotyls and the Suc and flavonoid contents of ripe fruits have been reported to be elevated in the hp-1 mutant compared with WT (Yen et al., 1997).
In this paper we characterize the effect of the hp-1 mutation on CAB, RBCS, and CHS gene expression at different developmental stages using the most extreme allele available (hp-1w).
MATERIALS AND METHODS
Plant Material and Growth Conditions
The tomato (Lycopersicon esculentum Mill.) genotypes used in the experiments were hp-1w (Peters et al., 1989); hp-1w,fri1 (far-red light insensitive), deficient in phyA (Kerckhoffs et al., 1997b); hp-1w,tri1 (temporarily red-light insensitive), deficient in phyB1 (Kerckhoffs et al., 1997b) in the genetic backgrounds MoneyMaker (MM) or breeding line GT.
For the experiments with seedlings, seeds were surface sterilized for 3 min in a 1% (v/v) dilution of commercial bleach and rinsed for 5 min in Milli-Q water (Milli-RO 8 water purification system, Millipore). Seeds were sown at noon on 0.6% (w/v) agar medium containing 0.46 g L−1 Murashige-Skoog basal salts (Murashige-Skoog, 1962) in plastic tissue culture containers (Plantcon, Flow Laboratories Inc., McLean, VA) and germinated in a FR-6113A growth chamber (Koito, Tokyo, Japan) at 25°C. To germinate seedlings in absolute darkness, tissue culture containers were wrapped in aluminum foil, put in a black velvet sack, and grown in a dark room at 25°C. In the light-pulse experiments R (27 μmol m−2 s−1) was obtained from FL20SRF fluorescent tubes (National, Osaka, Japan) filtered through a red, plastic filter (Shinkolite A no. 102, Mitsubishi Rayon Corp., Tokyo, Japan) and FR (33 μmol m−2 s−1) from FL20S–FR74 fluorescent tubes (Toshiba) wrapped with one layer of Polycolor no. 22 and one layer of Polycolor no. 72 film (Tokyo Butai Shomei Co., Tokyo, Japan). B (11 μmol m−2 s−1) was obtained from FL20S.B fluorescent tubes (Toshiba). WL-grown seedlings were germinated in 16-h WL (120 μmol m−2 s−1 PAR) 8-h dark cycles at 25°C. WL was obtained from FL20SD SDL fluorescent tubes (National).
For the experiments with adult plants and fruits, seeds were sown in the greenhouse in a 4:1 vermiculite/granular-clay-based compost mixture. After 1 month plants were transplanted to pots (19 cm [diameter] × 15 cm [height] for vegetative tissues of adult plants and 27.1 cm × 28.6 cm for fruits) containing 2:1 vermiculite/granular-clay based compost mixture and transferred to a phytotron KG-206HL-D (Koito) with 16-h WL (250 μmol m−2 s−1 PAR) 8-h dark cycles at 25°C. Vegetative plant material was harvested 2 months after sowing and frozen in liquid nitrogen. The frozen material was stored in a −135°C freezer until use. After the first fruit(s) on a plant became red, all fruits of that particular plant were harvested. Harvest was always at noon because of diurnal mRNA fluctuations of CAB genes in tomato fruits (Piechulla and Gruissem, 1987). Directly after harvest a picture was taken of the fruits from one plant. The age, diameter, length, and weight of each fruit were measured and samples were taken for the chlorophyll assay. The remaining material was separated into pericarp (the outer wall of the pericarp including the epidermis) and the inner section (radial and inner wall of the pericarp, placental tissue, and locular cavity with seeds), and frozen in liquid nitrogen. The frozen material was stored in a −135°C freezer until use.
Chlorophyll Assay
To determine the chlorophyll content in the fruits, samples were taken from the equator of the fruits using an 11-mm cork borer. From this sample the pericarp and about 5 mm of the inner section (for definition, see above) directly bordering the pericarp were cut. For each fruit the chlorophyll was extracted from one pericarp and one inner-section disc. The fruit discs were placed in 15-mL tubes (Falcon) and incubated in darkness for at least 48 h at 65°C in DMSO (after the work of Hiscox and Israelstam, 1979). Samples were re-extracted with DMSO until no extra chlorophyll could be extracted, and the samples were always kept in the dark. When the samples were cooled to room temperature, A649 and A665 were determined spectrophotometrically. Chlorophyll a and b were calculated on a gram fresh weight basis, using the equations for ethanol published by Lichtenthaler and Wellburn (1983).
Anthocyanin Assay
Anthocyanin was extracted from cotyledons and hypocotyls of seedlings with 0.6 mL of acidified (0.3% HCl, v/v) methanol for 48 h. The extraction was carried out by shaking the samples in darkness at room temperature for 48 h. At the end of the extraction 0.45 mL of water and 1.2 mL of chloroform were added. Samples were vortexed and centrifuged for 20 min at 4500g. The A535 of the upper anthocyanin-containing phase was determined spectrophotometrically (DU650, Beckman).
RNA Gel-Blot Analysis
Total RNA was isolated by a modification of the method of Loening (1969) and was previously described by Peters and Silverthorne (1995). RNA was electrophoresed in 1% (w/v) agarose gels containing Mops buffer (20 mm Mops, 1 mm EDTA, and 5 mm sodium acetate, pH 7.0) and 6.7% (v/v) formaldehyde. Gels were soaked in distilled water to remove the formaldehyde (three changes of 20 min each) and visualized by staining with ethidium bromide (1 μg/mL) prior to blotting onto Hybond N+ membranes (Amersham). Protocol no. 4 of the VacuGene XL blotting system (Pharmacia LKB Biotechnology, Bromma, Sweden) was used for vacuum transfer of RNA.
The blots were prehybridized overnight at 42°C in 50% (v/v) formamide, 5× Denhardt's reagent (1× Denhardt's reagent is 0.02% [w/v] Ficoll Type 400 Sigma, 0.02% [w/v] PVP, and 0.02% [w/v] bovine albumin Fraction V, Sigma), 0.1% (w/v) SDS, 5× SSC (1× SSC is 150 mm NaCl and 15 mm sodium citrate, pH 7.0), and 50 μg/mL salmon sperm DNA (0.5 mL per cm2 blot).
