Abstract
In the March 2012 issue of The Plant Cell we describe extensive alternative splicing (AS) of Arabidopsis circadian clock genes. Notably these distinct post-transcriptional events associate with different steady-state temperatures and also with plants undergoing temperature transitions leading us to propose that temperature-associated AS is an additional mechanism involved in the operation and control of the plant circadian clock. Here we show that temperature associated AS also extends to REVEILLE 8 (RVE8), demonstrating a hitherto unrecognized link between the expression of this clock associated gene and temperature. Finally we discuss our observations of the plastic nature of clock gene expression at the post-transcriptional level in the context of the ongoing fascination of how plants respond to temperature.
Keywords: circadian, clock, temperature, alternative splicing, temperature compensation, temperature perception
Alternative splicing (AS) – the regulated use of alternative splice sites within precursor mRNAs (pre-mRNAs) during gene transcription – is generally regarded as a major mechanism generating structural and functional diversity of gene products. Approximately 95% of human genes undergo AS,1 while a recent genome-wide study of the extent of AS in Arabidopsis estimated that 61% of intron-containing genes underwent AS,2 an increase on a previous estimate of 42%.3 Although this implies that AS is less extensive in plants compared with human genes, these estimates may overlook the contribution of various developmental, growth and stress conditions to plant AS. One commonality in AS between animals and plants seems to be the prevalence of AS in genes encoding receptor, signal transduction and transcription factor proteins, and increasingly alternatively spliced genes in plants seem to be associated with abiotic stress responses.4-6
Although there are reports of AS providing functional diversity of the encoded protein (for some recent examples see refs.7-9) it is not yet clear to what extent AS feeds through to proteome expansion in plants.10-12 Another role of regulated AS appears to be the precise modulation of levels of functional – or translatable – mRNA. This is achieved by differential selection of splices sites (ss) during transcription resulting in proportions of total mRNA gene message containing premature termination codons (PTCs) that, rather than being translated, are the subject of RNA quality control mechanisms. One such aberrant mRNA surveillance mechanism is the nonsense mediated decay (NMD) pathway which selectively degrades PTC-containing transcripts.12-14
We performed a system-wide analysis of AS in the Arabidopsis circadian clock15- analysis of 10 clock genes showed the presence of 73 splice variants of which 55 had not been described previously. Fifteen events were influenced by temperature since they either increased to between 10–50% of total transcripts in at least one phase of transition or acclimation to low temperature or were absent at ambient temperature and only visible at low temperature. We found that the splice variants encompassed the well established AS ‘types’ – intron retention (IR), exon skipping (ES) and the use of alternative 3′ and 5′ ss. The majority disrupted the open reading frame and introduced PTCs leading to non-productive mRNAs, some of which triggered NMD.
Importantly, our data provides strong evidence for the functional importance of AS in the clock, because in most cases the effect of temperature on AS complemented its effect on total transcript abundance. For example, for the clock genes LATE ELONGATED HYPOCOTYL (LHY), PSEUDO RESPONSE REGULATOR 7 (PRR7), TIMING OF CAB EXPRESSION 1 TOC1 (also known as PRR1) and PRR3, a reduction in temperature resulted in decreased expression of fully functional – or translatable – transcripts that was complemented with a concomitant increase of non-functional transcripts. Indeed for LHY the effect of cooling on transcript abundance was mediated at least partly by AS/NMD but not by promoter strength. We also showed that there were differences in the responses of two clock output genes, CATALASE 3 (CAT3) and CHLOROPHYLL A/B-BINDING PROTEIN 2 (CAB2), to temperature changes, resonating with previous work showing that CAT3 and CAB2 are controlled by distinct clocks that differ in their temperature sensitivities.16
