Abstract
Background: CRC caused more than 600,000 estimated deaths in 2008. Dysregulated signaling through the RAS/RAF/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway due to mutations in K-Ras and B-Raf are common events in CRC. Methods: Incidence of mutations in codons 12 and 13 of K-Ras and exons 11 and 15 of B-Raf were analyzed in amplified PCR products from primary tumors of 273 patients with CRC, and their prognostic and predictive significance was assessed. The prognostic role of clinical and pathological factors was also examined. Results: K-Ras mutations were present in 89 patients (32.6%), of whom 76 (85.4%) had mutations in codon 12 and 10 (11.2%) had mutations in codon 13. B-Raf gene mutations were present in 17 patients (6.9%), of whom 6 (35.3%) had mutations in exon 15. Multivariate analysis revealed a predictive significance for K-Ras mutations with respect to time to progression in patients treated with irinotecan and oxaliplatin as first-line chemotherapy. There was no predictive significance for B-Raf gene mutation status in these patients. The following risk factors were found to affect overall survival (OS) rates: primary tumor location, lymph node involvement grade, carcinoembryonic antigen (CEA) level before treatment, and performance status according to WHO criteria. Conclusions: Based on the results of this study, K-Ras mutation status may be a suitable indicator of patient eligibility and a prognostic indicator for responsiveness to anti-EGFR therapy alone, or in combination with chemotherapy. Also, K-Ras mutation status may predict time to progression in patients treated with irinotecan and oxaliplatin.
Keywords: B-Raf mutation, K-Ras mutation, RAS/RAF/mitogen-activated protein kinase, anti-EGFR therapy, colorectal cancer, irinotecan, oxaliplatin
Introduction
Colorectal cancer (CRC) is the third most common cancer in men (663,000 cases) and second in women (571,000) in the world, with more than one million newly diagnosed cases reported annually. Approximately 608,000 CRC deaths are estimated worldwide each year, accounting for 8% of all cancer deaths and making it the fourth most common cause of death from cancer.1
Ras proteins are proto-oncogenes that function as molecular switches. In response to various hormones, cytokines, mitogens, and differentiation and growth factors such as epidermal growth factor (EGF) acting via the EGF receptor (EGFR), GTP-bound RAS regulates a number of critical cellular processes, including gene expression, mitosis, embryogenesis, cell differentiation, movement, metabolism, and programmed death.2 RAS maintains these cellular phenotypes by regulating the activation of multiple downstream effector pathways, including the RAF/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway.3-6
Dysregulated signaling through this pathway due to mutations and genetic alterations in pathway components and/or upstream activators can lead to constitutive activation independent of EGFR signaling and uncontrolled cell proliferation. Indeed, constitutive activation of this pathway is found in many human cancers. Approximately 15–30% of all cancers have mutations in RAS family genes,7 with mutations in the K-Ras gene accounting for nearly 80% of these8 and 40% of all CRC.9,10 K-RAS codons 12 and 13 are the most common sites of oncogenic activation, with over 90% of mutations.11 Amino acid alterations at these codons, which are adjacent to the GDP/GTP binding pocket, reduce or abolish GTPase activity of K-RAS and lock the protein in an active, GTP-bound state. As a result, this “dominant active” mutant KRAS and its downstream effectors become independent of epidermal growth factor (EGFR), among others.
Somatic mutations in BRAF are associated with malignant melanomas,12 CRC,13 ovarian cancer,14 and papillary thyroid carcinomas.15 Over 30 single-site missense mutations in the B-Raf gene have been identified in human cancers, mostly within the kinase domain.16 These mutations likely insert a negatively charged residue adjacent to sites of regulatory phosphorylation, mimicking it in the activation segments of BRAF. A Glu for Val substitution at residue 599 in the activation segment accounts for over 90% of BRAF mutations in human cancers. This V599E BRAF mutant shows highly elevated kinase activity and stimulates ERK activity constitutively independent of RAS activation.16,17
The introduction of molecular biological techniques has facilitated the identification of hitherto unknown factors that influence both prognosis (prognostic markers) and response to previously administered anti-cancer therapy (predictive markers).
The aim of this study was to analyze the incidence of mutations in the K-Ras and B-Raf genes in patients with CRC, and to assess their significance as prognostic and predictive factors. Additionally, we also examined the potential role of selected clinical and pathological variables as prognostic factors.
