Abstract
Phosphoinositide-specific phospholipase C (PI-PLC) belongs to an important class of enzymes involved in signaling related to lipids. They hydrolyze a membrane-associated phospholipid, phosphatidylinositol-4,5-bisphosphate, to produce inositol-1,4,5-trisphosphate and diacylglycerol. The role of PI-PLC and the mechanism behind its functioning is well studied in animal system; however, mechanism of plant PI-PLC functioning remains largely obscure. Here, we attempted to summarize the understanding regarding plant PI-PLC mechanism of regulation, localization, and domain association. Using sedimentation based phospholipid binding assay and surface plasmon resonance spectroscopy, it was demonstrated that C2 domain of plant PI-PLC alone is capable of targeting membranes. Moreover, change in surface hydrophobicity upon calcium stimulus is the key element in targeting plant PI-PLC from soluble fractions to membranes. This property of altering surface hydrophobicity plays a pivot role in regulation of PI-PLC activity.
Keywords: C2 domain, membrane targeting, Phospholipase C
Based on substrate preference, two types of phospholipase activities have been identified in plants: non-specific phospholipase C hydrolyzes phosphatidylcholine (PC)1 and phosphoinositide-specific phospholipase C (PI-PLC) hydrolyzes phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2), a minor phospholipid producing diacylglycerol (DAG), and inositol-1,4,5-triphosphate (Ins(1,4,5)P3). In this review, PI-PLC will be discussed. In mammalian cells, Ins(1,4,5)P3 is known to trigger calcium influx by activating calcium channels,2 whereas DAG causes activation of protein kinase C (PKC).3 Plant PI-PLC is associated with a spectrum of processes concerning signal transduction in events such as guard cell signaling,4 salt stress, osmotic stress, acquired resistance,5,6 Nod factor signaling,7 drought,8 systemic acquired resistance,9 carbon fixation in C4 plants,10 and response to the pathogen.11 In this review, we have attempted to introduce current understanding of plant PI-PLC with respect to its localization, regulation, and activity. We also wish to highlight a model for PI-PLC activity based on change in surface hydrophobicity upon calcium stimulation.
Domain Organization
The domain organization of plant PI-PLC is similar to that of animal PI-PLC-ζ, which is a sperm-specific PI-PLC.12 The domains shared by all known PI-PLC are EF-hand, XY catalytic domain, and the phospholipid-binding C2 domain. These domains are common in all organisms without exception and may represent bare minimum requirement for functioning of PI-PLC.13 The other conserved domain present exclusively in all animal PI-PLCs is the Pleckstrin homology (PH) domain which is responsible for recognizing and binding to PtdIns(4,5)P2 in the membranes.14 Although this domain is absent in plant PI-PLC, it still appears to bind to PtdIns(4,5)P2 in the membranes.13 The exact mechanism behind this ability in plant PI-PLC requires further elucidation.
Classes of Plant PI-PLC
Plant PI-PLCs were classified into two classes based on localization of the enzyme in the membrane and cytosolic fractions. It was also noted that they require different concentrations of calcium for their activity and that they also differ in their substrate preference. The cytosolic PI-PLC was characterized as phosphatidylinositol (PI) hydrolyzing enzyme, which requires millimolar amounts of calcium for its activity. The membrane-associated PI-PLC on the other hand, was described as an enzyme that prefers PtdIns(4,5)P2 as the substrate and it requires micromolar amounts of calcium for its activity.15,16 It is interesting to note that most of the plant PI-PLCs are able to hydrolyze both the substrates (PtdIns(4,5)P2 and PI).17-19 However, the concentration of calcium can dictate the preference of substrates; high concentration favors PI and PIP, whereas low concentration is sufficient for hydrolysis of PtdIns(4,5)P2.20 On the contrary, the notion of existences of two classes of plant PI-PLC appears to be dubious for many reasons. From genome sequencing data, there is no report of plant PI-PLC with different sequences amounting for the presence of such distinct classes. At domain organization level, it is known that PI-PLC in animal system, which are membrane-associated shares unique domains; but such domain diversity, is absent in plant PI-PLC. The catalytic domains X and Y are conserved throughout biological systems; therefore, if there is any such isoforms of plant PI-PLC, it must have the conserved X and Y domains; such isoforms would be difficult to miss in the genome sequence database. All plant PI-PLCs cloned to date are thought to be belonging to the membrane-associated class.