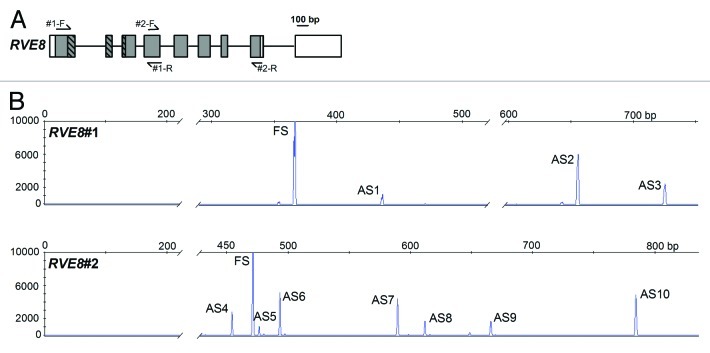
We have also recently looked for evidence of temperature associated AS for other genes that are thought to have roles in the circadian clock. One further example is RVE8 that, like CCA1 and LHY, is a member of the REVEILLE gene family encoding single Myb domain transcription factors. We used the strategy deployed in15 to examine RVE8 AS. First we used total RNA from a mixture of plant material grown at different temperatures and light regimes at various time points in order to cast a wide net on potential RVE8 AS events. Second, AS events were amplified using primer pairs (one of which was 6-FAM fluorescently labeled) that were designed to capture different splicing events across the whole RVE8 sequence (Fig. 1A). Products were subsequently resolved on a capillary ABI3730 sequencer and on the basis of the resultant electropherograms (Fig. 1B), ten RVE8 AS events were characterized (Table 1) (only one AS transcript is annotated in TAIR10). Two events, AS4 and AS6, show alternative 3′ splice site selection generating premature termination codons (PTCs) that are likely to trigger NMD; the other events were intron retentions or combinations of these events (Table 1).
Figure 1. Alternative Splicing of RVE8. (A) RVE8 (At3g09600) gene organization with exons depicted as rectangles, and introns as intervening horizontal lines. Protein coding exons/regions are shaded gray, UTR regions are depicted as clear rectangles. The region of RVE8 encoding the predicted single Myb domain is represented as hatched rectangles. Primer locations are denoted by arrows. Primer sequences are: RVE8#1-F GCTGGACAGAGGAAGAGCAC; RVE8#1-R TGCTCCACGAAGAGTAGCAA; RVE8#2-F GGGATATGCTTCATGGGATG; RVE8#2-R GCTGATTTGTCGCTTGTTGA. (B) Representative HR RT-PCR electropherograms for RVE8. AS isoforms were characterized from pooled RNAs representing plants harvested under both diurnal and constant light conditions at either normal ambient temperature or for plants undergoing cooling. Electropherograms show the size of detected peaks corresponding to RT-PCR products from alternatively spliced transcript variants. The x-axis is the size in bp and the y-axis represents arbitrary scales (relative fluorescent units) to reflect relative abundance of peaks. Individual products are indicated and AS variants are described in more detail in Table 1. FS – fully spliced product; ASn – alternative splicing variant where n is the number to identify the product. AS transcripts were detected on an ABI3730 automatic DNA sequencer along with GeneScan™ 1200 LIZ size standard. Amplicons were accurately sized using GeneMapper software. For more details of Methods see15
Table 1. Alternative splicing events in RVE8.
| Primer pair | AS event# | RT-PCR size (bp) |
Description | Outcome |
|---|---|---|---|---|
| |
|
|
|
|
|
RVE8#1 |
FS |
371 |
Fully spliced |
|
| |
AS1 |
440 |
I3R |
PTC |
| |
AS2 |
658 |
I1R |
PTC |
| |
AS3 |
727 |
I1R and I3R |
PTC |
| |
|
|
|
|
| |
|
|
|
|
|
RVE8#2 |
FS |
474 |
Fully spliced |
|
| |
AS4 |
457 |
Alternative 3′ss in exon 5 (removes 17 nt) |
PTC; downstream splice junctions; likely NMD |
| |
AS5 |
479 |
Alternative 3′ss in exon 5 (removes 17 nt) plus alternative 3′ss in intron 7 (adds 22 nt) |
PTC; downstream splice junctions; likely NMD |
| |
AS6 |
496 |
Alternative 3′ss in intron 7 (adds 22 nt) (TAIR.2) |
PTC; downstream splice junctions; likely NMD |
| |
AS7 |
592 |
I4R |
PTC |
| |
AS8 |
614 |
I4R plus alt 3′ss in intron 7 adds 22nt |
PTC; downstream splice junctions; likely NMD |
| |
AS9 |
668 |
I7R |
PTC |
| |
AS10 |
786 |
I4R+I7R |
PTC |
FS, fully spliced; AS, alternatively spliced; ss, splice site; INR, intron retention of intron “N”; PTC, premature termination codon; NMD, nonsense-mediated decay.