The coding region of the tomato CAB-1 (Pichersky et al., 1985) and RBCS-2 (Pichersky et al., 1986) and CHS1 (O'Neill et al., 1990) genes were used to synthesize DNA probes by random priming using the Rediprime DNA labeling system (Amersham). To remove unincorporated nucleotides 1 μL of 10% SDS and 2 μL of denatured salmon sperm (10 mg/mL) were added to the 50-μL reaction mix in an ultra-free microcentrifuge tube (Ultrafree-C3 TGC, Nihon Millipore Ltd., Tokyo, Japan) and spun at room temperature (5 min, 5000g). After washing the labeled DNA with 100 μL of sterile water, the probe was recovered from the upper part of the Millipore tube, denatured (by boiling for 5 min), and put on ice until use. For hybridization, the appropriate probe (specific activity 0.5 dpm/μg) was added to the hybridization buffer (0.1 mL of prehybridization buffer per cm2 blot). Hybridizations were carried out overnight at 42°C. As a loading control each blot was rehybridized with a 17-base oligonucleotide complementary to the 18S rRNA (Gallo-Meager et al., 1992). The oligonucleotide was labeled by phosphorylation with T4 polynucleotide kinase and the hybridization was carried out as described by Gallo-Meager et al. (1992).
Washes of nylon membranes (Hybond N+, Amersham) were performed in 2× SSC, 0.1% SDS (3 × 10 min at room temperature), and 0.1× SSC, 0.1% SDS (2 times for 30 min at 65°C). The signals were visualized and quantitated with a phosphor imager (Fujix BAS 2000, Fuji, Japan).
RESULTS
Effect of a Single R Pulse on CAB and RBCS Gene Expression
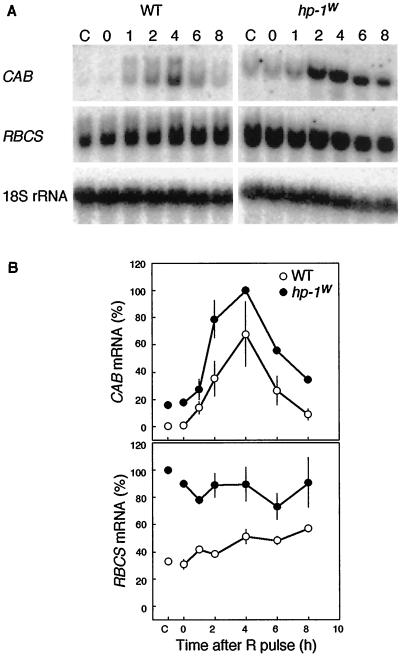
In tomato seedlings the expression of CAB genes is controlled by phytochrome (Sharrock et al., 1988; Wehmeyer et al., 1990). Wehmeyer et al. (1990) showed that CAB mRNA accumulation reached a maximum 4 h after a R pulse. In contrast to CAB mRNA, they could not easily demonstrate phytochrome regulation of RBCS mRNA. To determine whether the CAB and RBCS gene expression in the hp-1w-mutant seedlings is controlled by phytochrome, we first studied the kinetics of expression of both genes after a single R pulse. To this end, 4-d-old etiolated seedlings were irradiated with a 10-min saturating R pulse and returned to the dark. Samples were collected immediately (0 h) and at 1, 2, 4, 6, and 8 h after the R pulse. Control seedlings were maintained in continuous darkness and harvested simultaneously with the seedlings harvested immediately after the R pulse. Very low levels of CAB gene expression were detectable in dark-grown WT seedlings (Fig. 1). In contrast to WT, the hp-1w-mutant seedlings exhibited a substantial level of CAB gene expression in the dark. A R pulse induced CAB mRNA accumulation in both WT and hp-1w-mutant seedlings and maximum expression occurred 4 h after the light-pulse treatment. However, at this time point the hp-1w mutant accumulated more transcript than WT. The RBCS mRNA accumulation in dark-grown hp-1w-mutant seedlings was about 3-fold higher than the levels observed in WT. Although a R pulse slightly induced RBCS gene expression in WT seedlings, no induction was observed in hp-1w-mutant seedlings.
Figure 1.
Effect of a 10-min R pulse on the CAB and RBCS mRNA abundance in etiolated 4-d-old WT and hp-1w-mutant tomato seedlings. A, For the RNA blots shown, RNA was extracted directly (0 h), 1, 2, 4, 6, and 8 h after onset of the R pulse. The control (lanes C) represents the CAB mRNA amount in seedlings that did not receive a R pulse but were kept in continuous darkness. As a loading control the blots were probed with an 18S rRNA probe. B, The CAB and RBCS mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown for CAB and RBCS, respectively. A value of 100% on the ordinate represents the maximum steady-state mRNA detected within the experiment.
Phytochrome Regulation of CAB Gene Expression in Photomorphogenic Mutants
Figure 1 showed that in hp-1w-mutant seedlings CAB but not RBCS gene expression could be induced by a single R pulse. Therefore, we limited our experiment to study the involvement of phytochrome in light-regulated gene expression to CAB gene expression. Since various photomorphogenic mutants of tomato are available, we used phyA-deficient, hp-1w,fri1, and phyB1-deficient, hp-1w,tri1, double mutants in addition to the hp-1w mutant and WT. All genotypes were grown in continuous darkness for 4 d and exposed to a 10-min R pulse, 15-min FR pulse, or 10-min R pulse followed by a 15-min FR pulse. Control seedlings were kept in continuous darkness during the experiment. Whole seedlings were harvested 4 h after the light pulse(s) when maximum response occurs (Fig. 1).