Results
Patient characteristics
Patient characteristics are summarized in Table 1. The median age of the patients included in this study was 65 y (181 women, 92 men). Most underwent primary tumor resection (260/273 patients, 95.2%), while secondary metastatic disease was diagnosed in 194 patients (71.1%), of whom 70 (36.1%) underwent resection and 22 (11.3%) underwent thermoablation. The primary tumor was located in the colon in 112 patients (41.1%), sigmoid colon in 100 patients (36.6%) and rectum in 61 patients (22.3%). The metastases were located in the liver in 129 (66.5% of patients with metastases), in the lungs in 39 (20.1%), and in other organs in 126 patients (64.9%). Pretreatment carcinoembryonic antigen (CEA) levels were elevated above the normal range in 89 patients (32.6%).
Table 1. Patient and tumor characteristics.
| Age in years | |
|---|---|
| (median age, age range) | 65 (25–85) |
| Gender | |
|---|---|
| Female |
181 (66.3%) |
| Male | 92 (33.7%) |
| K-Ras gene mutation status | |
|---|---|
| Mutation |
89 (32.6%) |
| Codon 12 |
76 (27.8%) |
| Codon 13 |
10 (3.7%) |
| Undetermined localization |
3 (1.1%) |
| Wild-type | 184 (67.4%) |
| B-Raf gene mutation status | |
|---|---|
| Mutation |
17 (6.2%) |
| Exon 11 |
1 (0.3%) |
| Exon 15 |
16 (5.9%) |
| Undetermined status of mutation |
10 (3.7%) |
| Wild-type | 246 (90.1%) |
| Primary tumor localization | |
|---|---|
| Colon |
112 (41.1%) |
| Sigmoid colon |
100 (36.6%) |
| Rectum | 61 (22.3%) |
| Localization of metastases | |
|---|---|
| Liver |
129 (66.5%) |
| Lungs |
39 (20.1%) |
| Other localizations | 126 (64.9%) |
K-Ras and B-Raf gene mutation status
K-Ras gene mutations were present in 89 patients (32.6%), of whom 76 (85.4%) had mutations in codon 12 and 10 (11.2%) had mutations in codon 13. Women showed a higher incidence of K-Ras gene mutations relative to men (p = 0.0290). No significant differences were observed with respect to tumor size, lymph node involvement grade, histological grade, histopathological type, primary tumor localization, performance status, age, or pretreatment CEA level.
B-Raf gene mutations were present in 17 patients (6.9%), of whom 6 (35.3%) had mutations in exon 15. One patient had a mutation in exon 11, while mutation status was not determined in 10 patients (58.8%). A higher incidence of B-Raf gene mutations was detected in patients with low-grade neoplasm (p < 0.0001), primary tumor localization outside the sigmoid colon (p = 0.0467) and with non-tubular neoplasms (p = 0.0468). Other parameters assessed were not statistically different.
Prognostic significance of K-Ras and B-Raf gene mutation status
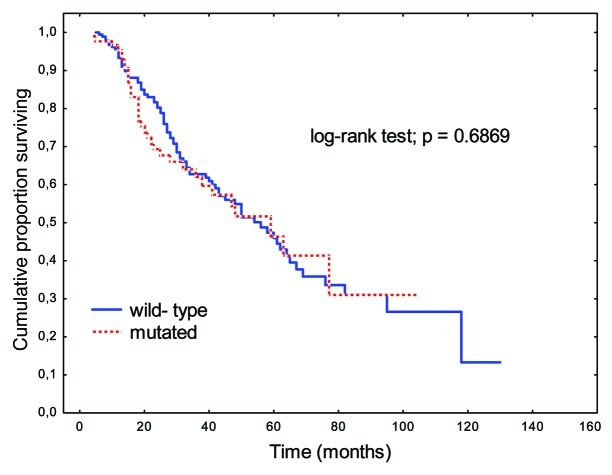
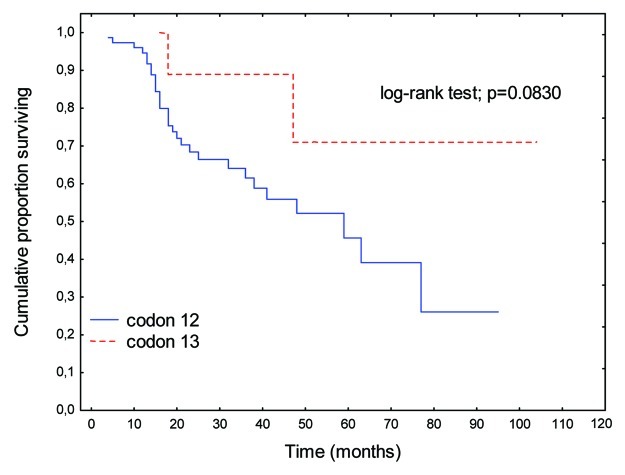
There were no significant differences in OS rates between patients with K-Ras mutations and wild-type K-Ras genes (p = 0.6869; Fig. 1). A perceptible trend to prolongation of OS was apparent when K-Ras mutations were present in codon 13 relative to codon 12 (p = 0.0830; Fig. 2).
