Abstract
Whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleryrodidae), is a serious pest of black gram, (Vigna mungo (L.) Hepper), an important legume pulse crop grown in north India. This research investigated the potential role of selected plant oxidative enzymes in resistance/susceptibility to whitefly in nine black gram genotypes. Oxidative enzyme activity was estimated spectrophotometrically from leaf samples collected at 30 and 50 d after sowing (DAS) from whitefly infested and uninfested plants. The enzymes showed different activity levels at different times after the infestation. The results indicated that in general, whitefly infestation increased the activities of peroxidase and decreased the catalase activity. Resistant genotypes NDU 5-7 and KU 99-20 recorded higher peroxidase and catalase activities at 30 and 50 DAS under whitefly-stress conditions as compared with non-stressed plants. The results suggest that the enhanced activities of the enzymes may contribute to bioprotection of black gram plants against B. tabaci infestation. The potential mechanisms to explain the correlation of resistance to whitefly in black gram genotypes with higher activities of oxidative enzymes are also discussed.
Keywords: Bemisia tabaci, host plant resistance, black gram, peroxidase, catalase
Introduction
Plants face several challenges in terms of biotic stress due to insect pests. However, plants make use of a wide range of physio-chemical mechanisms to protect themselves against such biotic stresses induced by insect pests.1-3 The resistance strategies could be constitutive, that is, they are always present in the plant independent of herbivore attack,4 or inducible, activated only when the plant is attacked.5
Accumulation of defensive compounds through physiological, morphological, and chemical changes is the most vital line of defense in plants against insect attack.3,6,7 Various biochemical constituents present in the cells and tissues of the host plant exert a profound influence on biology of insect pests.8-10 One of the prominent plant responses to insect herbivore attack is the induction and accumulation of oxidative enzymes, such peroxidase and catalase.3,11-18 These enzymes, because of their potential roles in plant signaling, synthesis of defense compounds, and/or in oxidative stress tolerance, have been implicated in plant resistance to insect herbivores. The potential role of these enzymes in the synthesis of defense compounds and/or in oxidative stress tolerance makes them an important weapon of plant resistance against insect herbivores.
Black gram (Vigna mungo (L.) Hepper) is an important legume crop cultivated worldwide in tropical and subtropical regions of the world and is valued for easily digestible protein in its seeds. India is the largest producer and consumer of black gram in the world. During 2008–09, black gram occupied an area of 2.02 million ha in India; with an annual production of 0.84 million tonnes and an average yield of 419 kg ha-1 19. The crop is damaged by a number of insect pests during successive stages of the growth right from root nodules to flowers and pods, however whitefly B. tabaci appears as a major pest on black gram, besides mungbean and soybean.20 The avoidable losses due to B. tabaci and other insect pests in black gram have been reported to range from 17.42 to 71 per cent at different locations.21-23
Screening of germplasm for resistance to insect pests has received considerable attention; however, there is limited progress in characterization of biochemical mechanisms conferring resistance to insects.2,24 Currently, the information on biochemical mechanisms and induced resistance in black gram in response to whitefly is scarce. The objective of this investigation was to compare the enzymatic responses of resistant vs. susceptible genotypes of black gram to feeding by whitefly with a purpose to decipher mechanisms that will facilitate efforts to breed black gram varieties with durable whitefly resistance. The present study focused on the induction of the oxidative enzymes, i.e., peroxidase and catalase in nine genotypes of black gram challenged by whitefly feeding.
Results
Multiple-Choice Test
Whitefly population
ANOVA for numbers of eggs indicated a significant difference among genotypes in both the years (p = 0.05). Seasonal means of whitefly egg numbers were lowest on KU 99-20 and NDU 5-7, and highest on KU 7-504 and KU 7-505 (Table 3). The moderately resistant genotypes KU 99-20 and NDU 5-7 supported lower nymphal population as compared with other genotypes, suggesting that these two genotypes were poorly colonized by B. tabaci. On the other hand, the highly susceptible genotypes KU 7-504 and KU 7-505 recorded higher nymphal population during the study period. The remaining genotypes viz., IPU 02-043, KU 7-618, KU 7-605, KU 7-602 and Mash 1-1 recorded intermediate number of eggs and nymphal population. As far as the colonization by whitefly adults was concerned, KU 99-20 and NDU 5-7 harbored significantly less number of whitefly adults per trifoliate leaf than all other genotypes and were the least attractive to adults (Table 4). On the other hand, genotypes KU 7-504 and KU 7-505 recorded more number of whitefly adults than all other genotypes.
Table 3.B. tabaci eggs and red-eyed nymphs across different V. mungo genotypes in multiple-choice test under screen-house conditions.
| Genotype | *Mean ± SEM number of eggs per trifoliate leaf | * Mean ± SEM number of red-eyed nymphs per trifoliate leaf |
|---|---|---|
| KU 7-504 |
20.43 ± 1.79 |
52.67 ± 9.37 |
| KU 7-505 |
20.00 ± 2.08 |
52.18 ± 9.83 |
| KU 7-602 |
11.27 ± 0.61 |
16.31 ± 1.88 |
| KU 7-605 |
10.89 ± 0.88 |
15.13 ± 2.36 |
| KU 7-618 |
11.04 ± 0.81 |
15.88 ± 2.26 |
| KU 99-20 |
3.87 ± 0.49 |
10.70 ± 2.08 |
| IPU 02-043 |
11.24 ± 0.60 |
16.40 ± 1.78 |
| NDU 5-7 |
4.19 ± 0.49 |
11.31 ± 1.79 |
| Mash 1-1 | 13.27 ± 1.57 | 39.69 ± 7.67 |
Mean of three replications at weekly intervals recorded over a period of five weeks. SEM, Standard error of mean.
