Abstract
The Arabidopsis B3-domain transcription factor FUSCA3 (FUS3) is a master regulator of seed maturation and also a central modulator of hormonal responses during late embryogenesis and germination. Recently, we have identified AKIN10, the Arabidopsis ortholog of Snf1 (Sucrose Non-Fermenting-1)–Related Kinase1 (SnRK1), as a FUS3-interacting protein. We demonstrated that AKIN10 physically interacts with and phosphorylates FUS3 at its N-terminal region, and genetically interacts with FUS3 to regulate developmental phase transition and lateral organ growth. Snf1/AMPK/SnRK1 kinases are important sensors of the cellular energy level, and they are activated in response to starvation and cellular stress. Here we present findings that indicate FUS3 and AKIN10 functionally overlap in ABA signaling, but play different roles in sugar responses during germination. Seeds overexpressing FUS3 and AKIN10 both display ABA-hypersensitivity and delayed germination. The latter is partly dependent on de novo ABA synthesis in both genotypes, as delayed germination can be partially rescued by the ABA biosynthesis inhibitor, fluridone. However, seeds and seedlings overexpressing FUS3 and AKIN10 show different sugar responses. AKIN10-overexpressing seeds and seedlings are hypersensitive to glucose, while those overexpressing FUS3 display overall defects in osmotic stress, primarily during seedling growth, as they show increased sensitivity toward sorbitol and glucose. Hypersensitivity to sugar and/or osmotic stress during germination are partly dependent on de novo ABA synthesis for both genotypes, although are likely to act through distinct pathways. This data suggests that AKIN10 and FUS3 both act as positive regulators of seed responses to ABA, and that AKIN10 regulates sugar signaling while FUS3 mediates osmotic stress responses.
Keywords: ABA, AKIN10, FUSCA3, germination, sugar signaling
Introduction
Seed formation is a critical adaptation in the plant life cycle, as it allows plants to temporarily cease growth in adverse conditions until the environment becomes favorable. During the maturation phase of embryogenesis, the embryo accumulates nutrient reserves, acquires desiccation tolerance and enters a stage of dormancy. The transition from embryonic to vegetative development (germination) is tightly regulated by the hormones abscisic acid (ABA), which promotes dormancy and inhibits germination, and gibberellic acid (GA), which has the opposite effect of breaking dormancy and stimulating germination.1,2 In Arabidopsis, B3-domain transcription factors of the AFL (ABSCISIC ACID INSENSITIVE3, FUSCA3, LEAFY COTYLEDON2) family act as master regulators of late embryogenesis, as loss- and gain-of-function mutations in these genes greatly affect seed maturation.3
Genetic and molecular analyses indicate FUSCA3 (FUS3) inhibits the transition from the embryonic to the vegetative phase of development by promoting ABA accumulation while inhibiting GA biosynthesis.4 The loss-of-function mutant, fus3–3, bypasses dormancy and enters postembryonic development prematurely due to a lower ABA/GA ratio.4-7 Conversely, ectopic expression of FUS3 post-embryonically (ML1:FUS3) delays seed germination and plant development by increasing ABA level while repressing GA biosynthesis.4,8 The stability of the FUS3 protein also appears to be tightly regulated by ABA, GA and the 26S proteasome.4,9
In our recent study, AKIN10, was identified as an interactor of FUS3 from yeast two-hybrid screens.10 AKIN10 belongs to the Sucrose-Non-Fermenting 1 (Snf1)-related kinase1 (SnRK1) family and acts as a central regulator of cellular energetics in plants.11 Our results indicate AKIN10 physically interacts with and phosphorylates FUS3 at its N-terminal region, and delays its degradation in a cell-free system. Overexpression of AKIN10 (35S:AKIN10) causes delays in developmental phase transitions (germination and flowering) and defects in lateral organ formation. These phenotypes can be partially rescued by the fus3–3 mutation, suggesting FUS3 and AKIN10 act in overlapping pathways to regulate developmental phase transitions and lateral organ development.10 Interestingly, SnRK kinases also regulate stress responses, hormonal and sugar signaling pathways by global transcriptional modulations.12-16 This prompted our investigation of the role of AKIN10 and FUS3 in ABA and sugar responses during germination.