Previously we established that splice site choices in the region immediately downstream and flanking the exons encoding the predicted Myb domain of LHY appeared to be particularly influenced by low temperature cooling.15 We therefore next focused on AS in the equivalent region of RVE8. Here we used exon-intron boundary spanning primers targeting the region flanking and 3′ to the predicted RVE8 Myb encoding exons (Fig. 2A) in order to PCR amplify transcripts from plants maintained in 12 h light:dark diurnal cycles at either 20°C, or for plants undergoing cooling (20 to 4°C), with the temperature transition initiated at dusk at 20°C. Levels of the canonically, or fully spliced RVE8 RT-PCR product (denoted by the arrow on the gel in Figure 2B), phased to around dawn in a 12 h dark:light cycle at 20°C, are dramatically reduced during a cold temperature transition (Fig. 2B and C). Notably there is a concomitant increase and peak broadening of a slower migrating PCR product (Fig. 2C; denoted by a star on the gel in Figure 2B) during cold transition that, based on the lengths of the introns bracketed by the regions targeted during PCR, equates to the transcript retaining intron 4 (assigned as AS7 in Table 1). We cannot entirely rule out that AS4 transcripts are amplified (this product may not be clearly resolved from the FS product since the difference in length is only 17 nt, Table 1), and the abundance of AS1 was very low in this experiment. Taken together this suggests that the splicing of transcripts retaining intron 4 (AS7) is specifically sensitive to temperature and future work will be necessary to establish if other AS transcripts identified in the HR RT-PCR screen (Table 1), that are not targeted by the primers used in Figure 2, are modulated by temperature.
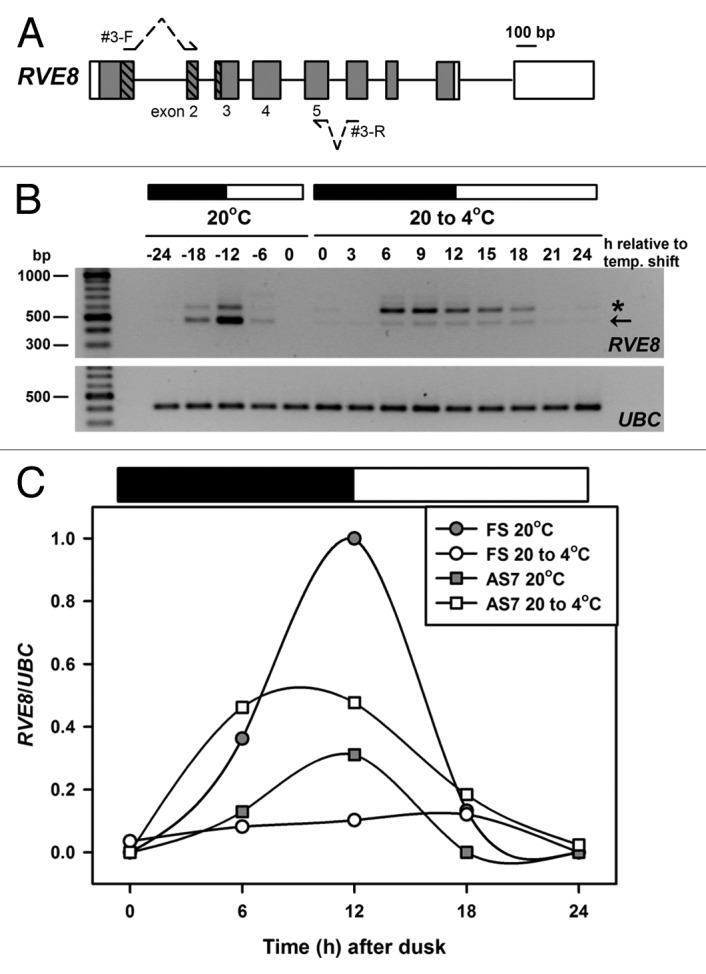
Figure 2. Temperature associated AS of RVE8. (A) RVE8 gene organization is as outlined in Figure 1A. To preclude potential amplification of gDNA during RT-PCR both forward and reverse primers – shown as arrows interrupted with dotted lines – spanned an intron and had the potential to amplify AS transcripts between exons 2 and 5. Primer sequences are: RVE8#3-F; TGAAGCACTTCAACTGTTTGATCGTGACTG and RVE8#3-R; ATCAGGAACACCGTGAAGCACTGGA. Bases in bold are separated by an intron in the RVE8 gDNA sequence. (B) RT-PCR amplification of RVE8, using the primer sequences described above, for plants harvested during diurnal cycles of 12 h dark and 12 h light (denoted by the dark and white bars, respectively) either at ambient temperature (20°C) or during a low temperature transition (20 to 4°C). The cooling temperature