Table 4.B. tabaci adult population across different V. mungo genotypes in multiple-choice test under screen-house conditions.
| Genotype | * Mean ± SEM adult whitefly population per trifoliate leaf |
Mean ± SEM | ||
|---|---|---|---|---|
| Upper canopy | Middle canopy | Lower canopy | ||
| KU 7-504 |
68.26 ± 2.77 |
69.53 ± 2.69 |
68.99 ± 2.91 |
68.93 ± 0.29 |
| KU 7-505 |
68.63 ± 2.66 |
69.09 ± 2.81 |
68.63 ± 2.79 |
68.78 ± 0.12 |
| KU 7-602 |
37.66 ± 2.86 |
37.76 ± 2.96 |
37.93 ± 9.26 |
37.78 ± 0.06 |
| KU 7-605 |
37.99 ± 2.99 |
37.86 ± 2.89 |
37.73 ± 2.97 |
37.86 ± 0.06 |
| KU 7-618 |
37.69 ± 2.85 |
38.52 ± 3.18 |
38.53 ± 2.89 |
38.25 ± 0.22 |
| KU 99-20 |
13.83 ± 0.78 |
13.62 ± 0.64 |
13.39 ± 0.71 |
13.61 ± 0.10 |
| IPU 02-043 |
40.33 ± 2.81 |
37.99 ± 2.79 |
38.43 ± 3.00 |
38.91 ± 0.58 |
| NDU 5-7 |
13.23 ± 0.68 |
13.26 ± 0.75 |
13.92 ± 0.65 |
13.47 ± 0.18 |
| Mash 1-1 | 51.96 ± 2.28 | 51.53 ± 2.38 | 52.02 ± 2.46 | 52.11 ± 0.14 |
Mean of five replications (each replication represents population at weekly interval). SEM, Standard error of mean.
Peroxidase enzyme
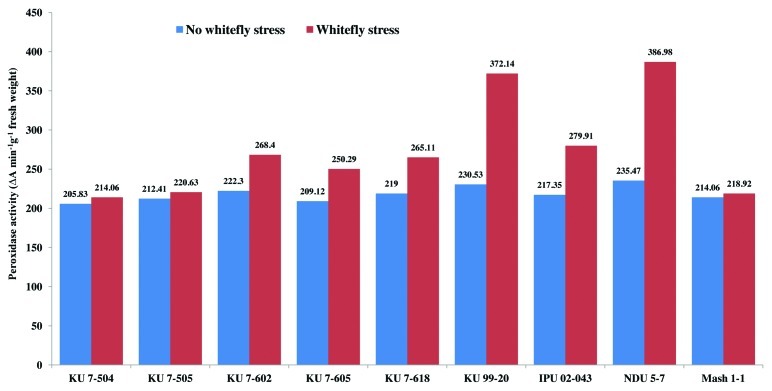
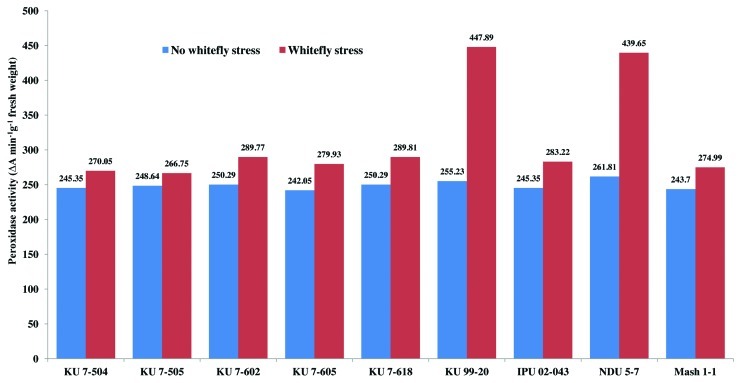
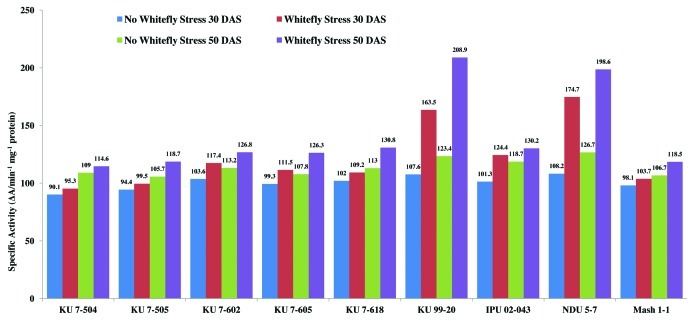
There was a non-significant variation in the peroxidase enzyme activity among all the non-stressed black gram genotypes at 30 and 50 d after sowing (DAS) during the study period (Figs. 1 and 2). However, peroxidase activity increased in all the genotypes under whitefly stress conditions. Resistant genotypes NDU 5-7 and KU 99-7 recorded higher peroxidase activity under whitefly stress conditions during both the years. The highly susceptible genotypes KU 7-504 and KU 7-505 recorded least increase in peroxidase activity upon whitefly feeding. The specific activity of peroxidase under stress conditions was also higher in genotype NDU 5-7 and lower in KU 7-505 (Fig. 3). The remaining genotypes viz., IPU 02-043, KU 7-618, KU 7-605, KU 7-602 and Mash 1-1 recorded intermediate peroxidase activity.