Results
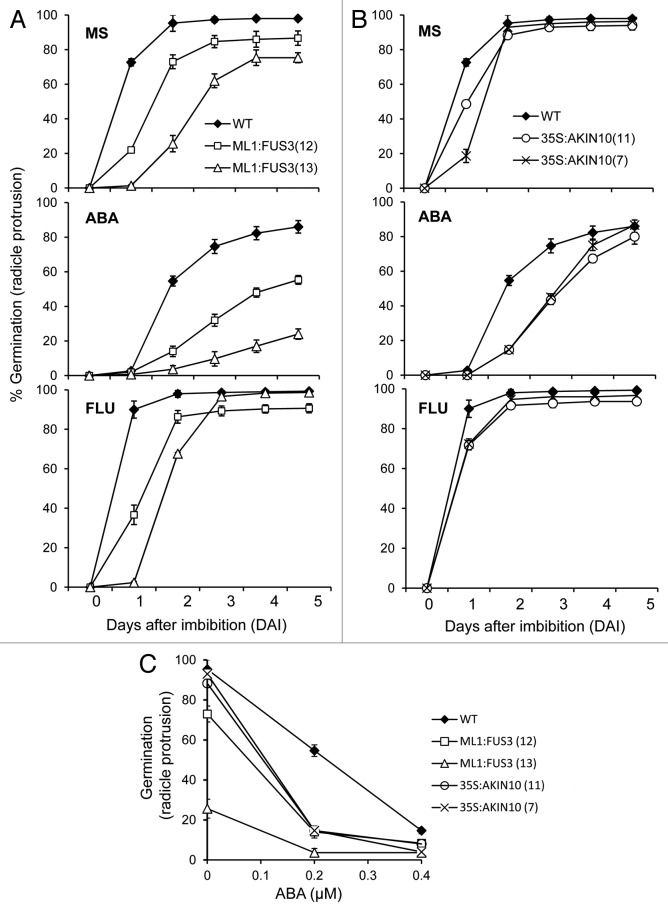
We first tested germination rates (radicle protrusion) of two independent lines overexpressing AKIN10 (35S:AKIN10) and FUS3 (ML1:FUS3) on ABA (see Material and Methods). Both 35S:AKIN10 and ML1:FUS3 transgenic plants were previously shown to delay seed germination on minimal MS medium,8,12 with ML1:FUS3 showing a greater germination delay than 35S:AKIN10 (Fig. 1). The delayed germination has been attributed to the heightened sensitivity to and level of ABA in ML1:FUS3 seeds,4,8 but the cause of delay remains unknown for 35S:AKIN10 seeds. To test whether altered ABA sensitivity contributes to the 35S:AKIN10 germination delay, wild type (WT), ML1:FUS3 and 35S:AKIN10 seeds were germinated on a low concentration of ABA (0.2 μM). ABA delayed WT germination, though germination rates recovered to approximately 80% in 5 d (Fig. 1). ML1:FUS3 germination was more sensitive to ABA compared with WT, and reached only 20–50% after 5 d of treatment (Fig. 1A). 35S:AKIN10 germination was also hypersensitive to ABA, but recovered to approximately 80% within 5 d of treatment (Fig. 1B). Germination rates tested on 0.4 μM ABA show that both genotypes are slightly hypersensitive to this concentration of ABA compare with WT (Fig. 1C). This suggests that both AKIN10 and FUS3 positively regulate ABA sensitivity during germination.
Figure 1. Overexpression of FUS3 or AKIN10 leads to ABA hypersensitivity and delayed germination, which is partly dependent on de novo ABA synthesis. Germination (radicle protrusion) kinetics of seeds from WT and two independent lines of ML1:FUS3 (A) or 35S:AKIN10 (B) on MS media or MS supplemented with 0.2 μM ABA or 10 µM fluridone (FLU). WT germination is significantly higher (p < 0.01) than 35S:AKIN10 on 10 µM FLU at 2 d after imbibition (DAI). (C) ABA dose-response curves for WT, ML1:FUS3 and 35S:AKIN10 seed germination (radicle protrusion) 2 d after imbibition. WT germination is higher (p < 0.001) than ML1:FUS3 and 35S:AKIN10 at 0.4 µM ABA. Averages from 3 experiments ± SD are shown. 100–150 seeds were used in each experiment.
In order to dissect the role of ABA in the delayed germination of FUS3- and AKIN10- overexpressing seeds, WT, ML1:FUS3 and 35S:AKIN10 germination rates were assayed in media supplemented with 10 μM fluridone, an inhibitor of phytoene desaturase which reduces ABA synthesis.17,18 If delayed germination of the transgenic lines is due to increased ABA synthesis, then fluridone should rescue this delay. Germination rates of WT, ML1:FUS3 and 35S:AKIN10 seeds were higher in the presence of fluridone compared with untreated seeds (Fig. 1A and B), suggesting de novo ABA synthesis negatively regulates germination in all genotypes. However, ML1:FUS3 and 35S:AKIN10 seeds still germinated later than WT on fluridone (Fig. 1A and B), and their rates did not increase even at a higher concentration of fluridone (50 μM; data not shown). This suggests de novo ABA synthesis alone cannot explain the delayed germination phenotype of ML1:FUS3 and 35S:AKIN10 seeds.