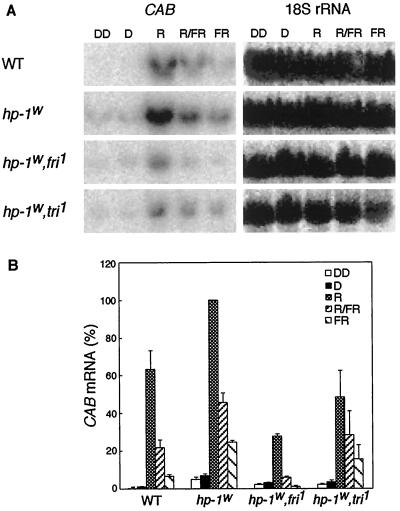
In WT, CAB mRNA accumulation was induced by a 10-min R pulse and could be partially reversed by a FR pulse (Fig. 2). This incomplete reversal can be explained by a partial escape from FR reversibility during the R pretreatment. The response after FR alone probably reflects the VLFR component of CAB mRNA accumulation (Sharrock et al., 1988). Recently, a similar VLFR component of CAB gene expression was shown for Arabidopsis (Hamazato et al., 1997). In agreement with the data in Figure 1, a substantial level of CAB gene expression was detected in dark-grown hp-1w-mutant seedlings (Fig. 2). Due to the hp-1w mutation, the hp-1w,fri1 and hp-1w,tri1 double mutants also showed CAB gene expression in the dark, although at a reduced level compared with the hp-1w monogenic mutant. To account for the possible effect of harvest under green safe light on CAB gene expression, the seedlings were also harvested in total darkness (in Fig. 2, DD) and compared with samples harvested under green safe light (Fig. 2, D). Figure 2 shows that when seedlings were harvested in complete darkness, the hp-1w mutation resulted in significant CAB mRNA accumulation. Moreover, the hp-1w mutant exhibited a higher response to all light-pulse treatments. The phyA-deficient hp-1w,fri1 double mutant exhibited approximately 30% of the CAB gene expression induced by a single R pulse in the hp-1w mutant. A similar reduction in CAB mRNA accumulation could be seen when the fri1 mutant was compared with WT (data not shown). This implies a role for phyA in the low fluence response, either directly or indirectly. The effect of FR and R/FR treatments are markedly reduced in the hp-1w,fri1 mutant compared with the hp-1w mutant, which verifies the role of phyA in the VLFR proposed previously (Casal et al., 1994; Botto et al., 1996; Shinomura et al., 1996). In the phyB1-deficient hp-1w,tri1 double mutant, the CAB gene expression induced by a R pulse was also reduced (about 50%) compared with the hp-1w mutant. This clearly indicates that phyB1 also plays a role in the regulation of CAB gene expression.
Figure 2.
Effect of no light pulse (lanes D), a 10-min R, 15-min FR, and 10-min R followed by 15-min FR pulse (R/FR) on the CAB mRNA abundance in etiolated 4-d-old tomato seedlings. The genotypes used were WT, hp-1w mutant, hp-1w,fri1, and hp-1w,tri1 double mutants. A, For the RNA gel blots shown, RNA was isolated 4 h after the light pulse(s) and probed with a CAB cDNA probe. As a loading control the blots were probed with an 18S rRNA probe. B, The CAB mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown. A value of 100% on the ordinate represents the maximum steady-state mRNA detected within the experiment.
Phytochrome Regulation of CHS Gene Expression
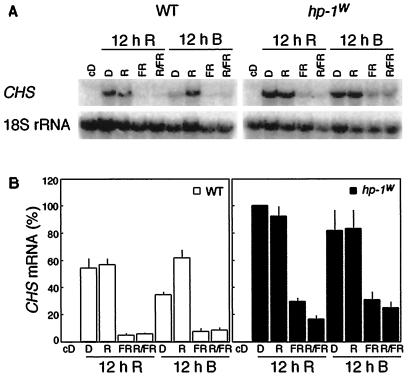
Although a single R pulse could induce CAB gene expression (Figs. 1 and 2), the same light treatment was ineffective for the induction of CHS gene expression (data not shown). Therefore, an irradiation schedule used by Peters et al. (1992b), which is efficient in inducing phytochrome regulation of anthocyanin biosynthesis, was applied to determine if phytochrome regulates CHS gene expression. The WT and hp-1w-mutant seedlings were grown in darkness for 4 d and exposed to a 12-h R or B pretreatment followed by no pulse, a 10-min R pulse, 15-min FR pulse, or a 10-min R pulse followed by a 15-min FR pulse. Control seedlings were kept in darkness during the experiment. Figure 3 shows that neither WT nor hp-1w-mutant seedlings accumulated detectable levels of CHS mRNA when grown in complete darkness. In seedlings that were given R and B pretreatments, a R pulse induced high levels of CHS expression in the WT. This effect could be reversed by FR. In the hp-1w mutant R and B pretreatments followed by a R pulse resulted in a significant amplification of CHS gene expression compared with WT, and a FR pulse could not reverse the level of expression to the same extent as in the WT. In summary, the total level of response is enhanced in the hp-1w mutant compared with WT for all treatments. Thus, by using light pretreatments, CHS gene expression was shown to be regulated by phytochrome in tomato. Moreover, the phytochrome-induced, R/FR reversible, CHS mRNA accumulation response is significantly enhanced in the hp-1w mutant compared with WT after R pretreatment, but not significantly affected after B pretreatment. This indicates that B alone can enhance the phytochrome response in WT to an amount similar to that in the hp-1w mutant.
Figure 3.
Effect of a 12-h R or B pretreatment terminated with no light pulse (lanes D), a 10-min R, 15-min FR, and 10-min R followed by 15-min FR pulse (R/FR) on the CHS mRNA abundance in etiolated 4-d-old WT and hp-1w-mutant seedlings of tomato. A, For the RNA gel blots shown, RNA was isolated 4 h after the light pulse(s) and probed with a CHS1 cDNA probe. As a loading control the blots were probed with an 18S rRNA probe. B, The CHS mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown. A value of 100% on the ordinate represents the maximum steady-state mRNA detected within the experiment.