Figure 1. Peroxidase activity (30 DAS) in V. mungo leaves as influenced by B. tabaci feeding.
Figure 2. Peroxidase activity (50 DAS) in V. mungo leaves as influenced by B. tabaci feeding.
Figure 3. Specific activity of peroxidase in V. mungo leaves as influenced by B. tabaci feeding.
Catalase enzyme
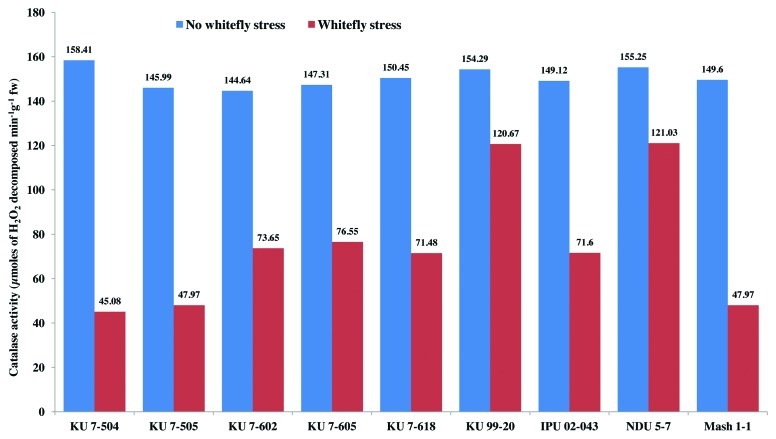
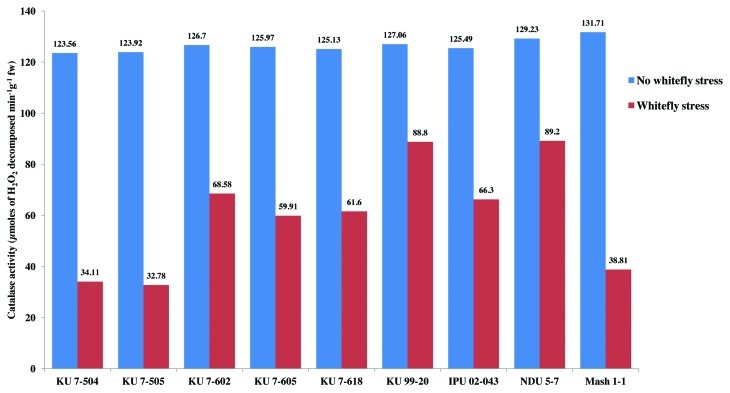
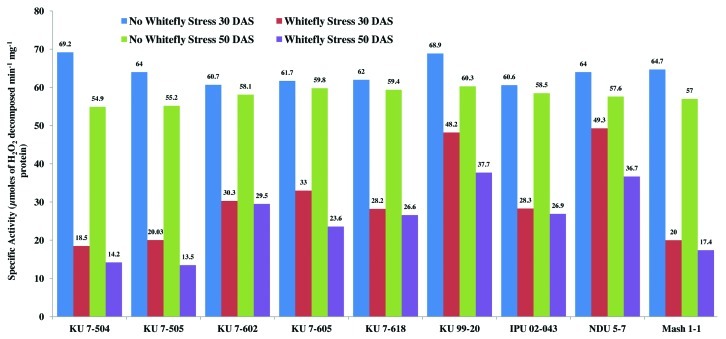
Among all the non-stressed black gram genotypes at 30 and 50 d after sowing, there was a non-significant variation in the catalase activity (Figs. 4 and 5). However, catalase activity declined in all the genotypes under whitefly stress conditions. In spite of a decline in catalase activity after whitefly feeding, the resistant genotypes NDU 5-7 and KU 99-7 recorded significantly high catalase activity during both the years. The maximum decline in catalase activity was observed in the highly susceptible genotypes KU 7-504 and KU 7-505 upon whitefly feeding. The specific activity of catalase under stress conditions was highest in resistant genotypes and lowest in susceptible ones (Fig. 6). The remaining genotypes viz., IPU 02-043, KU 7-618, KU 7-605, KU 7-602 and Mash 1-1 recorded intermediate catalase activity.
Figure 4. Catalase activity (30 DAS) in V. mungo leaves as influenced by B. tabaci feeding.
Figure 5. Catalase activity (50 DAS) in V. mungo leaves as influenced by B. tabaci feeding.
Figure 6. Specific activity of catalase in V. mungo leaves as influenced by B. tabaci feeding.
Correlation Studies
Peroxidase activity (at 30 DAS) also showed a significant negative correlation with whitefly nymphs (r = -0.70**) and adults (r = -0.87**) indicating that higher the peroxidase activity in the genotype, lesser would be the number of whitefly nymphs and adults (Table 5). The data recorded at 50 DAS also showed similar trend. There was a significant negative correlation between catalase activity (both at 30 and 50 DAS) with whitefly population (nymphs and adults). The correlation values of catalase (at 30 DAS) with whitefly nymphs and adults were r = -0.60** and -0.82**, respectively. Similar trend was observed at 50 DAS as well. This indicated that more the catalase activity less will be the number of nymphs and adult population. CAT activity had the maximum effect on reducing the adult whitefly population only at 50 DAS (R2 = 0.83 compared with 0.72 for peroxidase) while peroxidase activity has the maximum effect at 30 DAS (R2 = 0.77 compared with 0.68 for catalase).
