Abstract
Rice (Oryza sativa) represents one of the most important food crops in the world, since it feeds more than two billion people. The increased rice production can play significant roles in upgrading the economic status of countries like India and China. A great deal of research has been carried out in the recent past on the molecular biology, genomics and biotechnology of rice. By employing recombinant DNA technology, remarkable progress had been made towards production of rice plants with increase yield, improved nutritional quality and resistance to various diseases. Under these circumstances, the study of microRNAs can contribute to new discoveries in this field. The miRNAs are assign to modulate gene expression at the post-transcriptional level. They are small, non-coding, single stranded RNAs that are abundantly found in prokaryotic and eukaryotic cells and can trigger translational repression or gene silencing by binding to complementary sequences on target mRNA transcripts. In the recent years, miRNAs have been reported to control a variety of biological processes, such as plant development, differentiation, signal transduction or stress responses. The present review provides an up-date on microRNAs and their involvement in the stress response in rice. A section is specifically dedicated to the genetic engineering perspectives regarding the miRNAs applications in rice tolerance to stress conditions.
Keywords: Abiotic stress tolerance, amiRNAs, microRNAs, pri-miRNAs, siRNAs, stress response, rice
Rice (Oryza sativa) represents one of the most important food crops in the world, since it feeds more than two billion people. The increased rice production can play significant roles in upgrading the economic status of countries like India and China. A great deal of research has been performed in the recent past on the molecular biology, genomics and biotechnology of rice. By employing recombinant DNA technology, remarkable progress had been made toward production of rice plants with increase yield, improved nutritional quality and resistance to various diseases. Under these circumstances, the study of microRNAs can contribute to new discoveries in this field. The miRNAs are assigned to modulate gene expression at the post-transcriptional level. They are small, non-coding, single stranded RNAs that are abundantly found in prokaryotic and eukaryotic cells and can trigger translational repression or gene silencing by binding to complementary sequences on target mRNA transcripts. In the recent years, miRNAs have been reported to control a variety of biological processes, such as plant development, differentiation, signal transduction or stress responses. The present review provides an up-date on microRNAs and their involvement in the stress response in rice. A section is specifically dedicated to the genetic engineering perspectives regarding the miRNAs applications in rice tolerance to stress conditions.
Introduction
Crop productivity is strictly related to genome stability, an essential requisite for optimal plant growth and development. Several environmental stresses, such as UV light, ionizing radiations, heavy metals and other pollutants can induce severe injury within the cellular compartment, affecting crop productivity. Genotoxic agents influence genome integrity, inducing constant DNA damage which requires cells to activate proper repair responses.1-3
Rice, wheat, and maize are the three leading food crops in the world. Together they directly supply more than 50% of all calories consumed by the entire human population. Wheat is the leader in the harvested area, with 214 million ha/year, followed by rice with 154 million ha and maize with 140 million ha. Human consumption accounts for 85% of total production for rice, compared with 72% for wheat and 19% for maize (FAO, 2012). Besides its economical importance, rice also represents a model cereal system since it has a relatively small genome size as compared with other cereals, a vast germplasm collection, wide array of molecular genetic resources, and an efficient transformation protocol.4 The increase in rice production can play significant roles in upgrading the economic status, especially in developing countries from Asia and Africa. In the recent past, the molecular biology, genomics and biotechnology techniques greatly advantaged the rice research. By employing recombinant DNA technology, remarkable progress had been made toward production of rice plants with increase yield, improved nutritional quality and resistance to various diseases.5 However, rice is particularly sensitive to a number of different stress conditions among which excess salts, reduced or excess water supply and suboptimal temperature regimes are the most important.5 Within this context, the study of novel approaches designed for better understanding the stress-related mechanisms in rice, and possibly in other crops, are still of great interest.