ABA and sugar signaling pathways are intricately related and share common downstream signaling components.19-21 Since AKIN10 is known to partake in sugar signaling11,22 and since FUS3 expression is regulated by sugar,23 we investigated the role of FUS3 and AKIN10 in sugar signaling during germination. WT, ML1:FUS3 and 35S:AKIN10 germination rates were assayed two days after imbibition on 3% glucose or 3% sorbitol as an osmotic control. At the concentration tested glucose, but not sorbitol, significantly reduced WT seed germination, as previously described (Fig. 2A).21,24 35S:AKIN10 germination was similar to WT on sorbitol, but showed hypersensitivity on glucose (Fig. 2A). Surprisingly, ML1:FUS3 germination was reduced by sorbitol, while the effect of glucose varied between the two transgenic lines (Fig. 2A). To better understand the effect of exogenous glucose application, seedlings growth rates (cotyledon expansion) were assayed 4 d after imbibition in the presence of the sugars. In this case, cotyledon expansion of both ML1:FUS3 transgenic lines was hypersensitive to both sorbitol and glucose, whereas 35S:AKIN10 cotyledon expansion was inhibited specifically by glucose (Fig. 2B). These results indicate that overexpression of FUS3 during germination causes hypersensitivity toward osmotic stress, whereas overexpression of AKIN10 leads to hypersensitivity specifically toward glucose. We next tested whether the osmotic hypersensitivity of ML1:FUS3 and glucose hypersensitivity of 35S:AKIN10 seeds during germination are dependent on increased ABA synthesis. Germination rates were assayed 2 d after imbibition on 10 µM fluridone in the presence and absence of 3% glucose or sorbitol. In both cases, fluridone was able to partially restore the germination delay imposed by sorbitol and glucose on ML1:FUS3 and glucose on 35S:AKIN10 (Fig. 3). These results indicate the hypersensitivity of ML1:FUS3 seeds to osmotic stress and glucose hypersensitivity of 35S:AKIN10 seeds are both partially dependent on de novo ABA synthesis.
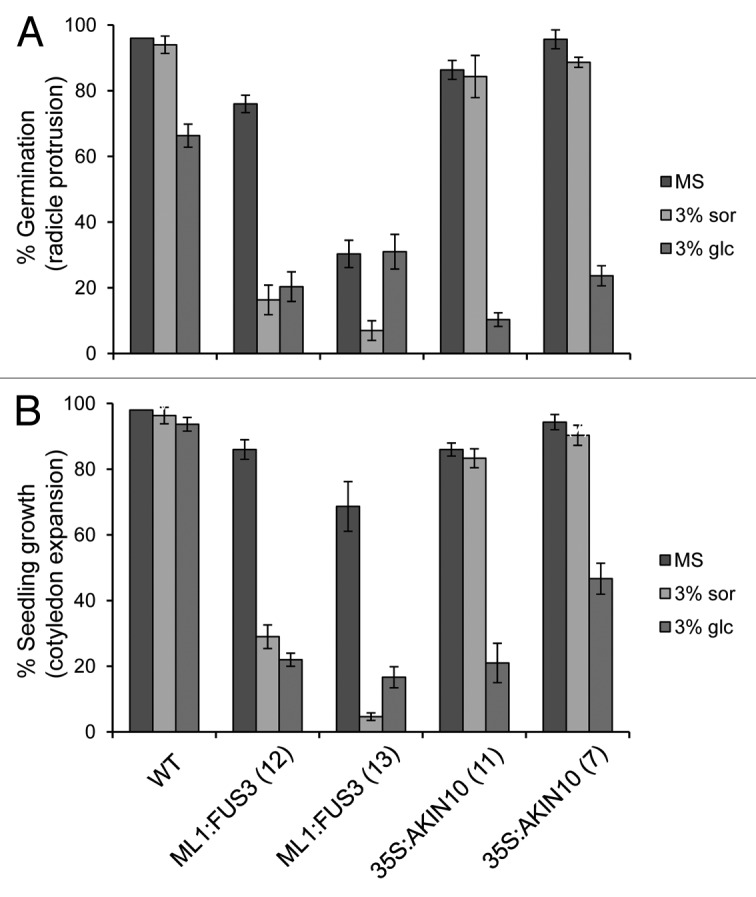
Figure 2. Overexpression of FUS3 or AKIN10 leads to different responses to sugar during seed germination and seedling growth. (A) Germination (radicle protrusion) of WT, ML1:FUS3 and 35S:AKIN10 seeds 2 d after imbibition on MS ± 3% sorbitol (sor) or 3% glucose (glc). (B) Seedling growth (cotyledon expansion) of WT, ML1:FUS3 and 35S:AKIN10 seeds 4 d after imbibition on MS ± 3% sor or 3% glc. Averages from 3 experiments ± SD are shown. 100–150 seeds were used in each experiment.