Localization and Membrane Targeting
Plant PI-PLCs are considered to be associated with plasma membrane even though it was also detected in the cytosol. Using FLAG-tagged PI-PLC fusion protein overexpression in tobacco cells, Shi et al.18 demonstrated that soybean PI-PLC is present in both soluble and membrane fractions. Interestingly, the same PI-PLC was identified by screening soybean cDNA expression library with anti-plasma membrane serum. Using antibodies, Cao et al.21 showed that AtPLC4 was localized in plasma membrane and cytosolic fractions prepared from Arabidopsis thaliana leaf extract. AtPLC4-specific antibody was prepared using the N-terminal peptides of AtPLC4; this antibody was unable to react with the other six heterologously expressed AtPLC isoforms. When Petunia GFP-tagged PI-PLC was coexpressed with either active site mutant or C2 domain of the same PI-PLC, it resulted in the displacement of GFP-tagged PI-PLC from the membranes to the cytosol.22 This also provided an indirect evidence that C2 domain could alone perform membrane targeting. Using yellow fluorescent protein (YFP) tag, it was demonstrated that EF-YFP-C2 domain was capable of binding to membranes, whereas C2-YFP construct did not show a similar binding.23 In vitro lipid binding assays with N-terminal deletion mutant corresponding to EF-hand in A. thaliana PI-PLC displayed membrane binding capacity but lacked catalytic activity.17 In our earlier study, using sedimentation based lipid binding assay, fluorescence spectroscopy, and surface plasmon resonance spectroscopy (SPR), we demonstrated that C2 domain could bind membranes independent of EF-hand or XY catalytic domain.13 Earlier, it was argued that PI-PLC activity in soluble fraction may be due to breaking-off of the membrane-associated PI-PLC during extraction processes. Contrary to popular notion, we argued that the cytosolic PI-PLC could be found in membranes because of their binding as a result of transient increase in calcium due to breaking of cellular membranes. Moreover, successful purification of PI-PLC from isolated membranes in the presence of chelating agents, as indicated in earlier reports,15,16 suggested that the bound PI-PLC could not be dissociated from the membranes merely by decreasing calcium levels. We have demonstrated the same in our earlier study using in vitro lipid binding assay and SPR.13
Mechanism of Membrane Targeting and Regulation
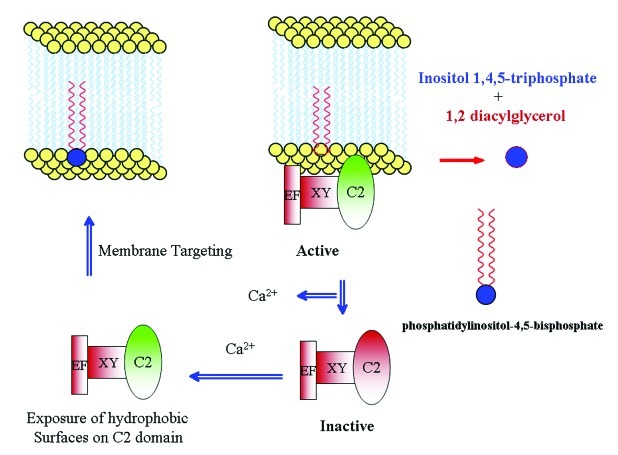
Mammalian pathway leading to the activation of PI-PLC is well studied and is proposed to be activated by heterotrimeric G-protein-coupled receptor, which helps in transduction of extracellular signal. The presence of a similar pathway in plants is unclear. However, there is a report on tobacco PI-PLC (NtPLC3), interacting with G-protein NtRac5.23 Furthermore, in Lilium davidii, PI-PLC activity was stimulated by the addition of G-protein activator, cholera toxin.24 In another study, overexpression of G-protein-coupled receptor was linked to the activation of PI-PLC.25 The regulation of plant PI-PLC by G-protein-coupled receptor is by and large obscure but activation by calcium is well established.10,16,22,23 There are two levels of regulation by calcium: (1) its requirement for the catalytic activity of XY domain and (2) its requirement for the membrane targeting activity of C2 domain.13 We attempted to explain the mechanism of membrane targeting of PI-PLC based on the ability of C2 domain to expose hydrophobic surfaces upon stimulation by calcium.13 C2 domain acts as a switch that is regulated by calcium levels, which in turn determines PI-PLC localization (Fig. 1). A certain type of membrane-associated protein in tobacco, designated as NtC7, was found to facilitate tobacco PI-PLC association with membranes.26 It will be interesting to further investigate the role of such proteins in plant PI-PLC signaling.