The development of high-throughput transcriptomic and proteomic technologies, have enabled the identification of hundreds of genes induced under stress conditions for a better understanding of the stress-related mechanisms. Out of the many mechanisms responsible for stress adaptation, transcriptional regulation mediated by specific transcription factors (TFs) that bind to conserved cis-acting promoter elements, is the most widely studied especially in the case of abiotic stress-induced changes in gene expression.6 Several studies have shown the importance of post-transcriptional regulation of gene expression under stress, but still little is known concerning this field. The precise control of transcription is critical for the regulation of gene expression during cell differentiation and plant development. At the transcriptional level, the TFs play a major role in regulating gene expression. By binding to specific regions, the TFs can control the transcription activities of target genes regulating the production of mRNA transcripts.7 Another intriguing aspect related to the transcription regulation process involves the activity of microRNAs (miRNAs). In multicellular organisms, TFs and miRNAs are the major families of gene regulators. Recent studies have suggested that these two kinds of molecules share similar regulatory logics and participate in cooperative activities in gene regulatory networks.8 There is increasing evidence that miRNAs contribute to the modulation of gene expression at the post-transcriptional level. miRNAs are small (18–22 nucleotides) non-coding, single stranded RNAs that are abundantly found in prokaryotic and eukaryotic cells. They can trigger translational repression or gene silencing by binding to complementary sequences on target mRNA transcripts, controlling in this way the regulation of their target genes at the post-transcriptional level.9,10 In the recent years, miRNAs have been reported to control many biological processes, such as plant development, differentiation, signal transduction or stress responses.11-13
MicroRNAs were first discovered in Caenorhabditis elegans14 and since then they have been also found in several other organisms, including plants.15 A database has been constructed through the Sanger Institute and annotated miRNA sequences are available (http://www.sanger.ac.uk/cgi-bin/Rfam/mirna/browse.pl and www.mir-base.org/index.shtml). Several miRNAs have been reported in Arabidopsis thaliana (199), Oryza sativa (447), Medicago truncatula (375), Zea mays (170), Sorghum bicolor (148), Vitis vinifera (137) and the numbers are still increasing. Although miRNA sequences are not conserved between animals and plants, they are highly conserved within each kingdom, and this evolutionary conservation is one of their defining characteristics.16,17
The present review will provide an up-date on microRNAs and their involvement in the stress responses in rice. A section will be specifically dedicated to the genetic engineering perspectives regarding the miRNAs applications in rice tolerance to stress conditions.
microRNA biogenesis in plants
Even though plant miRNAs are mostly similar to animal miRNAs, they show several substantial differences. For example, plant pre-miRNAs have larger and more variable stem-loop structures. The mature plant miRNAs pair their target sites with near-perfect complementarity. In animals, miRNAs usually recognize several target sequences in the 3′ UTR of mRNAs and cause translation inhibition, while plant miRNAs often recognize a single target site in the coding region and guide the mRNA to cleavage. The specificity of plant miRNA targeting coding sequences with fewer mismatches suggests that they may act more like small interfering RNAs (siRNAs).18 siRNAs (~21 nt) also play crucial roles in post-transcriptional gene silencing (i.e., RNA silencing) in plants.19 The function of siRNA and its biogenesis is thought to be highly conserved in all the eukaryotes including plants.