Circadian Rhythm of CAB Gene Expression
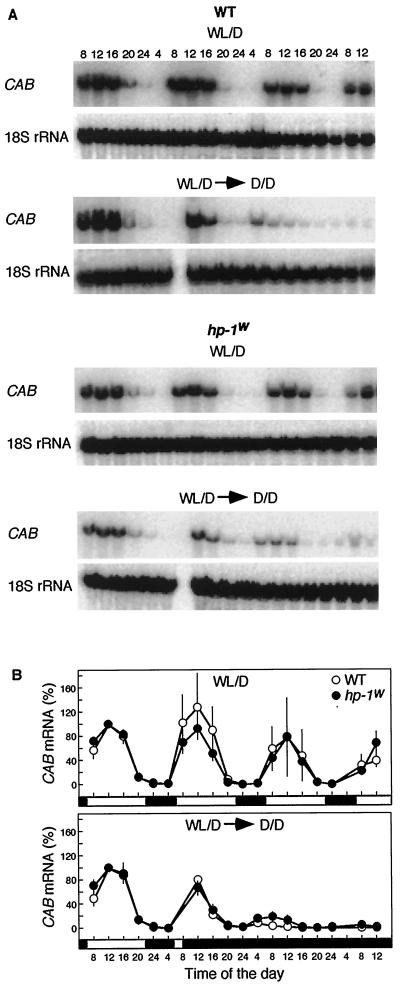
Accumulation of CAB mRNA shows a circadian rhythm in tomato with a maximum level of expression at noon (Kellman et al., 1993). A difference in the circadian rhythm of CAB mRNA accumulation of WT and hp-1w-mutant plants could result in misleading conclusions when studying tomato plants grown in light/dark cycles. Therefore, we studied the effect of the hp-1w mutation on the circadian rhythm. Seedlings were grown in 16-h WL, 8-h dark cycles (lights on at 6 am; light off at 10 pm) and transferred to continuous darkness at 8 am of d 5 (Fig. 4, WL/D → D/D). Control seedlings were kept in WL/D cycles and showed diurnal oscillations (Fig. 4, WL/D). Whole seedlings were harvested every 4 h for 3 d, starting at 8 am on the 4th d after sowing. No differences in the diurnal oscillations of CAB gene expression of WT and hp-1w-mutant seedlings were observed (Fig. 4, WL/D). The brief, 2-h exposure to WL before the transfer to continuous darkness resulted in a phase shift, and the subsequent peak of CAB gene expression occurred at 8 am instead of at noon in both WT and hp-1w mutant (Fig. 4, WL/D → D/D). Although a slight difference in dampening of the CAB gene expression between WT and hp-1w-mutant seedlings may exist, the pattern and quantitative level of CAB mRNA accumulation in the two genotypes is very similar.
Figure 4.
Circadian rhythmic CAB mRNA accumulation in WT and hp-1w-mutant seedlings of tomato. A, For the RNA gel blots shown, seedlings were grown in 16-h WL (WL, 6 am–10 pm), 8-h dark (lanes D, 10 pm–6 am) cycles. To describe the diurnal CAB transcript oscillations (WL/D), samples were collected every 4 h starting with 4-d-old seedlings at 8 am. To study the circadian rhythm of CAB genes (WL/D → D/D), seedlings were transferred to darkness on d 5 at 8 am. As a loading control the blots were probed with an 18S rRNA probe. B, The CAB mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown. A value of 100% on the ordinate represents the mRNA abundance detected on d 4 at noon. {/ANNT;;;left;top}
Organ-Specific Expression of CAB, RBCS, and CHS Genes
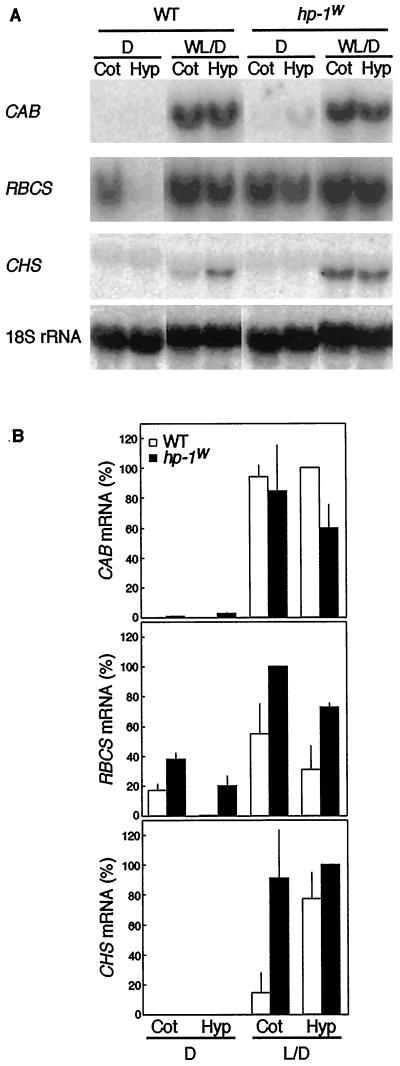
At least one of the light-regulated genes studied, the RBCS gene family, exhibits organ-specific expression in tomato (Sugita and Gruissem, 1987; Wanner and Gruissem, 1991). Since the hp-1w-mutant seedlings showed elevated responses for the genes studied, it is important to compare the organ-specific expression patterns of these genes in seedlings and adult plants of the WT and hp-1w mutant. Such a comparison can answer the question of whether more mRNA accumulates in the same organs or whether the distribution patterns also change due to the hp-1w mutation. Seedlings were grown in either continuous darkness or in 16-h WL, 8-h dark cycles for 4 d. The CAB, RBCS, and CHS mRNA accumulation in cotyledons and hypocotyls is shown in Figure 5. None of the genes were expressed in the roots (data not shown). The patterns of mRNA accumulation in hp-1w-mutant seedlings differ from that of WT in several aspects. CAB mRNA in dark-grown hypocotyls and cotyledons and RBCS mRNA in dark-grown hypocotyls accumulated to a much higher level in the hp-1w mutant than WT (Fig. 5). Moreover, a higher CHS mRNA accumulation was observed in WL/D-grown hp-1w-mutant seedlings compared with WT. The CHS mRNA accumulation was significantly increased in cotyledons of the hp-1w mutant compared with WT (Fig. 5), which correlates well with the anthocyanin accumulation data for the WT and hp-1w mutant (A535 per three cotyledon pairs was 0.065 ± 0.004 and 0.449 ± 0.015 for WT and hp-1w mutant, respectively; A535 per three hypocotyls was 0.323 ± 0.022 and 0.600 ± 0.038 for WT and hp-1w mutant, respectively). The RBCS gene expression was always higher in the hp-1w mutant, irrespective of the light conditions under which the plants were grown (Fig. 5). As in the R and B pretreatment experiment (Fig. 3), no CHS mRNA accumulation was observed in the dark. Accumulation of CHS mRNA reached a higher level in WL/D-grown hp-1w-mutant seedlings than in WT. In contrast to RBCS and CHS, CAB gene expression was only higher in seedlings grown in darkness.
Figure 5.