shift was initiated at dusk at 20°C and samples are labeled as relative to the temperature shift. Arrow denotes the fully spliced or canonical RT-PCR product (predicted size 481 bp), and the star an AS product, representing transcripts retaining intron 4 (AS7, predicted size 599 bp, see also Figure 1B and Table 1). Note that samples are every 6 h at 20°C and every 3 h during the cold transition. For more details of Methods, and for the UBC primer sequences, see15 (C) RVE8 FS and AS7 RT-PCR band intensities (normalized to the UBC positive controls) from panel B were plotted across the diurnal cycles of 12 h dark and 12 h light (denoted by the dark and white bars, respectively) either at ambient temperature (20°C) or during a low temperature transition (20 to 4°C). Band intensities were determined using a BioRad GelDoc 2000 imaging system and QuantityOne software.
Retention of the intron downstream of the Myb encoding exons is a familiar scenario for both CCA1 and LHY, and intriguingly recent work17 has demonstrated that retention of intron 4 in CCA1, a transcript that we found not to be sensitive to NMD,15 was template for the synthesis of a peptide that retained the dimerization domain and functioned in a dominant-negative fashion to interfere with the transactivation potential of CCA1. The intron 4 sequence of RVE8 contains an in-frame start codon and this might suggest that a similar interfering micro-protein (miP) could be synthesized from RVE8 AS7 transcripts in plants adjusting to low temperatures. Further work will be required to clarify this, or indeed whether the AS transcripts are instead subjected to the plant’s NMD surveillance pathways or produce an N-terminal peptide. Notably however, all of these scenarios could result in reduced amounts functional RVE8, and how modulation of RVE8 protein levels would fit with the current clock model is unclear. RVE8 is thought to operate centrally in the clock by forming a negative feedback loop with PRR5,18 and is also proposed to have a role in regulating TOC1 expression.19 RVE8 has been shown to bind evening element (EE) sequences in the promoters of TOC1 and PRR5 and although overexpression of RVE8 increases the trough levels of PRR5, levels of PRR5 were not affected in rve8 mutants.18 We found that peak levels of PRR5 increased during low temperature transition (see Figure 7A and B in reference 15). Whether this modulation of PRR5 levels involves the proposed RVE8-PRR5 negative feedback loop has yet to be established but clearly modulation of RVE8 expression levels by AS is an important aspect of the response of the clock to temperature.