A schematic representation of miRNA biogenesis in showed in Figure 1. miRNAs are generated from primary miRNA transcripts (pri-miRNAs) that are generally transcribed by RNA polymerase II and they form characteristic hairpin structure by intramolecular pairing.20 In animals a pri-miRNA can contain several different pre-miRNAs, but in plants each transcript usually contains a single pre-miRNA. Two sequential RNase III enzyme-mediated cleavages are required to produce mature miRNAs. First, DICER-LIKE1 (DCL1) in plants, or Drosha, in animals, cleaves near the base of the pri-miRNA stem-loop to produce a miRNA precursor (pre-miRNA). Then DCL1 (in plants) or Dicer (in animals) cleaves at a second position near the pre-miRNA loop to generate the miRNA/miRNA* duplex.18 In plants, the two-step processing of pri-miRNAs into mature miRNAs occurs entirely in the nucleus. In addition to DCL1, genetic analysis revealed that HYL1, a dsRNA-binding protein, and SE, a C2H2-type zinc finger, are also required for processing pri-miRNAs and for accumulation of mature miRNAs. The importance of DCL1, HYL1, and SE in plant growth and development was evidenced, but only recently these genes were found to be required for miRNA accumulation in plants.21 After the procession in the nucleus, the miRNA/miRNA* duplex is transported to the cytoplasm by HASTY, an EXP-5 homolog, incorporated into the RISC complex (RNA-induced silencing complex) and guided to the target mRNA.22
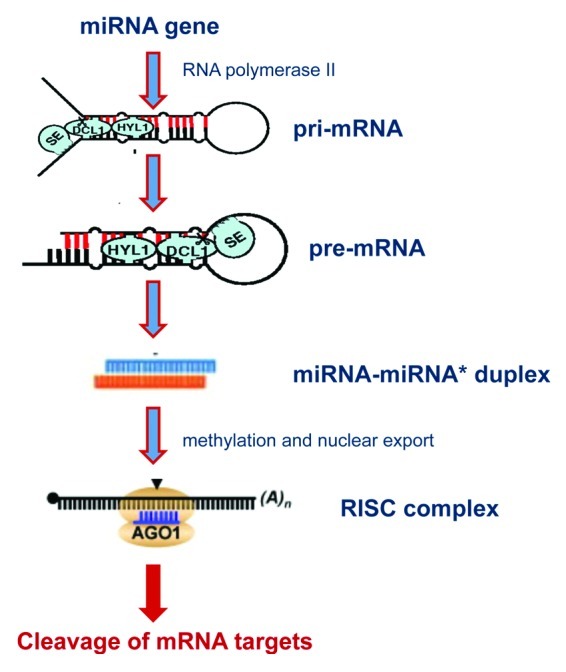
Figure 1. Schematic representation of miRNA biogenesis plants. The primary miRNA transcript is processed by Dicer-like 1 (DCL1) and is associated RNA-binding cofactors HYL1 and SE to generate a miRNA, which is then methylated, exported to the cytoplasm and incorporated into the Agonaute 1 (AGO1)-containing RNA-induced silencing complex (RISC) to silence mRNA targets important for development, diseases, and stress responses.
Experimental and computational analysis indicates that many plant miRNAs and their targets are conserved between monocot (rice) and dicot (Arabidopsis) plants, which suggests that miRNAs arose early in eukaryotic evolution, before the divergence of monocots and dicots. The interaction between miRNAs and their binding sites was studied in rice. The results of this study revealed that these interactions are part of a dynamic process since some conserved miRNAs could lose their putative target genes, while others acquired new target genes which are usually unrelated to those ancestral targets.23 Differently from animals, only the mature miRNAs, and not the pre-miRNAs, are conserved in plants.17 The high degree of complementarity between plant miRNAs and the target mRNAs allowed the target prediction by using specific algorithms that scan the genome for mRNA-miRNA complementarity. Since one miRNA can target multiple mRNAs, a great number of genes were predicted as candidate targets.24 However, the bioinformatic prediction must be supported by experimental validation. Some of the most used validation techniques are the in vitro cleavage assay, in which radiolabelled target RNAs are used, and the RNA-ligase mediated rapid amplification of cDNA ends, or shortly RLM-5′RACE technique, which involves the ligation of a RNA adaptor to the uncapped RNA and specific amplification of the 5′- cleavage products by 5′RACE.25 Usually, miRNA directed cleavage occurs in the center of miRNA complementarity between the tenth and eleventh nucleotides.26
Stress-responsive microRNAs in rice
The study of rice miRNAs started with the work of Wang et al.,27 who first reported on the identification of 20 miRNAs by using an improved cDNA cloning procedure derived from experimental RNomics. One year after, Sunkar et al.,8 published a detailed study on rice miRNAs. In this paper, the authors reported on the identification of new miRNAs that were difficult to predict by in silico analysis and verified the previously predicted miRNAs. Sequencing of small RNA libraries and subsequent analysis led to the identification of 14 new miRNAs belonging to14 families, 13 of which were not present in Arabidopsis. Based on the sequence complementarity, they were able to predict 46 rice genes as putative targets of the new miRNAs. The predicted targets included transcription factors and genes involved in various physiological processes.28
Even if the investigations of plant miRNAs are a little behind of those from animals, the versatile functions of plant miRNAs are becoming clearer with the passing years. miRNAs functions are thoroughly investigated by ectopic expression. The role of miRNAs in controlling developmental processes has been widely studied, while the emerging roles of miRNAs in plant stress responses are still less discussed.