CAB, RBCS, and CHS mRNA abundance in cotyledons (lanes Cot) and hypocotyls (lanes Hyp) of 4-d-old WT and hp-1w-mutant seedlings of tomato. A, For the RNA gel blots shown, RNA was isolated from seedlings grown in dark (D) or 16-h WL, 8-h dark cycles (WL/D). As a loading control the blots were probed with a 18S rRNA probe. B, The CAB, RBCS, and CHS mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown. A value of 100% on the ordinate represents the maximum steady-state mRNA detected within the experiment.
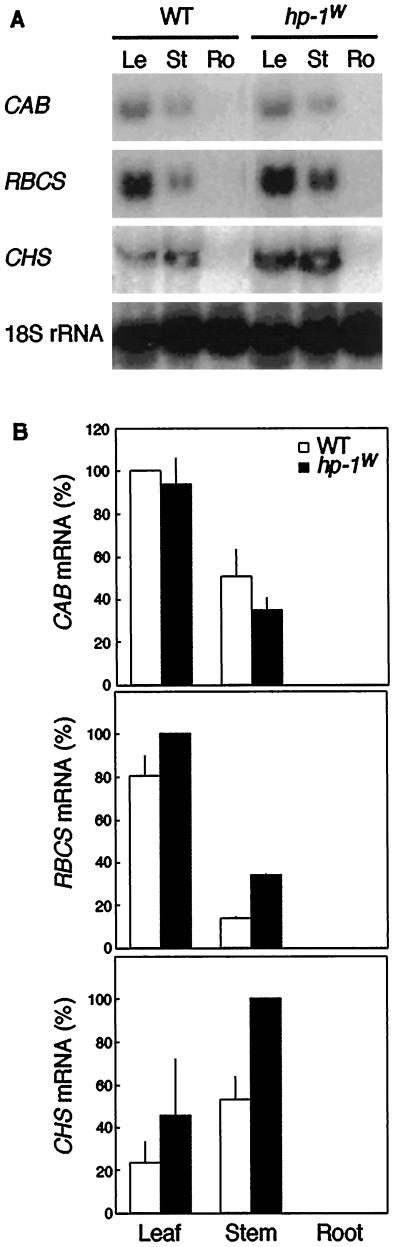
To investigate the CAB, RBCS, and CHS mRNA levels in the leaves, stems, and roots of adult plants, plants were grown under 16-h WL, 8-h dark cycles for 8 weeks. None of the genes were expressed in the roots of adult WT and hp-1w-mutant plants (Fig. 6). As in 4-d-old, WL/D-grown seedlings (Fig. 5), the CAB mRNA accumulation in adult plant parts was not higher in the hp-1w mutant than WT. However, both RBCS and CHS gene expression were significantly higher in stems of the hp-1w mutant compared with WT. These data on the organ-specific expression of light-regulated genes in seedling and adult plants show that the three genes studied are differentially affected by the hp-1w mutation.
Figure 6.
Abundance of CAB, RBCS, and CHS mRNA in young leaves (lanes Le), stems (lanes St), and roots (lanes Ro) of 8-month-old WT and hp-1w-mutant tomato plants. A, For the RNA gel blots shown, RNA was isolated from adult plants grown in 16-h WL, 8-h dark cycles. As a loading control the blots were probed with a 18S rRNA probe. B, The CAB, RBCS, and CHS mRNA abundance was quantified using a phosphor imager and the mean values (±se) are shown. A value of 100% on the ordinate represents the maximum steady-state mRNA detected within the experiment.
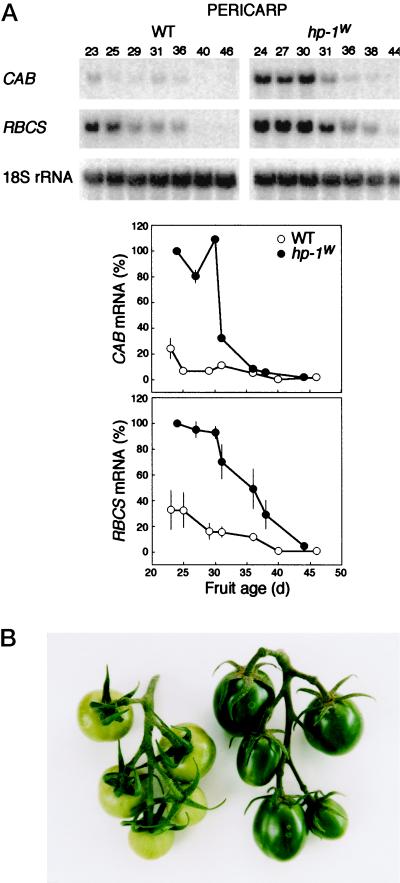
To investigate CAB and RBCS gene expression and chlorophyll content during fruit development, we analyzed fruits at seven different physiological ages. The levels of CAB and RBCS expression in the pericarp were found to be highest in young fruits and they both gradually declined during fruit development (Fig. 7A). The expression level of both CAB and RBCS was amplified in the hp-1w-mutant fruits compared with those of WT. The increased level of CAB and RBCS expression in the pericarp correlates well with its approximate 5-fold higher chlorophyll content (25-d-old fruits contain 100 and 25 μg chlorophyll g−1 fresh weight in the hp-1w mutant and WT, respectively; Fig. 7B).
Figure 7.
A, The CAB and RBCS mRNA accumulation in the pericarp of the hp-1w-mutant and WT tomato fruit during fruit ripening. For both CAB and RBCS mRNA accumulation, the amount of mRNA in the pericarp of the youngest hp-1w-mutant fruit was set at 100%. As a loading control the blots were probed with a 18S rRNA probe. B, A truss of WT (left) and hp-1w-mutant (right) immature tomato fruits. The fruits of the hp-1w mutant are darker green and have an elongated shape when compared with WT.