Figure 1. Model for the plant PI-PLC functioning. Plant PI-PLC under low calcium levels are rendered inactive by disassociation from the membrane. Upon calcium stimulus, the C2 domain acts as a switch by exposing hydrophobic surfaces and causing binding with the membrane.
Perspective
It will be of immense interest to understand how plant PI-PLC is regulated; more conclusive studies are required to establish this regulation by G-protein. Although the regulation by calcium appears to be sufficient for their regulation, considering that plant PI-PLC will be inactive once dissociated from membranes. It will be of interest to understand the mechanism by which plant PI-PLC are dissociated from the membrane because decrease in calcium levels alone may not be a condition sufficient for there dissociation. The regulation at membrane level by either G-proteins or other components of signal transduction pathway will help in refining downstream function or localization of PI-PLC activity. The exact role of DAG and Ins(1,4,5)P3 need to be investigated in depth owning to the absence of Ins(1,4,5)P3 sensitive calcium channels27 and a lack of concrete evidence for DAG-activating plant PKC homologs. Recently, we had demonstrated in Saccharomyces cerevisiae that PI-PLC can regulate lipid biosynthesis through upstream activation sequence inositol (UASINO).28 It will be of paramount interest to investigate existence of a similar type of regulation by PI-PLC in plants or higher organisms.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21436
References
- 1.Dowd PE, Gilroy S. The emerging roles of phospholipase C in plant growth and development. In: Munnik T, ed. Lipid signaling in plants. Berlin, Germany: Springer 2010; 23-28. [Google Scholar]
- 2.Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–25. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 3.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–14. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Choi YB, Suh S, Lee J, Assmann SM, Joe CO, et al. Abscisic acid-induced phosphoinositide turnover in guard cell protoplasts of Vicia faba. Plant Physiol. 1996;110:987–96. doi: 10.1104/pp.110.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X. Lipid signaling. Curr Opin Plant Biol. 2004;7:329–36. doi: 10.1016/j.pbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Cho MH, Shears SB, Boss WF. Changes in phosphatidylinositol metabolism in response to hyperosmotic stress in Daucus carota L. cells grown in suspension culture. Plant Physiol. 1993;103:637–47. doi: 10.1104/pp.103.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charron D, Pingret JL, Chabaud M, Journet EP, Barker DG. Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 2004;136:3582–93. doi: 10.1104/pp.104.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munnik T, Musgrave A. Phospholipid signaling in plants: holding on to phospholipase D. Sci STKE 2001; 2001:pe42. [DOI] [PubMed]
- 9.Reggiani R, Laoreti P. Evidence for the involvement of phospholipase C in the anaerobic signal transduction. Plant Cell Physiol. 2000;41:1392–6. doi: 10.1093/pcp/pcd073. [DOI] [PubMed] [Google Scholar]
- 10.Coursol S, Pierre JN, Vidal J. Role of the phosphoinositide pathway in the light-dependent C4 phosphoenolpyruvate carboxylase phosphorylation cascade in Digitaria sanguinalis protoplasts. Biochem Soc Trans. 2000;28:821–3. doi: 10.1042/BST0280821. [DOI] [PubMed] [Google Scholar]
- 11.de Jong CF, Laxalt AM, Bargmann BO, de Wit PJ, Joosten MH, Munnik T. Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 2004;39:1–12. doi: 10.1111/j.1365-313X.2004.02110.x. [DOI] [PubMed] [Google Scholar]