A study by Zhang et al.,29 based on EST analyses in Arabidopsis, showed that 25.8% of the ESTs containing miRNAs were found in stress-induced plant tissues, suggesting that miRNAs play an important role in plant responses to environmental stress. Several recent studies have provided supporting evidence for this hypothesis in different plant species, but mostly concentrated on Arabidopsis thaliana as a model organism.11,24,30
In the present review, the miRNAs involvement in stress response is treated in relation to rice as a model cereal system. In Table 1 there are summarized some miRNAs that were proven to be implicated in various stress responses in rice plants. Zhao et al.31 reported on the miR169 family which regulates the CBF/DREBs (dehydration-responsive element) transcriptions factors and they suggested that miR169 g might play a role under drought stress in rice. The miR393, which targets an auxin transporter gene (OsAUX1) and a rice tiller inhibitor gene (OsTIR1), was also found to be induced by drought and salinity stress. Transgenic rice plants overexpressing miR393 showed an increase in tillers and early flowering, together with a decrease tolerance to salt and hypersensitiveness to auxin.32 Even if few data has been reported on auxin signaling in rice compared with Arabidopsis, some miRNAs have been showed to be involved in auxin signaling. miR160 and miR167, whose targets are auxin response transcription factors (ARFs), play an important role in early auxin response. By using the auxin-resistant rice mutant osaxr, Meng et al.33 demonstrated that the number of auxin-sensitive miRNAs was dramatically reduced in the mutant plant compared with WT, thus indicating the involvement of miRNAs in the auxin-resistant phenotype of this mutant. Further research also showed that miR167 was involved in adventitious root development in rice by regulating its downstream targets.33
Table 1. Rice microRNAs involved in stress response.
| MicroRNA | Stress conditions | Response | Validated/putative targets | References |
|---|---|---|---|---|
|
miR169 |
Drought |
Upregulated |
CBF/DREBs transcription factors (TFs) |
Zhao et al. 200731 |
|
miR393 |
Salt and drought |
Upregulated |
Auxin receptors TIR1,AFB2, AFB3 |
Xia et al. 201232 |
|
miR398 |
Oxidative stress |
Upregulated |
Copper SOD enzyme |
Li et al. 201034 |
|
miR169 |
Oxidative stress |
Upregulated |
HAP2 like transcription factor |
Li et al. 201034 |
|
miR397 |
Oxidative stress |
Upregulated |
Laccase |
Li et al. 201034 |
|
miR827 |
Oxidative stress |
Upregulated |
SPX domain protein |
Li et al. 201034 |
|
miR1425 |
Oxidative stress |
Upregulated |
Pentatricopeptide repeat (PPR) protein |
Li et al. 201034 |
|
miR528 |
Oxidative stress Cadmium |
Downregulated Upregulated |
F-box containing protein, Dicer-like1 |
Liu et al. 201034 Ding et al. 201135 |
|
miR167 |
Auxin signaling |
Upregulated |
Auxin response factor |
Meng et al. 201033 |
|
miR160 |
Auxin signaling |
Upregulated |
Auxin response factor |
Meng et al. 201033 |
|
miR162 |
Cadmium |
Downregulated |
Dicer-like1 |
Ding et al. 201135 |
|
miR168 |
Cadmium |
Downregulated |
ARGONAUTE |
Ding et al. 201135 |
|
miR166 |
Cadmium |
Downregulated |
HD-Zip TFs |
Ding et al. 201135 |
|
miR171 |
Cadmium |
Downregulated |
Scarecrow-like TFs |
Ding et al. 201135 |
|
miR396 |
Cadmium |
Downregulated |
Rhodenase-like protein, kinesin-like protein B |