More generally, is the modulation of the clock splicing events simply a response to low temperature stress? We established several features of the dynamic changes in splicing patterns that suggest that this is not the case. First, many of the changes that we detected in plants experiencing a large drop in temperature (from 20°C to 4°C) are also detected in plants undergoing more moderate low temperature transitions and acclimations (from 20°C to 12°C). Second, where a change in splicing pattern is observed on temperature reduction, the opposite effect is seen in a temperature increase. Lastly, specific introns seem to be targets for temperature associated splicing – clearly many other introns, including introns within the same gene, are spliced normally during the same temperature changes. Therefore it is not simply that the splicing machinery operates differently, or is dysfunctional, at lower temperatures, but that the changes in splicing patterns we observe represent subtle and specific modulation of functional clock gene expression levels. Most previous work on circadian systems employed constant conditions, for example constant light or dark and temperature regimes such that the organism experiences a single constant, or acclimated, temperature. However, in the natural environment, plants are exposed to fluctuating temperatures, there is a constant requirement to adapt to, at times, rapid changes in the environmental temperatures – indeed temperature gradients of > 30°C have been reported to be tolerated in Arabidopsis.20,21 Our deliberate use of temperature transients has clearly proved useful for extending our understanding of the dynamic response plants have to temperature - continued use of transients and simulated natural conditions22 in experimental systems promises to yield further insight into the plasticity of plant responses with fluctuating environmental conditions.
There are several scenarios in which mutual interaction between temperature and the clock are important, including stalling of the clock at low temperatures,23-26 temperature entrainment,27 temperature compensation of the clock,28-31 and clock-mediated gating changes in temperature to downstream ‘cold-response’ signaling.32,33 The extent to which temperature associated AS might be involved in any of these scenarios is not as yet clear. A key aspect common to each of these situations is that they must rely on the perception of temperature and the identity of the primary mechanism – the plant thermometer - remains one of the great mysteries in plant science.20 It is unlikely that there is actually just one plant thermometer34,35 and several candidates have been put forward (see Box 2 in reference 20), including the involvement of RNA in the sensing mechanism. Whether AS in the clock is the thermometer, part of a thermometer, or is downstream of the thermometer remains to be seen but given the regulatory dominance of the clock on global transcription36 it may be more intuitive to regard temperature associated AS as orchestrating temperature perception with downstream processes rather than the perception mechanism per se. Thus, although probably not the equivalent of a thermometers ‘mercury in the bulb’ it seems plausible that AS in the clock is integral to the ‘read-out’ thereby aligning temperature with physiologically relevant downstream processes. However future work will explore the possible role of AS in the clock as a thermo-sensory mechanism or how close to the primary temperature perception event the clock is positioned and by what mechanism the fluctuating temperature stimulus feeds into the clock.
As touched upon in the previous paragraph the clock and temperature has also been of great interest in respect of the classical ‘temperature compensation’ conundrum - the observation that the period of circadian outputs varies little over a range of physiologically relevant temperatures. Identification of the mechanisms underlying temperature compensation has proved challenging. Interestingly we found that for clock genes that have been previously implicated in temperature compensation – CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LHY,30 and temperature responsiveness - PRR9 and PRR727 - levels of functional transcripts responded in opposite directions to transient cold conditions (with CCA1 and PRR9 upregulated, and LHY and PRR7 downregulated), suggesting that their effects are balanced, or compensated, in the clock. It may now emerge that the divergent response of PRR5 and RVE8 expression to cold temperature transition (up- and downregulation, respectively) is also part of this mechanism. An intriguing parallel of these ‘balance and checks’ of clock gene expression mediated by AS lies with compensating bimetallic gridiron pendulums invented by the famous 18th century English clock maker John Harrison.37 Use of these gridirons in timepieces such as chronometers revolutionised sea travel by permitting the accurate measurement of longitude.38 Gridirons rely on the relative expansion of two metals – iron and brass for example – and combined the two metals work mechanically to cancel temperature error.37 Future work will aim to clarify the possible role of AS in mediating temperature effects via ‘molecular gridirons’ in temperature compensated clocks.