Table 5. Correlation of biochemical leaf characteristics of V. mungo with B. tabaci population.
| Biochemical character | Red-eyed nymphs |
Adults |
|||
|---|---|---|---|---|---|
| r | R2 | r | R2 | ||
| Peroxidase |
30 DAS |
-0.70* |
0.49 |
-0.87* |
0.77 |
| 50 DAS |
-0.60* |
0.37 |
-0.85* |
0.72 |
|
| Catalase | 30 DAS |
-0.60* |
0.36 |
-0.82* |
0.68 |
| 50 DAS | -0.87* | 0.76 | -0.91* | 0.83 | |
Significant at 1 per cent. R, Correlation coefficient; R2, Coefficient of determination; DAS, Days after sowing.
Discussion
Induced resistance is an important component of plant defense that allows plants to face different stresses and is economical, environment friendly, and effective. Increased levels of defense-related proteins are a common phenomenon occurring in plants on account of biotic and abiotic stress.7 Peroxidase activity is known to increase with herbivore damage in many crop plants.25-28
Induction of resistance by feeding of whitefly nymphs resulting in the increase of defense-specific peroxidases has also been reported in tomato29 and cucumber seedlings.30 The enhanced activity of peroxidase enzyme with insect feeding has been recorded in different plant species.16,31,32 A number of reports have suggested that peroxidases play an important role in herbivore resistance in crop plants.33,34 It might be possible that the increased peroxidase activity observed in the present studies is triggered by the cellular damage caused by whitefly infestation. Increased phenol concentration might also increase the peroxidase reaction by acting as other substrate along with H2O2, leading to enhanced oxidation of phenolics.35 It is also possible that increase in the peroxidase activities interrupted the signals generated by the increase in the reactive oxygen species (ROS) and thus, might be involved in imparting resistance to black gram against whitefly. Hence, the abnormalities in the antioxidant defense may be related to the resistance of black gram genotypes to B. tabaci. In addition, the activity of peroxidase on some phenolics can produce phenoxy and other oxidative radicals that can directly deter feeding by insect herbivores and/or produce toxins that reduce plant digestibility.13
Present studies showed increased peroxidase activity in whitefly stressed plants than the uninfested ones. Enzyme activities were activated by whitefly infestation in all the black gram genotypes, however, the expression rhythm of activities varied among the genotypes. Peroxidase is an important defensive enzyme in plants against a number of biotic and abiotic stresses.36 Induction of such enzyme in response to insect herbivory is a common phenomenon.24,36,37 Observed patterns of different peroxidase activities in different genotypes could be due to the differences in levels of resistance against insect pests. Similarly, infestation by insect pests significantly increased peroxidase activities in the leaves of three chrysanthemum varieties, with activity levels being higher in more resistant genotypes.38 Induction in peroxidase activity has been implicated as an immediate response of plants in response to biotic stresses including insect attack.39 A 9-fold increase in peroxidase activity was observed in Russian wheat aphid infestation- resistant wheat cultivar, but only a 3-fold increase in the susceptible cultivar after infestation.31 Increase in peroxidase activity on account of insect infestation in plants detoxifies the peroxides, thus reducing plant tissue damage.36 Moreover, the role of peroxidase in cell wall toughening and toxic secondary metabolite production and its simultaneous oxidant and antioxidant properties enables it to play an important role in integrated defense response of plants to a variety of stresses.28,37,40
The results of present studies are in conformity with previous studies where insect infestation has been reported to induce peroxidases.3,24,30,36-38,41,42 The ability of the resistant genotypes to increase peroxidase activities suggest that genotypes with a higher level of resistance would either have a higher upregulation capacity for defensive enzymes or have a more sensitive upregulation response or both.36
To combat with the biotic and abiotic stresses, plants produce a number of defense-related enzymes and other protein-based defensive compounds.42 Higher induction of secondary metabolites and other defensive compounds in the insect resistant genotypes on account of insect damage is a common phenomenon.42-44 Moreover, higher activity of peroxidase has been linked with reduced insect growth and development in many plants.16,32,45
There was a significant decrease in the catalase activity upon infestation of whitefly irrespective of the resistance or susceptibility status of the black gram genotypes. However, decline in the catalase activity was more pronounced in the susceptible and whitefly-stressed genotypes as compared with the resistant and non-stressed genotypes. Under whitefly stress conditions, the resistant genotypes still exhibited higher catalase activity than the susceptible ones. Such contrasting differences between the whitefly-resistant and susceptible black gram genotypes may be the result of genetic differences in their metabolic pathways to scavange oxidative radicals.
Whitefly stress in susceptible black gram genotypes results in dramatic changes in plant biochemistry. Leaves of black gram having resistance to whitefly contain higher activities of catalase than do susceptible leaves. This enzyme may function in concert with other antioxidant enzymes to quench whitefly-induced free radical damage and thus impart resistance. Similar trend in the catalase activity was also observed in herbaceous weed, Mikania micrantha due to B. tabaci feeding.18 The decline in the catalase activity due to insect feeding has also been documented in other crops.24,46 The role of oxidative enzymes (peroxidase and catalase) as anti-nutritive and/ or toxicological defenses against insect herbivores has been documented by several authors.13,47-49
Therefore, the elevation of peroxidase might be associated with whitefly resistance in black gram. Resistant black gram genotypes exhibiting high activities of peroxidase and catalase can be used in wide hybridization to increase the levels and diversify the bases of resistance to B. tabaci. These results show the differential defensive responses of the nine black gram genotypes against B. tabaci and offer a perspective on plant resistance in insect plant interaction. Such information may also be used to develop enzyme markers for identifying whitefly resistance and/or susceptibility in black gram germplasm. This research will also help to develop a fundamental understanding of the biological pathways that contribute to plant resistance to insects, allow comparison of gene expression between resistant and susceptible genotypes, and serve to identify genes contributing to the resistance. The identification of genes unique to the resistant black gram genotypes provides a baseline against which to screen other black gram germplasm for the presence of these genes, and thus may provide useful markers for tolerance.