Ding et al. 201135 |
|
miR390 |
Cadmium |
Downregulated |
Leucine-rich repeat receptor-like protein kinase |
Ding et al. 201135 |
|
miR156 |
Cadmium |
Downregulated |
Squamosa-promoter-binding protein TFs |
Ding et al. 201135 |
|
miR1432 |
Cadmium |
Downregulated |
EF-handproteins |
Ding et al. 201135 |
| miR444 | Cadmium | Downregulated | MADS-box TFs | Ding et al. 201135 |
In a recent study, Li et al.34 used high throughput sequencing techniques to identify miRNAs from rice seedlings grown under normal conditions and treated with hydrogen peroxide (H2O2) in order to induce oxidative stress. They identified seven miRNAs families that were differentially expressed under H2O2 stress. Out of these, miR169, miR397, miR1425, miR827, miR319a.2 and miR408–5p were found to be upregulated in response to oxidative stress, while miR528 was downregulated. The validated targets of the H2O2-responsive miRNAs were demonstrated to play important roles in transcriptional regulation, nutrient transport, auxin homeostasis, cell proliferation and programmed cell death.35
MicroRNAs playing specific roles in the response and adaptation to heavy metal stress in rice, have been recently identified by Ding et al.35 They used a microarray assay in order to investigate the miRNAs functions under cadmium (Cd) stress and they have identified 19 Cd-responsive miRNAs. Out of these, miR528 was upregulated under Cd-treatments, while miR162, miR168, miR166, miR171, miR396, miR390, miR156, miR1432 and miR444 were downregulated. The predicted target genes for these Cd-responsive miRNAs encode transcription factors and proteins associated with metabolic processes or stress responses (Table 1).
High-throughput expression profiling analysis through one-tube stem-loop reverse transcription quantitative PCR (ST-RT qPCR) under normal and stress conditions, was also reported in rice.35 With this method, 41 rice miRNAs were quantified for their relative expression levels after drought, salt, cold, or ABA treatments, and 32 miRNAs showed induced or suppressed expression. Some of the predicted target genes of these microRNAs were also related to abiotic stresses.36
Umate and Tuteja (2010)37 identified by in silico analysis 12 miRNAs that potentially target different helicase genes in rice. Out of these, miR414, miR164 and miR408 were shown to be downregulated under salinity stress. These miRNAs were experimentally validated as targeting three putative DEAD-box helicases whose expression was upregulated in response to salt treatments (Macovei and Tuteja, unpublished observation).
In consequence, many rice miRNAs play important roles in the tolerance to stress conditions. So, understanding the regulatory networks guided by miRNAs under stress can help elucidate the mechanisms of tolerance and adaptation to stress and provide new tools for improving crop resistance.
microRNAs: useful applications for rice crop
Traditional breeding has been most successful in improving stress tolerance in plants, but this process is time-consuming and the genetic resources available are limited.38 In the last decades, these possibilities have been enriched by genetic engineering and gene transfer technologies, as well as through the identification of genome sequences in model plants. However, detailed understanding of plant metabolic pathways and regulatory genes is still required.