Figure 1. Overall survival (OS) rates in patients with K-Ras gene mutations relative to those with wild-type gene.
Figure 2. OS rates in patients with K-Ras gene mutations in codon 12 relative to codon 13.
Similarly, mutations in the B-Raf gene showed no prognostic significance (Fig. 3). Patients with disseminated CRC (M+) and B-Raf gene mutations tended toward shorter OS relative to those with wild-type B-Raf genes (p = 0.06723).
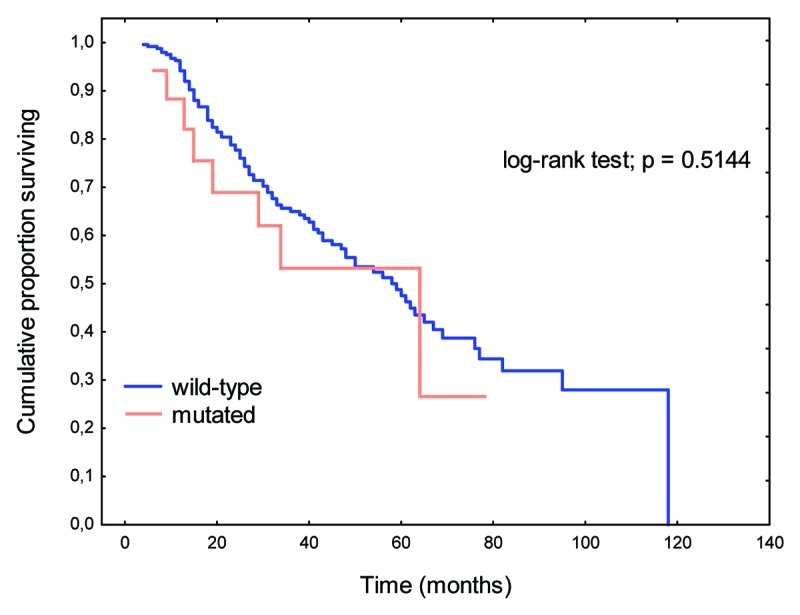
Figure 3. OS rates in patients with B-Raf gene mutations relative to those with wild-type gene.
Clinical and pathological variables identified by univariate analysis as potential prognostic factors for OS rate
These results are summarized in Table 2. Univariate analysis identified the following prognostic factors as influencing OS rate in this patient cohort: Age, patients 75 y and older lived for 36.7 mo relative to those younger than 75 (58.9 mo) (p = 0.0472); Gender, female patients lived for 62.7 mo relative to 42.6 mo for male patients (p = 0.0328); Primary tumor localization, patients with primary tumors in the sigmoid colon lived for 68.0 mo compared with 43.5 mo in patients with primary tumors located in the colon or rectum (p = 0.0039); Performance status, patients with a good performance score e.g., WHO 0–1 (58.4 mo) and Karnofsky status 81–100% (58.1 mo) lived longer relative to those with poor performance status (19.0 and 19.4 mo for patients with WHO 2–3 and Karnofsky status ≤ 80%, respectively) (p = 0.0027 and p = 0.0036, respectively); Lymph node involvement grade, survival in patients without lymph node metastases was 65.3 mo relative to 46.3 mo in patients presenting with metastases (p = 0.0031); Pretreatment CEA level, median time to progression in patients with normal pretreatment CEA level ≤ 5 ng/ml was 76.3 mo relative to 25.6 mo in patients with increased pretreatment CEA level (p < 0.0001; Table 2).
Table 2. Univariate and multivariate analysis of OS rate (log-rank test).