DISCUSSION
The results presented indicate that CAB, RBCS, and CHS gene expression is up-regulated in the hp-1w mutant compared with WT. Two of these genes, CAB and CHS, were shown to be phytochrome regulated in the hp-1w mutant (Figs. 2 and 3). However, their pattern of up-regulation is different and dependent on the stage of development and tissue studied. For instance, if we had only investigated gene expression in WL-grown seedlings, we would have only seen up-regulation of RBCS and CHS, but not CAB, compared with WT in the hp-1w mutant (Figs. 4–6). In other words, in WL-grown seedlings, CAB gene expression appears to be saturated and is comparable in WL-grown WT and hp-1w-mutant seedlings (Figs. 4–6). In contrast, dark-grown hp-1w-mutant seedlings accumulate higher levels of CAB (and RBCS) transcripts than WT. In dark-grown seedlings of hp-1, up-regulation of enzyme activity for Phe ammonia lyase (Goud et al., 1991), nitrate reductase, nitrite reductase, and amylase (Goud and Sharma, 1994) have been previously reported. All of these enzymes have been shown to be phytochrome regulated (Goud et al., 1991; Goud and Sharma, 1994). Taken together, these findings indicate that the hp-1 mutation causes changes in the dark at the molecular level. Unlike the dark “de-etiolated” Arabidopsis mutants such as cop, det, and fus, there are no visible differences between dark-grown WT and hp-1-mutant seedlings. Therefore, the hp-1 mutation appears to affect only a subset of responses regulated by cop, det, and fus genes. The CAB mRNA measured in dark-grown hp-1w-mutant seedlings could be due to an amplification of the response to the residual Pfr available in the seeds, and probably reflects a phyA-mediated VLFR, which is very difficult to test experimentally. The results with the phyA-deficient hp-1w,fri1 double mutant suggest that this is the case.
R induction of CAB was similar in the hp-1w-mutant and WT seedlings (Fig. 1) and confirms the observation of Wehmeyer et al. (1990). However, the difference between hp-1w and WT observed in darkness was retained. The RBCS gene expression level was always higher in hp-1w seedlings than those of WT, regardless of the conditions under which they were grown. In fact, the high RBCS gene expression in the dark could not be further enhanced by a single pulse of R, whereas in the WT a gradual elevation of expression was observed (Fig. 1), which never attained the level in hp-1w. We can therefore conclude that the hp-1w mutation affects both CAB and RBCS expression (Figs. 1, 2, 5, and 6). Studies involving doc and cue1 mutants suggested that different biochemical pathways downstream of phytochrome regulate CAB and RBCS gene expression (Li et al., 1994, 1995). Therefore, the differential effect of the hp-1 mutation on the expression of CAB and RBCS genes may be related to the differences in the role of HP-1 in these pathways.
The hp-1-mutant has high anthocyanin levels in both seedlings and adult plants (Kerckhoffs et al., 1997a) and increased flavonoid accumulation in ripe fruits (Yen et al., 1997). The CHS transcript accumulation of the enzyme that is the first committed step of flavonoid biosynthesis also shows a higher level in the hp-1 mutant than WT (Figs. 3, 5, and 6). Since no CHS gene expression could be observed in dark-grown or single pulse-treated seedlings, a 12-h R or B pretreatment irradiation schedule known to be effective in anthocyanin production (Peters et al., 1989) was used to study CHS gene expression (Fig. 3). Irrespective of whether a R or B pretreatment was given, significantly higher levels of CHS transcripts accumulated in the hp-1w-mutant seedlings compared with WT (Fig. 3). These CHS mRNA accumulation data correlate reasonably well with the anthocyanin accumulation data under the same irradiation conditions (Peters et al., 1989). When WL-grown seedlings were examined, we also found a strong correlation between CHS abundance and anthocyanin content (results given in text and Fig. 5).
The expression of CAB and RBCS genes decreased with increasing fruit age in both WT and hp-1w-mutant fruits (Fig. 7). In the pericarp these data show a positive correlation with the chlorophyll data (data not shown). The pericarp of green fruits is known to be photosynthetically active, and this activity decreases during chloroplast/chromoplast differentiation (Piechulla and Gruissem, 1987). Earlier work of Piechulla et al. (1985) showed that mRNA for photosynthetic polypeptides disappear during fruit ripening. These changes of mRNA levels correlated with alterations that occur at the photosynthesis and polypeptide level (Piechulla and Gruissem, 1987). Work of Meehan et al. (1996) using Arabidopsis transgenics that expressed a CAB promotor fused to a GUS reporter gene shows that GUS activities were positively correlated with chlorophyll content and cell size. Therefore, the transcription of nuclear genes for chloroplast components could be modulated by chloroplast numbers, which increase with cell size.
Our results support the proposal that the hp-1 mutation amplifies phytochrome responses (for review, see Kerckhoffs and Kendrick, 1997): the R induction of both CAB and CHS gene expression was higher in the hp-1 mutant compared with WT and was shown to be mediated by phytochrome (Figs. 2 and 3). Since the R induction of CAB gene expression is considerably lower in the phyA-deficient, hp-1w,fri1 mutant than the hp-1w mutant (Fig. 2), phyA appears to modulate the magnitude of the low fluence response. Phytochrome-mediated CAB gene expression in tomato has a VLFR component (Sharrock et al., 1988) and this response, as indicated by the level induced by FR alone, is much reduced in the hp-1w,fri1 mutant compared with the hp-1w mutant (Fig. 2). These data support the conclusion that phyA mediates the VLFR (Casal et al., 1994; Botto et al., 1996; Shinomura et al., 1996). In addition, phyB1 plays a role in CAB gene expression. The induction of CAB transcript accumulation is reduced in the phyB1-deficient, hp-1w,tri1 mutant compared with the hp-1w mutant (Fig. 2). Thus, both phyA and phyB1 play a role in CAB gene expression in tomato. Reed et al. (1994) and Hamazato et al. (1997) came to a similar conclusion when they studied phyA and phyB mutants in Arabidopsis.
The data presented support the hypothesis that the HP-1 protein has a repressive role in phytochrome-signal transduction. The pattern of up-regulation observed for CAB, RBCS, and CHS gene expression depends on the light conditions, stage of development, and tissue studied. To date, hp-like mutations have not been described in other plant species. The dark phenotype of the hp-1 mutant is more subtle compared with de-etiolated mutants such as cop, det, and fus. Considering the higher levels of anthocyanin responses and CHS mRNA accumulation, the tomato hp-1 mutant is somewhat similar to the icx1 mutant of Arabidopsis (Jackson et al., 1995). The major difference between the two mutations is that, whereas the icx1 mutation affects only the signal transduction processes leading to the regulation of CHS expression, the tomato hp-1 mutation also affects the expression of genes (CAB and RBCS) encoding proteins for the photosynthetic apparatus. This suggests that the hp-1 mutation acts on an upstream signal transduction event(s) that leads to the altered pattern of gene expression. Therefore, HP-1 is proposed to be a fundamental phytochrome signal transduction regulator, and the cloning of its gene and molecular characterization is eagerly awaited.