miRNAs downregulate gene expression by mRNA cleavage or by repressing mRNA translation. As consequence, it is possible to use miRNAs for the suppression of target gene expression in order to study gene functions in a way similar to the use of antisense mRNA and RNA interference (RNAi). Another possibility is the use of miRNA to improve plant yields, quality, or resistance to various environmental stresses including insect and pathogen infection.39
A recent study reported on the application of a microRNA microarray system to examine the expression of annotated rice miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids in order to exploit the heterosis mechanism. The authors found that from the 1141 tested miRNAs, 157 were differentially expressed in hybrids, suggesting that small RNAs might play critical roles in heterosis.40
The development and maturation of rice seeds involves detailed and solid gene regulation at the transcriptional and post-transcriptional levels. Novel miRNAs, identified through integrated bioinformatics analysis, were showed to play important roles in the regulatory network of rice seed development. For example, it is known that mutation of AUXIN RESPONSE FACTOR 2 (ARF2) leads to an increase in seed size and weight. The miR167, which targets the ARF factors, was preferentially expressed in rice seeds and was induced by auxin.41 High-throughput sequencing technology was also used to investigate the roles of known miRNAs in rice seed development and to identify potentially seed-specific miRNAs .The presence of a large set of miRNAs and miRNA-like molecules in developing rice seeds suggests that many processes are under miRNA regulation during seed development.42
Gene silencing opened a gate for new opportunities in molecular breeding. The attempts to improve agronomic performance in crops have mostly focused on gene overexpression. Nonetheless, there are several examples proving that loss-of-function mutations and gene silencing through RNAi present great potential for agriculture. It has been recently shown that reduction or the loss of gene function often underlies varietal differences and important traits in rice and other grasses.39
MicroRNAs can be easily used for the induction of gene silencing since they work as target gene suppressors. The most common techniques for improving stress tolerance through the use of microRNA are: artificial miRNA (amiRNAs), siRNA-directed DNA silencing and the transient miRNA (tasiRNA). Among these, the amiRNAs present a new and highly specific technology for gene silencing. The fold back structure of pre-miRNA is the key to efficient processing of miRNA and this led to the idea that synthetic or artificial miRNAs may be designed to suppress expression of specific genes.43 Successful experiments of amiRNAs-mediated gene silencing have been conducted in Arabidopsis, tomato, tobacco, rice, mosses (Physcomitrella patens), and algae (C. reinhardtii).44 A new method for easy and rapid construction of rice artificial miRNA vector was described by Wang et al.45 The procedure involved the use of osa-amiR528 in a modified pCAMBIA1300-UR vector. The authors showed that the method was highly efficient, and greatly reduced the time needed for vector construction, making it suitable for use in a systems biology approach to functional genomic research. Custom-made amiRNAs introduced into Arabidopsis-based endogenous pre-miRNA backbones were able to target genes that are not naturally regulated by miRNA.46 The amiRNAs were highly expressed in a tissue-specific manner in Arabidopsis thaliana and could effectively downregulate the predicted targets.46,47 Recently Arabidopsis miR159 precursor was engineered to express amiRNA targeting viral mRNA sequences encoding two gene silencing suppressors, P69 of tobacco yellow mosaic virus (TYMV) and HCPro of turnip mosaic virus (TuMV), resulting in transgenic plants resistant to TYMV and TuMV.48 This technique was successfully applied also in rice crops. Based on an endogenous rice miRNA precursor, artificial miRNA constructs were designed to target three different endogenous rice genes. Specific suppression of the target genes was achieved in both Nipponbare (japonica) and IR64 (indica) genotypes.49 So, amiRNAs could be considered one of the most suitable strategy to generate transgenic plants and improve crops.50
Other ways for improving stress tolerance through miRNAs are the use of siRNA-directed silencing and transient RNA silencing (tasiRNA). Endogenous siRNAs are synthesized from long double stranded RNAs (dsRNAs). The endogenous sources of dsRNAs could be the miRNA-directed cleavage products of noncoding transcripts, the dsRNAs formed from the mRNAs encoded by natural cis-antisense gene pairs and the dsRNAs generated from heterochromatin and DNA repeats.51 The siRNAs produced by miRNA-directed cleavage of mRNAs are referred to as trans-acting, or transient siRNAs (tasiRNAs), while the siRNAs derived from dsRNAs formed from the mRNAs encoded by natural cis-antisense gene pairs are called natural antisense transcript-derived siRNAs (nat-siRNAs). Lu et al.52 uncovered a group of seven nat-miRNAs in rice (miR444, miR1433, miR1426, miR1425, miR160, miR166 and miR10), which originate from the natural antisense strand of target genes.53 Intron processing from the overlapping primary miRNA transcripts generates a hairpin structure that is further processed by DCL1, suggesting for an additional pathway for miRNA evolution, biogenesis, and function. Work on nat-siRNAs derived from a cis-NAT gene pair of SRO5 (Similar to RCD One 5-like) and P5CDH (Δ1-pyrroline-5-carboxylate dehydrogenase) genes, demonstrated an important role of nat-siRNAs in osmoprotection and oxidative stress management under salt stress in Arabidopsis. The SRO5 gene is similar to RADICAL INDUCED CELL DEATH 1 (RCD1) gene, which prevents ROS-induced cell death, considering that rcd1 mutant plants are hypersensitive to ROS-induced cell death.53
Another intriguing application of miRNA was recently evidenced by Burklew et al.54 in a study involving the use of nanoparticles in tobacco plants. The authors studied the response of different concentrations of aluminum oxide nanoparticles (0%, 0.1%, 0.5%, and 1% Al2O3) on microRNA expression in tobacco and noted that miR395, miR397, miR398, and miR399 showed an extreme increase in expression during exposure to 1% Al2O3 nanoparticles. They concluded that these miRNAs may play a key role in mediating plant stress responses to nanoparticle stress in the environment.54
Thus, miRNA-mediated approaches provide powerful tools for the study of gene functions and expand the applicability in producing stress tolerant plants.