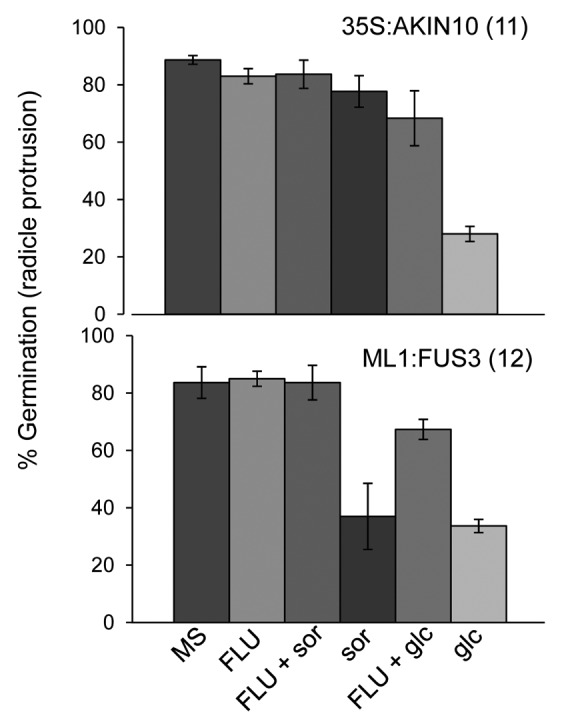
Figure 3. Osmotic stress hypersensitivity of ML1:FUS3 seeds and glucose hypersensitivity of 35S:AKIN10 seeds are both partially dependent on de novo ABA synthesis. Germination (radicle protrusion) of 35S:AKIN10 and ML1:FUS3 seeds 2 d after imbibition on MS, 10 µM fluridone (FLU), 3% sorbitol (sor) ± 10 µM FLU, and 3% glucose (glc) ± 10 µM FLU. Averages from 3 experiments ± SD are shown. 100–150 seeds were used in each experiment.
Discussion
Collectively, the data shown here demonstrate a positive role of AKIN10 and FUS3 in ABA responses during germination, as well as distinct roles in sugar and osmotic stress responses during seed germination and seedling growth. Indeed, FUS3 plays an inhibitory role under osmotic stress, while AKIN10 inhibitory role is specific for glucose. These data complement previous findings showing that plants overexpressing AKIN10 display defects in post-embryonic development and root elongation on exogenous ABA and glucose.12,14 Notably, the germination of seeds overexpressing AKIN10 was previously shown to be unaffected by 3 µM ABA.14 This is likely due to the high concentration of ABA used by Jossier et al.,14 and that we recorded germination rates over multiple time points after imbibition.
Although fluridone was able to elevate the germination kinetics in seeds of both genotypes on MS media, it was not sufficient to fully recover the delayed germination of ML1:FUS3 and 35S:AKIN10 seeds to the WT level. This suggests de novo ABA synthesis is only partially responsible for ML1:FUS3 and 35S:AKIN10 delayed germination. It is possible that overexpression of FUS3 or AKIN10 already increases ABA level during embryogenesis, thus increasing seed dormancy. This is likely the case for FUS3, considering that a short activation of FUS3 indeed increases ABA level in ML1:FUS3-GR seedlings, while loss-of-function fus3–3 embryos contain less ABA.4,5 The germination delay of 35S:AKIN10 seeds was more closely, but not completely, inhibited by fluridone. Although no differences in ABA level were previously found between WT and AKIN10-overexpressing seedlings,14 this does not exclude the possibility of higher ABA accumulation in 35S:AKIN10 embryos compared with WT. In addition, ABA accumulation prior to germination may also explain why fluridone was unable to fully restore ML1:FUS3 and 35S:AKIN10 reduced germination in the presence of glucose. Alternatively, ABA-independent pathways may be activated by FUS3 and AKIN10 during germination.