ACKNOWLEDGMENTS
We thank Ferenc Nagy for the CAB-1 cDNA clone, William Gruissem for the RBCS-2 cDNA clone, John Yoder for the CHS1 clone, and Shannon Frances for reading the manuscript and making suggestions. The WT and hp-1w-mutant seeds were supplied by Maarten Koornneef and colleagues of the Department of Genetics, Wageningen Agricultural University, The Netherlands.
Abbreviations:
- B
blue light
- CAB
chlorophyll a/b-binding protein
- CHS
chalcone synthase
- FR
far-red light
- R
red light
- RBCS
ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit
- VLFR
very low fluence response
- WL
white light
- WT
wild type
Footnotes
This work was supported by a Science and Technology Agency fellowship of the Japan International Science and Technology Exchange Center to J.L.P.
LITERATURE CITED
- Ahmad M, Cashmore AR. The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 1996;10:1103–1110. doi: 10.1046/j.1365-313x.1996.10061103.x. [DOI] [PubMed] [Google Scholar]
- Botto JF, Sánchez RA, Whitelam GC, Casal JJ. Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 1996;110:439–444. doi: 10.1104/pp.110.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994a;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Bowler C, Yamagata H, Neuhaus G, Chua N-H. Phytochrome signal transduction pathways are regulated by reciprocal control mechanisms. Genes Dev. 1994b;8:2188–2202. doi: 10.1101/gad.8.18.2188. [DOI] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. Oat phytochrome is biologically active in transgenic tomatoes. Plant Cell. 1989;1:765–773. doi: 10.1105/tpc.1.8.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Sánchez RA, Vierstra RD. Avena phytochrome A overexpressed in transgenic tobacco seedlings differentially affects red/far-red reversible and very-low-fluence responses (cotyledon unfolding) during de-etiolation. Planta. 1994;192:306–309. [Google Scholar]
- Frances S, White MJ, Edgerton MD, Jones AM, Elliot RC, Thompson WF. Initial characterization of a pea mutant with light-independent photomorphogenesis. Plant Cell. 1992;4:1519–1530. doi: 10.1105/tpc.4.12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Meager M, Sowinski DA, Elliot RC, Thompson WF. Both internal and external regulatory elements control expression of the pea Fed-1 gene in transgenic tobacco seedlings. Plant Cell. 1992;4:389–395. doi: 10.1105/tpc.4.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud KV, Sharma R. Retention of photoinduction of cytosolic enzymes in aurea mutants of tomato (Lycopersicon esculentum) Plant Physiol. 1994;105:643–650. doi: 10.1104/pp.105.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud KV, Sharma R, Kendrick RE, Furuya M. Photoregulation of phenylalanine ammonia lyase is not correlated with anthocyanin induction in photomorphogenetic mutants of tomato (Lycopersicon esculentum) Plant Cell Physiol. 1991;32:1251–1258. [Google Scholar]
- Hamazato F, Shinomura T, Hanzawa H, Chory J, Furuya M. Fluence and wavelength requirements for Arabidopsis CAB gene induction by different phytochromes. Plant Physiol. 1997;115:1533–1540. doi: 10.1104/pp.115.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JP, Israelstam GF. A method for extraction of chlorophyll from leaf tissue without maceration. Can J Bot. 1979;57:1332–1334. [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H, Tanaka A, Tano S, Nagatani A (1996) Isolation and characterization of photomorphogenic mutants of Arabidopsis which promote transduction of light-signal. In Programme and Abstract of Seventh International Conference on Arabidopsis Research, June 23–27, 1996. University of East Anglia, Norwich, UK, p 190
- Jackson JA, Fuglevand GF, Brown BA, Shaw MJ, Jenkins GI. Isolation of Arabidopsis mutants altered in light-regulation of chalcone synthase gene expression using a transgenic screening approach. Plant J. 1995;8:369–380. doi: 10.1046/j.1365-313x.1995.08030369.x. [DOI] [PubMed] [Google Scholar]
- Jarret RL, Sayama H, Tigchelaar EC. Pleiotropic effects associated with the chlorophyll intensifier mutations high pigment and dark green in tomato. J Am Soc Hortic Sci. 1984;109:873–878. [Google Scholar]
- Kellman J-W, Merforth N, Wiese M, Pichersky E, Piechulla B. Concerted circadian oscillations in transcript levels of nineteen Lha/b (cab) genes in Lycopersicon esculentum (tomato) Mol Gen Genet. 1993;237:439–448. doi: 10.1007/BF00279449. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs LHJ (1996) Physiological functions of phytochromes in tomato: a study using photomorphogenic mutants. PhD thesis. Wageningen Agricultural University, Wageningen, The Netherlands
- Kerckhoffs LHJ, de Groot NAMA, van Tuinen A, Schreuder MEL, Nagatani A, Koornneef M, Kendrick RE. Physiological characterization of exaggerated-photoresponse mutants of tomato. J Plant Physiol. 1997a;150:578–587. [Google Scholar]
- Kerckhoffs LHJ, Kendrick RE. Photocontrol of anthocyanin biosynthesis in tomato. J Plant Res. 1997;110:141–149. doi: 10.1007/BF02506853. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs LHJ, Schreuder MEL, van Tuinen A, Koornneef M, Kendrick RE. Phytochrome control of anthocyanin biosynthesis in tomato seedlings: analysis using photomorphogenic mutants. Photochem Photobiol. 1997b;65:374–381. [Google Scholar]
- Kerr EA. Identification of high-pigment, hp, tomatoes in the seedling stage. Can J Plant Sci. 1965;45:104–105. [Google Scholar]
- Li H-M, Altschmied L, Chory J. Arabidopsis mutants define downstream branches in the phototransduction pathway. Genes Dev. 1994;8:339–349. doi: 10.1101/gad.8.3.339. [DOI] [PubMed] [Google Scholar]
- Li H-M, Culligan K, Dixon RA, Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans. 1983;11:591–592. [Google Scholar]