Materials and Methods
Black gram genotypes
Nine black gram genotypes were used in the multiple-choice test conducted inside the screen-house. These test genotypes had shown different levels of resistance to B. tabaci based on the whitefly resistance index (WRI) (Taggar and Gill, unpublished data). Based upon the kind and intensity of leaf injury symptoms due to the feeding of B. tabaci in black gram, the leaf-injury grades (I-V) were categorized as follows (Table 1):
Table 1.
| Leaf injury grade | Symptoms |
|---|---|
| I |
No damage |
| II |
Appearance of yellow chlorotic spots |
| III |
Starting of black sooty mold |
| IV |
Severe blackening of leaves |
| V | Complete drying of leaves |
Test genotypes were examined for the appearance of leaf injury grades at weekly interval up to six weeks after the release of whitefly and grouped into different categories of resistance/susceptibility after working out WRI as under:
Whitefly Resistance Index (WRI) =
 |
where,
G = Number of the leaf-injury grade (in numerics)
p = Number of plants falling under that grade
Each grade was given a range of score for grouping different genotypes into resistant/susceptible categories as follows (Table 2):
Table 2.
| Leaf injury grade | Symptoms | Score | Category/Reaction |
|---|---|---|---|
| I |
No damage |
< 1.00 |
Resistant (R) |
| II |
Appearance of yellow chlorotic spots |
1.01-1.50 |
Moderately Resistant (MR) |
| III |
Starting of black sooty mold |
1.51-2.50 |
Moderately Susceptible (MS) |
| IV |
Severe blackening of leaves |
2.51-3.50 |
Susceptible (S) |
| V | Complete drying of leaves | > 3.50 | Highly Susceptible (HS) |
Based on WRI, genotypes KU 99-20 and NDU 5-7 were moderately resistant, whereas genotypes KU 7-504 and KU 7-505 were highly susceptible to B. tabaci under screen-house conditions. Other black gram genotypes evaluated were susceptible to B. tabaci and included IPU 02-043, KU 7-602, KU 7-605, KU 7-618 and Mash 1-1 (Taggar and Gill, unpublished data).
Whitefly colonies
Field collected B. tabaci adults were released on disease-free black gram variety Mash 1-1 grown in earthen pots under unsprayed, natural conditions simulated under screen-house. Non-viruliferous whiteflies were transferred to the insect-transfer cabin for sexing. For raising the insect culture, paired individuals were aspirated from the glass pan and released on brinjal plants (Solanum melongela L.) grown in pots and covered with split-cages. Whiteflies were collected from these colonies and re-introduced on black gram variety Mash 1-1 for conditioning. The whiteflies were allowed to develop and multiply on these plants till newly emerged adults appeared on the plants. Old plants were replaced with new ones, whenever needed. The whiteflies from this culture were further used to conduct the experiments.
Multiple-Choice Test
Test genotypes, each replicated thrice, were sown in plastic pots (10 cm in diameter) with single plant per pot and arranged in completely randomized design (CRD) inside the screen-house during July 2009 and July 2010. Five pots (one plant/pot) of each genotype represented one replication. Whitefly adults were aspirated and released @ 100 adults per plant on the test genotypes at 3rd trifoliate leaf stage (21 d old) randomly at different places inside the screen-house for their uniform distribution. For studying the biochemical basis of resistance, leaf samples representing fully-formed trifoliate leaf from the upper canopy of 30 and 50 d old plants were used in each replication. Test genotypes kept free from whitefly infestation (sown in a separate screen house) served as control.
Pest population counts
Whitefly population counts were made from three randomly tagged plants from each genotype per replication. The total number of eggs laid per trifoliate leaf was recorded from an area of 1 cm2 of each leaflet from the lower surface of the top fully-formed trifoliate leaf at weekly interval using binocular stereomicroscope (40x) for a period of five weeks. The fourth instar red-eyed nymphs (RENs) were recorded from an area of 2 cm2 of each leaflet from the lower surface of the trifoliate leaf in the middle canopy using magnifying lens at weekly interval for a period of five weeks. Adult whiteflies were counted from the lower surface of three fully-formed trifoliate leaves, one each from the upper, middle and lower canopy using an ordinary mirror at weekly interval for a period of five weeks.
Biochemical characteristics
The leaf biochemical investigations were performed from leaves of nine test genotypes of black gram from both whitefly-stressed (initial stress of 100 whitefly adults per plant) and non-stressed (no whitefly infestation) plants.
Extraction and assay of antioxidative enzymes
Fresh leaves (whitefly-stressed as well as non-stressed) were plucked and immediately transferred to ice. The leaf samples were, then, brought to the laboratory for enzymatic analysis. Both peroxidase and catalase were extracted with relevant extraction buffers at 4°C and assayed at 30°C.
Extraction of peroxidase
Fresh leaf (100 mg) of each genotype was extracted with 2 ml of ice-cold 0.1M potassium phosphate buffer (pH 7.5) containing 1.0 mM EDTA, 1% polyvinylpyrrolidone (PVP), 10 mM β-mercaptoethanol using pre-chilled pestle and mortar. There were three replications for each genotype. Homogenate was centrifuged at 10,000 g at 4°C for 20 min and clear supernatant was used for enzyme assay.