Our work therefore shows that AS in the plant clock is much more widespread and quantitatively important than has previously been appreciated. Here we add RVE8 to the list of clock genes that show AS in response to temperature in the normal ambient range, and by examining AS in the clock as a system we draw attention to the apparent plasticity of the clock network in its response to temperature including the response to temperature transitions. Together with recent work demonstrating a link between AS and the circadian clock - mutants of PROTEIN ARGININE METHYL TRANSFERASE 5 (PRMT5) show a longer circadian period and dramatic changes in the abundance of AS transcripts of PRR939,40 - it promises to be an interesting ‘time’ for alternative splicing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21491
References
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Marquez Y, Brown JW, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–95. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazan K. Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 2003;8:468–71. doi: 10.1016/j.tplants.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, et al. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32:5096–103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duque P. A role for SR proteins in plant stress responses. Plant Signal Behav. 2011;6:49–54. doi: 10.4161/psb.6.1.14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo PJ, Kim MJ, Ryu JY, Jeong EY, Park CM. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat Commun. 2011;2:303. doi: 10.1038/ncomms1303. [DOI] [PubMed] [Google Scholar]
- 8.Lamberto I, Percudani R, Gatti R, Folli C, Petrucco S. Conserved alternative splicing of Arabidopsis transthyretin-like determines protein localization and S-allantoin synthesis in peroxisomes. Plant Cell. 2010;22:1564–74. doi: 10.1105/tpc.109.070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XN, Mount SM. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009;150:1450–8. doi: 10.1104/pp.109.138180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Severing EI, van Dijk AD, Stiekema WJ, van Ham RC. Comparative analysis indicates that alternative splicing in plants has a limited role in functional expansion of the proteome. BMC Genomics. 2009;10:154. doi: 10.1186/1471-2164-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severing EI, van Dijk AD, van Ham RC. Assessing the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana using proteomics data. BMC Plant Biol. 2011;11:82. doi: 10.1186/1471-2229-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW. Alternative splicing in plants - coming of age. Trends Plant Sci. 2012 doi: 10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–69. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–81. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michael TP, Salome PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci U S A. 2003;100:6878–83. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, et al. A Self-Regulatory Circuit of CIRCADIAN CLOCK-ASSOCIATED1 Underlies the Circadian Clock Regulation of Temperature Responses in Arabidopsis. Plant Cell. 2012 doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–29. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 20.McClung CR, Davis SJ. Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr Biol. 2010;20:R1086–92. doi: 10.1016/j.cub.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–4. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- 22.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, et al. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature. 2012;484:371–5. doi: 10.1038/nature10991. [DOI] [PubMed] [Google Scholar]
- 23.Ramos A, Pérez-Solís E, Ibáñez C, Casado R, Collada C, Gómez L, et al. Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci U S A. 2005;102:7037–42. doi: 10.1073/pnas.0408549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–79. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibañez C, Ramos A, Acebo P, Contreras A, Casado R, Allona I, et al. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One. 2008;3:e3567. doi: 10.1371/journal.pone.0003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibáñez C, Kozarewa I, Johansson M, Ogren E, Rohde A, Eriksson ME. Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 2010;153:1823–33. doi: 10.1104/pp.110.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170:387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–50. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18:1177–87. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salomé PA, Weigel D, McClung CR. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell. 2010;22:3650–61. doi: 10.1105/tpc.110.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–8. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–39. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- 34.Penfield S. Temperature perception and signal transduction in plants. New Phytol. 2008;179:615–28. doi: 10.1111/j.1469-8137.2008.02478.x. [DOI] [PubMed] [Google Scholar]
- 35.Ruelland E, Zachowski A. How plants sense temperature. Environ Exp Bot. 2010;69:225–32. doi: 10.1016/j.envexpbot.2010.05.011. [DOI] [Google Scholar]
- 36.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 37.Matthys RJ. A short history of temperature compensation. In: Matthys RJ, ed. Accurate clock pendulums. Oxford; New York: Oxford University Press, 2004:7-12. [Google Scholar]
- 38.Sobel D. Longitude: The True Story of a Lone Genius Who Solved the Greatest Scientific Problem of His Time. New York: Penguin, 1995. [Google Scholar]
- 39.Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, McClung CR. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:21211–6. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–6. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]