- Loening UE. The determination of the molecular weight of ribonucleic acid by polyacrylamide gel electrphoresis. Biochem J. 1969;113:131–138. doi: 10.1042/bj1130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan L, Harkins K, Chory J, Rodermel S. Lchb transcription is coordinated with cell size and chlorophyll accumulation. Plant Physiol. 1996;112:953–963. doi: 10.1104/pp.112.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Kamimura S. Photoselective method for selection of hp at the cotyledon stage. Tomato Genet Coop Rpt. 1985;35:12–13. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua N-H. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- O'Neil SD, Tong Y, Spörlein B, Forkmann G, Yoder JI. Molecular genetic analysis of chalcone synthase in Lycopersicon esculemtum and an anthocyanin-deficient mutant. Mol Gen Genet. 1990;224:279–288. doi: 10.1007/BF00271562. [DOI] [PubMed] [Google Scholar]
- Peters JL, Schreuder MEL, Heeringa G, Wesselius JC, Kendrick RE, Koornneef M. Analysis of the response of photomorphogenetic tomato mutants to end-of-day far-red light. Acta Hortic. 1992a;305:67–77. [Google Scholar]
- Peters JL, Schreuder MEL, Verduin SJW, Kendrick RE. Physiological characterization of a high-pigment of tomato. Photochem Photobiol. 1992b;56:75–82. [Google Scholar]
- Peters JL, Silverthorne J. Organ-specific stability of two Lemna rbcS mRNAs is determined primarily in the nuclear compartment. Plant Cell. 1995;7:131–140. doi: 10.1105/tpc.7.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Van Tuinen A, Adamse P, Kendrick RE, Koornneef M. High pigment mutants of tomato exhibit high sensitivity of phytochrome action. J Plant Physiol. 1989;134:661–666. [Google Scholar]
- Pichersky E, Bernatzky R, Tanksley SD, Breidenbach RB, Kausch AP, Cashmore AR. Molecular characterization and genetic mapping of two clusters of genes encoding chlorophyll a/b-binding proteins in Lycopersicon esculentum (tomato) Gene. 1985;40:247–258. doi: 10.1016/0378-1119(85)90047-2. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Bernatzky R, Tanksley SD, Cashmore AR. Evidence for selection as a mechanism in the concerted evolution of Lycopersicon esculentum (tomato) genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1986;83:3880–3884. doi: 10.1073/pnas.83.11.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B, Chonoles-Imlay KR, Gruissem W. Plastid gene expression during fruit ripening in tomato. Plant Mol Biol. 1985;5:373–385. doi: 10.1007/BF00037558. [DOI] [PubMed] [Google Scholar]
- Piechulla B, Gruissem W. Diurnal mRNA fluctuations of nuclear and plastid genes in developing tomato fruits. EMBO J. 1987;6:3593–3599. doi: 10.1002/j.1460-2075.1987.tb02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard GB. Origin of the Webb Special (Black Queen) tomato. Tomato Genet Coop Rpt. 1956;6:22. [Google Scholar]
- Sanders DC, Pharr DM, Konsler TR. Chlorophyll content of a dark green mutant of ‘Manapal’ tomato. HortScience. 1975;10:262–264. [Google Scholar]
- Sharrock RA, Parks BA, Koornneef M, Quail PH. Molecular analysis of the phytochrome deficiency in an aurea mutant of tomato. Mol Gen Genet. 1988;213:9–14. [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furaya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:289–315. [Google Scholar]
- Soressi GP. New spontaneous or chemically-induced fruit-ripening tomato mutants. Tomato Genet Coop Rpt. 1975;25:21–22. [Google Scholar]
- Sugita M, Gruissem W. Developmental, organ-specific, and light-dependent expression of the tomato ribulose-1,5-bisphosphate carboxylase small subunit gene family. Proc Natl Acad Sci USA. 1987;84:7104–7108. doi: 10.1073/pnas.84.20.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AE. A comparision of fruit quality constituents of normal and high pigment tomatoes. Proc Am Soc Hortic Sci. 1962;78:464–473. [Google Scholar]
- Traas J, Laufs P, Jullien M, Caboche M. A mutation affecting etiolation and cell elongation in Nicotiana plumbaginifolia causes abnormal division plane alignement and pattern formation in root meristem. Plant J. 1995;7:785–796. [Google Scholar]
- van Tuinen A, Cordonnier-Pratt M-M, Pratt LH, Verkerk R, Zabel P, Koornneef M. The mapping of phytochrome genes and photomorphogenic mutants of tomato. Theor Appl Genet. 1997;94:115–122. doi: 10.1007/s001220050389. [DOI] [PubMed] [Google Scholar]
- van Tuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M. Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol Gen Genet. 1995a;246:133–141. doi: 10.1007/BF00294675. [DOI] [PubMed] [Google Scholar]
- van Tuinen A, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M. A temporarily red light-insensitive mutant of tomato lacks a light-stable, B-like phytochrome. Plant Physiol. 1995b;108:939–947. doi: 10.1104/pp.108.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Hoecker U, Quail PH. Red1 is necessary for phytochrome B-mediated red light-specific signal transduction in Arabidopsis. Plant Cell. 1997;9:731–743. doi: 10.1105/tpc.9.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Gruissem W. Expression dynamics of the tomato rbcS gene family during development. Plant Cell. 1991;3:1289–1303. doi: 10.1105/tpc.3.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer B, Cashmore AR, Schäfer E. Photocontrol of the expression of genes encoding chlorophyll a/b binding proteins and small sununit of ribulose-1,5-bisphosphate carboxylase in etiolated seedlings of Lycopersicon esculentum (L.) and Nicotiana tabacum (L.) Plant Physiol. 1990;93:990–997. doi: 10.1104/pp.93.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng X-W. The role of COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Nagatani A, Kendrick RE, Murfet IC, Reid JB. New lv mutants of pea are deficient in phytochrome B. Plant Physiol. 1995;108:525–532. doi: 10.1104/pp.108.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Shelton BA, Howard LR, Lee S, Vrebalov J, Giovannoni JJ. The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor Appl Genet. 1997;95:1069–1079. [Google Scholar]