Peroxidase Assay
Peroxidase was assayed as per the method of Shannon et al.50 with slight modification. The assay system contained 3 ml of 0.05 M guaiacol in 0.1 M phosphate buffer (pH 6.5), 0.02 ml of enzyme extract and 0.1 ml of 0.8 M H2O2. The reaction mixture without H2O2 was measured as a blank. The reaction was initiated by adding H2O2 and rate of change in absorbance was recorded at 470 nm using spectrophotometer for 3 min at an interval of 30 sec. Peroxidase activity has been defined as change in absorbance min−1 g−1 fresh weight. Specific activity of the enzyme was calculated and expressed as change in absorbance min−1 mg−1 protein.
Extraction of catalase
Fresh leaf (100 mg) of each genotype was extracted with 2 ml of ice-cold 50 mM potassium phosphate buffer (pH 7.5) containing 1% polyvinyl pyrrolidone using pre-chilled pestle and mortar. There were three replications for each genotype. Homogenate was centrifuged at 10,000 g at 4°C for 20 min and clear supernatant was used for enzyme assay.
Catalase Assay
Catalase activity was estimated as per the method of Chance and Maehly.51 In spectrophotometric cuvette, 1 ml of 0.12 per cent H2O2 solution and 1.8 ml of 50 mM potassium phosphate buffer (pH 7.5) was added. To this assay mixture, 0.2 ml of enzyme extract was added and utilization of H2O2 for 3 min was recorded at intervals of 30 sec by measuring the decrease in absorbance at 240 nm using UV spectrophotometer. Catalase activity was expressed as μmoles of H2O2 decomposed min−1 g−1 fresh weight. Specific activity of the enzyme was calculated and expressed as µmoles of H2O2 decomposed min−1 mg−1 protein. Proteins were estimated by the method of Lowry et al.52 using Folin-phenol reagent.
Statistical Analysis
The replication data were pooled together and means were calculated. The data were subjected to analysis of variance (ANOVA) to compare the differences between treatments within a genotype and among the genotypes using CPCS1. The significance level was set at p = 0.05 and the treatment means and whitefly population on different genotypes were compared using the completely randomized design. Mean and standard error of mean were also calculated. Data on biochemical characteristics were subjected to analysis of variance and were compared across genotypes and correlation analysis along with coefficient of determination were used to identify a possible relationship between whitefly nymph or adult density and biochemical leaf characteristics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21435
References
- 1.Rasmann S, Agrawal AA. Plant defense against herbivory: progress in identifying synergism, redundancy, and antagonism between resistance traits. Curr Opin Plant Biol. 2009;12:473–8. doi: 10.1016/j.pbi.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Sharma HC, Sujana G, Rao DM. Morphological and chemical components of resistance to pod borer, Helicoverpa armigera in wild relatives of pigeonpea. Arthropod-Plant Interact. 2009;3:151–61. doi: 10.1007/s11829-009-9068-5. [DOI] [Google Scholar]
- 3.Usha Rani P, Jyothsna Y. Biochemical and enzymatic changes in rice as a mechanism of defense. Acta Physiol Plant. 2010;32:695–701. doi: 10.1007/s11738-009-0449-2. [DOI] [Google Scholar]
- 4.Franceschi VR, Krokene P, Christiansen E, Krekling T. Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol. 2005;167:353–75. doi: 10.1111/j.1469-8137.2005.01436.x. [DOI] [PubMed] [Google Scholar]
- 5.Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal AA, Fishbein M, Jetter R, Salminen JP, Goldstein JB, Freitag AE, et al. Phylogenetic ecology of leaf surface traits in the milkweeds (Asclepias spp.): chemistry, ecophysiology, and insect behavior. New Phytol. 2009;183:848–67. doi: 10.1111/j.1469-8137.2009.02897.x. [DOI] [PubMed] [Google Scholar]
- 7.Broz AK, Broeckling CD, De-la-Peña C, Lewis MR, Greene E, Callaway RM, et al. Plant neighbor identity influences plant biochemistry and physiology related to defense. BMC Plant Biol. 2010;10:115. doi: 10.1186/1471-2229-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck SD. Resistance of plants to insects. Annu Rev Entomol. 1965;10:207–32. doi: 10.1146/annurev.en.10.010165.001231. [DOI] [Google Scholar]
- 9.Smith CM. Plant Resistance to Insects: A Fundamental Approach. Wiley, New York, USA, 1989. [Google Scholar]
- 10.Sharma H. Applications of Biotechnology in Pest Management and Ecological Sustainability. CRC Press Taylor and Francis, Boca Raton, USA, 2009: 526. [Google Scholar]