In conclusion, the data presented here indicate both FUS3 and AKIN10 act as positive regulators of ABA signaling during germination, although showing different sensitivity to the hormone, but they play different roles in sugar and osmotic stress signaling. AKIN10 is involved in glucose-specific pathway(s), while FUS3 modulates osmotic stress responses. Both sugar and osmotic responses regulated by AKIN10 and FUS3 are partly dependent on de novo ABA synthesis, likely through distinct pathways. It remains to determine if phosphorylation of FUS3 by AKIN10 is required to modulate seed sensitivity to ABA.
Material and Methods
Arabidopsis seeds of WT (Col-0), FUS3-overespressing (ML1:FUS3-GFP;4,9) and AKIN10-overexpressing (35S:AKIN10-HA;10) lines were vernalized and germinated as previously described.9 ABA, fluridone, sorbitol and glucose supplemented in the media at the concentrations specified in each experiment. Germination was considered positive when the radicle had emerged from the seed, while seedling growth was scored positive when the cotyledons were expanded. The ML1:FUS3 construct was previously shown to rescue the fus3–3 embryonic phenotypes, including desiccation intolerance of the seeds, and to ectopically express FUS3 post-embryonically.4,9 In contrast, 35S:FUS3 does not rescue the fus3–3 mutant and causes co-suppression when transformed in a WT background (data not shown). Therefore, the 35S promoter could not be used to overexpress FUS3. Conversely, 35S:AKIN10 has been shown to overexpress AKIN10 post-embryonically.10 Since ML1 and AKIN10 expression levels are very similar throughout development, as measured in several microarrays (eNorthern; http://bar.utoronto.ca), we did not attempt to overexpress AKIN10 using the ML1 promoter.
Glossary
Abbreviations:
- ABA
Abscisic acid
- GA
Gibberellic acid
- FUS3
FUSCA3
- SNF1
Sucrose Non-Fermenting 1
- AKIN10
Arabidopsis Snf1-Related Kinase homolog 10
- SnRK1
Snf1-Related Kinase 1
- ML1:FUS3
FUS3 overexpression
- 35S:AKIN10
AKIN10 overexpression
- WT
wild type
- FLU
fluridone
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21549
References
- 1.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5:33–6. doi: 10.1016/S1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol. 2008;11:548–53. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell. 2004;7:373–85. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, et al. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol. 2000;220:412–23. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- 6.Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, et al. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004;136:3660–9. doi: 10.1104/pp.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith K, Kraml M, Dengler NG, McCourt P. fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu RS, Nahal H, Provart NJ, Gazzarrini S. The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 2012;12:15. doi: 10.1186/1471-2229-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu QS, Paz JD, Pathmanathan A, Chiu RS, Tsai AY, Gazzarrini S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 2010;64:100–13. doi: 10.1111/j.1365-313X.2010.04307.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsai AY, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012;69:809–21. doi: 10.1111/j.1365-313X.2011.04832.x. [DOI] [PubMed] [Google Scholar]
- 11.Halford NG, Hey SJ. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J. 2009;419:247–59. doi: 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- 12.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–42. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 13.Bradford KJ, Downie AB, Gee OH, Alvarado V, Yang H, Dahal P. Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol. 2003;132:1560–76. doi: 10.1104/pp.102.019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, et al. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009;59:316–28. doi: 10.1111/j.1365-313X.2009.03871.x. [DOI] [PubMed] [Google Scholar]
- 15.Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006;140:263–78. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, et al. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2010;61:324–38. doi: 10.1111/j.1365-313X.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- 17.Chamovitz D, Sandmann G, Hirschberg J. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J Biol Chem. 1993;268:17348–53. [PubMed] [Google Scholar]
- 18.Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M. Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta. 2000;210:279–85. doi: 10.1007/PL00008135. [DOI] [PubMed] [Google Scholar]
- 19.Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol. 2001;4:387–91. doi: 10.1016/S1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- 20.Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell. 2002;14:2723–43. doi: 10.1105/tpc.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson SI. Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol. 2005;8:93–102. doi: 10.1016/j.pbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 23.Tsukagoshi H, Morikami A, Nakamura K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci U S A. 2007;104:2543–7. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekkers BJW, Schuurmans JAMJ, Smeekens SCM. Glucose delays seed germination in Arabidopsis thaliana. Planta. 2004;218:579–88. doi: 10.1007/s00425-003-1154-9. [DOI] [PubMed] [Google Scholar]