- 12.Swann K, Saunders CM, Rogers NT, Lai FA. PLCzeta(zeta): a sperm protein that triggers Ca2+ oscillations and egg activation in mammals. Semin Cell Dev Biol. 2006;17:264–73. doi: 10.1016/j.semcdb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Rupwate SD, Rajasekharan R. C2 domain is responsible for targeting rice phosphoinositide specific phospholipase C. Plant Mol Biol. 2012;78:247–58. doi: 10.1007/s11103-011-9862-1. [DOI] [PubMed] [Google Scholar]
- 14.Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, et al. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 15.Helsper JPFG, Heemskerk JWM, Veerkamp JH. Cytosolic and particulate phosphatidylinositol phospholipase C activities in pollen tubes of Lilium longiflorum. Physiol Plant. 1987;71:120–6. doi: 10.1111/j.1399-3054.1987.tb04628.x. [DOI] [Google Scholar]
- 16.Drøbak BK. The plant phosphoinositide system. Biochem J. 1992;288:697–712. doi: 10.1042/bj2880697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otterhag L, Sommarin M, Pical C. N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 2001;497:165–70. doi: 10.1016/S0014-5793(01)02453-X. [DOI] [PubMed] [Google Scholar]
- 18.Shi J, Gonzales RA, Bhattacharyya MK. Characterization of a plasma membrane-associated phosphoinositide-specific phospholipase C from soybean. Plant J. 1995;8:381–90. doi: 10.1046/j.1365-313X.1995.08030381.x. [DOI] [PubMed] [Google Scholar]
- 19.Kopka J, Pical C, Gray JE, Müller-Röber B. Molecular and enzymatic characterization of three phosphoinositide-specific phospholipase C isoforms from potato. Plant Physiol. 1998;116:239–50. doi: 10.1104/pp.116.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munnik T, Irvine RF, Musgrave A. Phospholipid signalling in plants. Biochim Biophys Acta. 1998;1389:222–72. doi: 10.1016/S0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]
- 21.Cao Z, Zhang J, Li Y, Xu X, Liu G, Bhattacharrya MK, et al. Preparation of polyclonal antibody specific for AtPLC4, an Arabidopsis phosphatidylinositol-specific phospholipase C in rabbits. Protein Expr Purif. 2007;52:306–12. doi: 10.1016/j.pep.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Dowd PE, Coursol S, Skirpan AL, Kao TH, Gilroy S. Petunia phospholipase c1 is involved in pollen tube growth. Plant Cell. 2006;18:1438–53. doi: 10.1105/tpc.106.041582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helling D, Possart A, Cottier S, Klahre U, Kost B. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell. 2006;18:3519–34. doi: 10.1105/tpc.106.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan YY, Wang X, Ma LG, Sun DY. Characterization of phosphatidylinositol-specific phospholipase C (PI-PLC) from Lilium daviddi pollen. Plant Cell Physiol. 2005;46:1657–65. doi: 10.1093/pcp/pci181. [DOI] [PubMed] [Google Scholar]
- 25.Apone F, Alyeshmerni N, Wiens K, Chalmers D, Chrispeels MJ, Colucci G. The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol. 2003;133:571–9. doi: 10.1104/pp.103.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura K, Sano H. A plasma-membrane linker for the phosphoinositide-specific phospholipase C in tobacco plants. Plant Signal Behav. 2009;4:26–9. doi: 10.4161/psb.4.1.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krinke O, Novotná Z, Valentová O, Martinec J. Inositol trisphosphate receptor in higher plants: is it real? J Exp Bot. 2007;58:361–76. doi: 10.1093/jxb/erl220. [DOI] [PubMed] [Google Scholar]
- 28.Rupwate SD, Rupwate PS, Rajasekharan R. Regulation of lipid biosynthesis by phosphatidylinositol-specific phospholipase C through the transcriptional repression of upstream activating sequence inositol containing genes. FEBS Lett. 2012;586:1555–60. doi: 10.1016/j.febslet.2012.04.022. [DOI] [PubMed] [Google Scholar]