Conclusions
Extensive efforts over the past decades have identified a great number of stress-regulated genes. With the recent identification of miRNAs, siRNAs and amiRNAs as components of stress response, another level of gene regulation has been uncovered. As a result, important roles are attributed for these small RNAs in stress response. The extent of microRNAs involvement in abiotic stress response in rice and other crops should become clearer in the next years if sufficient efforts will be directed to this field. In summary, the study of post-transcriptional gene regulation by microRNAs under abiotic stress is crucial for understanding and improving stress tolerance in rice as well as in other relevant crop plants.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21586
References
- 1.Tuteja N, Ahmad P, Panda BB, Tuteja R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutat Res. 2009;681:134–49. doi: 10.1016/j.mrrev.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–30. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Balestrazzi A, Confalonieri M, Macovei A, Donà M, Carbonera D. Genotoxic stress and DNA repair in plants: emerging functions and tools for improving crop productivity. Plant Cell Rep. 2011;30:287–95. doi: 10.1007/s00299-010-0975-9. [DOI] [PubMed] [Google Scholar]
- 4.Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–50. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- 5.Grover A, Minhas D. Towards the production of abiotic stress tolerant transgenic rice plants: Issues, progress and future research needs. Proc Indian Nat Sci Acad. 2000;1:13–32. [Google Scholar]
- 6.Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008;174:420–31. doi: 10.1016/j.plantsci.2008.02.005. [DOI] [Google Scholar]
- 7.Grasser KD. Emerging role for transcript elongation in plant development. Trends Plant Sci. 2005;10:484–90. doi: 10.1016/j.tplants.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Lin CC, Chen YJ, Chen CY, Oyang YJ, Juan HF, Huang HC. Crosstalk between transcription factors and microRNAs in human protein interaction network. BMC Syst Biol. 2012;6:18. doi: 10.1186/1752-0509-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 10.Yang T, Xue L, An L. Functional diversity of miRNA in plants. Plant Sci. 2007;172:423–32. doi: 10.1016/j.plantsci.2006.10.009. [DOI] [Google Scholar]
- 11.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–19. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin HJ, Zhang ZM, Shen YO, Gao SB, Pan GT. Review of plant miRNAs in environmental stressed conditions. Res J Agr Biol Sci. 2009;5:803–14. [Google Scholar]
- 13.Trindade I, Capitão C, Dalmay T, Fevereiro MP, Santos DM. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta. 2010;231:705–16. doi: 10.1007/s00425-009-1078-0. [DOI] [PubMed] [Google Scholar]
- 14.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 15.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–6. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 16.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng C, Wang W, Zheng Y, Chen X, Bo W, Song S, et al. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2010;38:981–95. doi: 10.1093/nar/gkp1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–89. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–42. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu JK. Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:9851–2. doi: 10.1073/pnas.0805207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra NS, Mukherjee SK. A peep into the plant miRNA world. Plant Sci. 2007;1:1–9. [Google Scholar]
- 23.Guo X, Gui Y, Wang Y, Zhu QH, Helliwell C, Fan L. Selection and mutation on microRNA target sequences during rice evolution. BMC Genomics. 2008;9:454–64. doi: 10.1186/1471-2164-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–99. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Zheng J, Li QQ. A novel plant in vitro assay system for pre-mRNA cleavage during 3′-end formation. Plant Physiol. 2011;157:1546–54. doi: 10.1104/pp.111.179465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–20. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH. Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 2004;32:1688–95. doi: 10.1093/nar/gkh332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–60. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 30.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–65. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 31.Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, et al. Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun. 2007;354:585–90. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, et al. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Y, Chen D, Ma X, Mao C, Cao J, Wu P, et al. Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant Signal Behav. 2010;5:252–4. doi: 10.4161/psb.5.3.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li T, Li H, Zhang YX, Liu JY. Identification and analysis of seven H2O2 –responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica) Nucleic Acids Res. 2010;27:1–13. doi: 10.1093/nar/gkq1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y, Chen Z, Zhu C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa) J Exp Bot. 2011;62:3563–73. doi: 10.1093/jxb/err046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen J, Xie K, Xiong L. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics. 2010;284:477–88. doi: 10.1007/s00438-010-0581-0. [DOI] [PubMed] [Google Scholar]
- 37.Umate P, Tuteja N. microRNA access to the target helicases from rice. Plant Signal Behav. 2010;5:1171–5. doi: 10.4161/psb.5.10.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoisington D, Khairallah M, Reeves T, Ribaut JM, Skovmand B, Taba S, et al. Plant genetic resources: what can they contribute toward increased crop productivity? Proc Natl Acad Sci U S A. 1999;96:5937–43. doi: 10.1073/pnas.96.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang G, Galili G, Zhuang X. RNAi and microRNA: breacktrough technologies for the improvement of plant nutritional value and metabolic engineering. Metabolomics. 2007;3:357–69. doi: 10.1007/s11306-007-0073-3. [DOI] [Google Scholar]
- 40.Chen F, He G, He H, Chen W, Zhu X, Liang M, et al. Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol. 2010;52:971–80. doi: 10.1111/j.1744-7909.2010.00985.x. [DOI] [PubMed] [Google Scholar]
- 41.Xue LJ, Zhang JJ, Xue HW. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009;37:916–30. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, et al. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–65. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Quintero AL, López C. Artificial microRNAs and their applications in plant molecular biology. Agronomia Columbiana. 2010;28:373–81. [Google Scholar]
- 45.Wang X, Yang Y, Yu C, Zhou J, Cheng Y, Yan C, et al. A highly efficient method for construction of rice artificial MicroRNA vectors. Mol Biotechnol. 2010;46:211–8. doi: 10.1007/s12033-010-9291-4. [DOI] [PubMed] [Google Scholar]
- 46.Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–51. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Zhang L, Sun J, Luo Y, Wang MB, Fan YL, et al. A simple artificial microRNA vector based on ath-miR169d precursor from Arabidopsis. Mol Biol Rep. 2010;37:903–9. doi: 10.1007/s11033-009-9713-1. [DOI] [PubMed] [Google Scholar]
- 48.Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol. 2006;24:1420–8. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 49.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–90. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Chen YQ. A new mechanism in plant engineering: the potential roles of microRNAs in molecular breeding for crop improvement. Biotechnol Adv. 2010;28:301–7. doi: 10.1016/j.biotechadv.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38(Suppl):S31–6. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 52.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc Natl Acad Sci U S A. 2008;105:4951–6. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahlfors R, Lång S, Overmyer K, Jaspers P, Brosché M, Tauriainen A, et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925–37. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burklew CE, Ashlock J, Winfrey WB, Zhang B. Effects of aluminum oxide nanoparticles on the growth, development, and microRNA expression of tobacco (Nicotiana tabacum) PLoS One. 2012;7:e34783. doi: 10.1371/journal.pone.0034783. [DOI] [PMC free article] [PubMed] [Google Scholar]