- 11.Green TR, Ryan CA. Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science. 1972;175:776–7. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand DF, Rodriguez JG, Brown GC, Luu KT, Volden CS. Peroxidase responses of leaves in two soybean genotypes injured by two-spotted spider mites (Acari: Tetranychidae). J econ Ent 1986; 79:1459-65. http://digitalcommons.unl.edu
- 13.Felton GW, Donato K, Del Vecchio RJ, Duffey SS. Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. J Chem Ecol. 1989;15:2667–94. doi: 10.1007/BF01014725. [DOI] [PubMed] [Google Scholar]
- 14.Felton GW, Bi J, Summers CB, Mueller AJ, Duffey SS. Potential role of lipoxygenases in defense against insect herbivory. J Chem Ecol. 1994;20:651–66. doi: 10.1007/BF02059605. [DOI] [PubMed] [Google Scholar]
- 15.Felton GW, Summers CB, Mueller AJ. Oxidative responses in soybean foliage to herbivory by bean leaf beetle and three-corned alfalfa leafhopper. J Chem Ecol. 1994;20:639–50. doi: 10.1007/BF02059604. [DOI] [PubMed] [Google Scholar]
- 16.Stout MJ, Fidantsef AL, Duffey SS, Bostock RM. Signal interactions in pathogen and insect attack: systemic plant mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol. 1999;54:97–114. doi: 10.1006/pmpp.1998.0193. [DOI] [Google Scholar]
- 17.Constabel CP, Yip L, Patton JJ, Christopher ME, Christopher ME. Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiol. 2000;124:285–95. doi: 10.1104/pp.124.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang LL, Wen DZ. Photosynthesis, chlorophyll fluorescence, and antioxidant enzyme responses of invasive weed Mikania micrantha to Bemisia tabaci infestation. Photosynthetica. 2008;46:457–62. doi: 10.1007/s11099-008-0078-9. [DOI] [Google Scholar]
- 19.Anonymous. All India Coordinated Research Project on MULLaRP (ICAR): Project Coordinator’s Report. Indian Institute of Pulses Research, Kanpur; 2011. [Google Scholar]
- 20.Chhabra KS, Kooner BS. Insect pest management in mungbean and black gram: Status and strategies. In: Upadhyay RK, Mukherji KG, Rajak RL, ed. IPM System in Agriculture, Vol. 4, Pulses, Aditya Books Pvt. Ltd.,New Delhi, 1998: 233-310. [Google Scholar]
- 21.Saxena HP. Losses in black gram due to insect-pests. In: Crop losses due to insect-pests. Indian J Ent (Spl. Issue) 1983; 294-97. [Google Scholar]
- 22.Chhabra KS. Advances in management of insect pests of Vigna species. In: Sachan JN, ed. Proc Nat Symp New Frontiers in Pulses Res and Dev. Directorate of Pulses Research, Kanpur, 1992: 178-86. [Google Scholar]
- 23.Mansoor-ul-Hassan Akbar R, Latif A. Varietal response of mung and mash beans to insect attack. Pak Ent. 1998;20:43–6. [Google Scholar]
- 24.Heng-Moss TM, Sarath G, Baxendale F, Novak D, Bose S, Ni X, et al. Characterization of oxidative enzyme changes in buffalograsses challenged by Blissus occiduus. J Econ Entomol. 2004;97:1086–95. doi: 10.1603/0022-0493(2004)097[1086:COOECI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Bi JL, Murphy JB, Felton GW. Antinutritive and oxidative components as mechanisms of induced resistance in cotton to Helicoverpa zea. J Chem Ecol. 1997;23:97–117. doi: 10.1023/B:JOEC.0000006348.62578.fd. [DOI] [Google Scholar]
- 26.Cipollini DF., Jr. Wind-induced mechanical stimulation increases pest resistance in common bean. Oecologia. 1997;11:84–90. doi: 10.1007/s004420050211. [DOI] [PubMed] [Google Scholar]
- 27.Tscharntke T, Thiessen S, Dolch R, Boland W. Herbivory, induced resistance and interplant signal transfer in Alnus glutinosa. Biochem Syst Ecol. 2001;29:1025–47. doi: 10.1016/S0305-1978(01)00048-5. [DOI] [Google Scholar]
- 28.Allison SD, Schultz JC. Differential activity of peroxidase isozymes in response to wounding, gypsy moth, and plant hormones in northern red oak (Quercus rubra L.) J Chem Ecol. 2004;30:1363–79. doi: 10.1023/B:JOEC.0000037745.66972.3e. [DOI] [PubMed] [Google Scholar]
- 29.Murugan M, Dhandapani N. Induced systemic resistance activates defense responses to interspecific insect infestations on tomato. J Veg Sci. 2007;12:43–62. doi: 10.1300/J484v12n03_05. [DOI] [Google Scholar]
- 30.Zhang SZ, Hau BZ, Zhang F. Induction of the activities of antioxidative enzymes and the levels of malondialdehyde in cucumber seedlings as a consequence of Bemisia tabaci (Hemiptera: Aleyrodidae) infestation. Arthropod-Plant Interact. 2008;2:209–13. doi: 10.1007/s11829-008-9044-5. [DOI] [Google Scholar]
- 31.Ni X, Quisenberry SS, Heng-Moss T, Markwell J, Sarath G, Klucas R, et al. Oxidative responses of resistant and susceptible cereal leaves to symptomatic and nonsymptomatic cereal aphid (Hemiptera: Aphididae) feeding. J Econ Entomol. 2001;94:743–51. doi: 10.1603/0022-0493-94.3.743. [DOI] [PubMed] [Google Scholar]
- 32.Chaman ME, Corcuera LJ, Zuniga GE, Cardemil L, Argandona VH. Induction of soluble and cell wall bound peroxidases by aphid infestation in barley. J Agric Food Sci. 2001;49:2249–53. doi: 10.1021/jf0011440. [DOI] [PubMed] [Google Scholar]
- 33.Chittoor JM, Leach JE, White FF. Induction of peroxidases during defense against pathogen. In: Datta SK, Muthukrishnan S, ed. Pathogenesis Related Proteins in Plants. CRC, Boca Raton, FL, 1999: 171-193. [Google Scholar]
- 34.Constabel CP. A survey of herbivore-inducible defensive proteins and phytochemicals. In: Agrawal AA, Tuzun S, Bent E, ed. Induced Plant Defenses against Pathogens and Herbivores: Chemistry, Ecology and Agriculture. The American Phytopathological Society, St. Paul, MN, 1999: 137-166. [Google Scholar]
- 35.Hammerschmidt R, Kuc J. Lignification as a mechanism for induced systemic resistance in cucumber. Physiol Plant Pathol. 1982;20:61–71. doi: 10.1016/0048-4059(82)90024-8. [DOI] [Google Scholar]
- 36.Gulsen O, Eickhoff T, Heng-Moss T, Shearman R, Baxendale F, Sarath G, et al. Characterization of peroxidase changes in resistant and susceptible warm-season turf grasses challenged by Blissus occiduus. Arthropod-Plant Interact. 2010;4:45–55. doi: 10.1007/s11829-010-9086-3. [DOI] [Google Scholar]
- 37.Han Y, Wang Y, Bi JL, Yang XQ, Huang Y, Zhao X, et al. Constitutive and induced resistance in aphid-resistant and aphid-susceptible cultivars of wheat. J Chem Ecol. 2009;35:176–82. doi: 10.1007/s10886-009-9589-5. [DOI] [PubMed] [Google Scholar]
- 38.He J, Chen F, Chen S, Lv G, Deng Y, Fang W, et al. Chrysanthemum leaf epidermal surface morphology and antioxidant and defense enzyme activity in response to aphid infestation. J Plant Physiol. 2011;168:687–93. doi: 10.1016/j.jplph.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Moloi MJ, van der Westhuizen AJ. The reactive oxygen species are involved in resistance responses of wheat to the Russian wheat aphid. J Plant Physiol. 2006;163:1118–25. doi: 10.1016/j.jplph.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Moore JP, Paul ND, Whittakar JB, Taylor JE. Exogenous jasmonic acid mimics herbivore-induced systemic increase in cell wall bound peroxidase activity and reduction in leaf expansion. Funct Ecol. 2003;17:549–54. doi: 10.1046/j.1365-2435.2003.00767.x. [DOI] [Google Scholar]
- 41.Huang W, Zhikuan J, Qingfang H. Effects of herbivore stress by Aphis medicaginis Koch on the malondialdehyde contents and activities of protective enzymes in different alfalfa varieties. Acta Ecol Sin. 2007;27:2177–83. doi: 10.1016/S1872-2032(07)60048-1. [DOI] [Google Scholar]
- 42.Chen Y, Ni X, Buntin GD. Physiological, nutritional, and biochemical bases of corn resistance to foliage-feeding fall armyworm. J Chem Ecol. 2009;35:297–306. doi: 10.1007/s10886-009-9600-1. [DOI] [PubMed] [Google Scholar]
- 43.Sharma HC, Pampapathy G, Dwivedi SL, Reddy LJ. Mechanisms and diversity of resistance to insect pests in wild relatives of groundnut. J Econ Entomol. 2003;96:1886–97. doi: 10.1603/0022-0493-96.6.1886. [DOI] [PubMed] [Google Scholar]
- 44.Bhonwong A, Stout MJ, Attajarusit J, Tantasawat P. Defensive role of tomato polyphenol oxidases against cotton bollworm (Helicoverpa armigera) and beet armyworm (Spodoptera exigua) J Chem Ecol. 2009;35:28–38. doi: 10.1007/s10886-008-9571-7. [DOI] [PubMed] [Google Scholar]
- 45.Sethi A, McAuslane HJ, Rathinasabapathi B, Nuessly GS, Nagata RT. Enzyme induction as a possible mechanism for latex-mediated insect resistance in romaine lettuce. J Chem Ecol. 2009;35:190–200. doi: 10.1007/s10886-009-9596-6. [DOI] [PubMed] [Google Scholar]
- 46.Khattab H. The defense mechanism of cabbage plant against phloem-sucking aphid (Brevicoryne brassicae L.). Aust. J Basic Appl Sci. 2007;1:56–62. [Google Scholar]
- 47.Duffey SS, Felton GW. Enzymatic and nutritive defenses of the tomato plants against insects. In: Hedin PA, ed. Naturally occurring pest bioregulators. American Chemical Society, Washington, DC, 1991: 167-197. [Google Scholar]
- 48.Duffey SS, Stout MJ. Antinutritive and toxic compounds of plant defense against insects. Arch Insect Biochem Physiol. 1996;32:3–37. doi: 10.1002/(SICI)1520-6327(1996)32:1<3::AID-ARCH2>3.0.CO;2-1. [DOI] [Google Scholar]
- 49.Gill RS, Gupta AK, Taggar GK, Taggar MS. Role of oxidative enzymes in plant defenses against insect herbivory. Acta Phytopathol Entomol Hung. 2010;45:277–90. doi: 10.1556/APhyt.45.2010.2.4. [DOI] [Google Scholar]
- 50.Shannon LM, Kay E, Lew JY. Peroxidase isozymes from horseradish roots. I. Isolation and physical properties. J Biol Chem. 1966;241:2166–72. [PubMed] [Google Scholar]
- 51.Chance B, Maehly AC. Assay of catalases and peroxidases. In: Colowick SP, Kaplan NO, ed. Methods in Enzymology, Vol. 2. Academic Press, New York, 1955: 764-75. [Google Scholar]
- 52.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]