| Univariate analysis |
|
|
|
|---|---|---|---|
| Clinical parameter | n | Median OS (months) | p value |
|
Age |
|
|
|
| < 75 y |
231 |
58.9 |
0.0472 |
| ≥ 75 y |
38 |
36.7 |
|
|
Gender |
|
|
|
| Male |
92 |
42.6 |
0.0328 |
| Female |
181 |
62.7 |
|
|
Primary tumor localization |
|
|
|
| Sigmoid colon |
100 |
68 |
0.0039 |
| Colon/Rectum |
173 |
43.5 |
|
|
WHO performance status |
|
|
|
| 0–1 |
258 |
58.4 |
0.0027 |
| 2–3 |
15 |
19 |
|
|
Karnofsky performance status | |||
| ≤ 80 |
16 |
19.4 |
0.0036 |
| > 80 |
257 |
58.1 |
|
|
Lymph node involvement grade |
|
|
|
| Involved lymph nodes |
60 |
65.3 |
0.0031 |
| Uninvolved lymph nodes |
231 |
46.3 |
|
|
Pretreatment CEA level (ng/ml) |
|
|
|
| ≤ 5 |
170 |
25.6 |
< 0.0001 |
| > 5 | 89 | 76.3 | |
| Multivariate analysis |
|
|
|
|---|---|---|---|
| Clinical parameter | HR (95% CI) | p value | |
| Primary tumor localization |
|
|
|
| Sigmoid colon vs. Rectum/Colon |
0.53 (0.35–0.81) |
0.0032 |
|
|
Lymph node involvement grade |
|
|
|
| Involved vs. Uninvolved |
1.94 (1.17–3.24) |
0.0107 |
|
|
WHO performance status |
|
|
|
| 0–1 vs. 2 |
0.34 (0.18–0.64) |
0.0008 |
|
|
Karnofsky performance status |
|
|
|
| ≤ 80 vs. > 80 |
NS |
> 0.05 |
|
|
Pretreatment CEA level (ng/ml) |
|
|
|
| ≤ 5 vs. > 5 |
2.68 (2.09–3.44) |
< 0.0001 |
|
|
Age |
|
|
|
| ≤ 75 vs. > 75 y |
NS |
> 0.05 |
|
|
Gender |
|
|
|
| Male vs. female | NS | > 0.05 |
NS, not significant.
Other clinical parameters such as histological differentiation grade and primary tumor size showed no significant differences between groups.
Clinical and pathological variables identified by multivariate analysis as potential prognostic factors for OS rate
These results are summarized in Table 2. Multivariate analysis identified the following independent prognostic factors affecting OS rates: Primary tumor localization (HR 0.53; p = 0.0032); Pretreatment CEA level (HR 2.68; p < 0.0001); WHO performance status (HR 0.34; p = 0.0008); Lymph node involvement grade (HR 1.94; p = 0.0107).
Other clinical parameters such as age, gender, and Karnofsky performance status showed no significant differences in this analysis.
Predictive roles of K-Ras and B-Raf mutations on time to progression in CRC patients treated with irinotecan-based first-line palliative chemotherapy on the basis of univariate analysis
These results are summarized in Table 3. Patients with higher pretreatment levels of CEA (> 5 ng/ml) showed a median time to progression of 9.0 mo relative to 13.0 mo in patients with normal levels (≤ 5 ng/ml, p = 0.0085). Patients without resection of metastases showed a median time to progression of 9.0 mo relative to 14.0 mo in patients who underwent resection (p = 0.0131). Patients with K-Ras gene mutations showed a median time to progression of 9.0 mo relative to 11.0 mo in those with the wild-type K-Ras gene (p = 0.05883). Other clinical parameters including histological differentiation grade, primary tumor location and size, lymph node involvement grade, and B-Raf gene mutation status showed no predictive significance in this analysis.
Table 3. Univariate and multivariate analysis of time to progression (log-rank test) for irinotecan-based chemotherapy.
| Univariate analysis |
|
|
|
|---|---|---|---|
| Clinical parameter | n | Median time to progression (months) | p value |
|
Age |
|
|
|
| < 75 y |
79 |
11 |
0.9099 |
| ≥ 75 y |
1 |
- |
|
|
Gender |
|
|
|
| Male |
48 |
12 |
0.1598 |
| Female |
32 |
9 |
|
|
Primary tumor localization |
|
|
|
| Sigmoid colon |
27 |
11 |
0.644 |
| Colon/Rectum |
53 |
10.1 |
|
|
WHO performance status |
|
|
|
| • 0–1 |
79 |
11 |
0.3185 |
| • 2–3 |
1 |
- |
|
|
Karnofsky performance status |
|
|
|
| ≤ 80 |
79 |
- |
0.3185 |
| > 80 |
1 |
11 |
|
|
B-Raf gene mutation status |
|
|
|
| Mutation |
4 |
10.5 |
0.2909 |
| Wild-type |
69 |
11 |
|
|
K-Ras gene mutation status |
|
|
|
| Mutation |
4 |
9 |
0.05883 |
| Wild-type |
76 |
11 |
|
|
Pretreatment CEA level (ng/ml) |
|
|
|
| ≤ 5 |
38 |
13 |
0.0085 |
| > 5 |
40 |
9 |
|
|
Resection of metastases |
|
|
|
| Yes |
27 |
14 |
0.0131 |
| No | 53 | 9 |
| Multivariate analysis |
|
|
|
|---|---|---|---|
| Clinical parameter | HR (95% CI) | p value | |
|
Histological type |
|
|
|
| Tubular vs. others |
NS |
> 0.05 |
|
|
K-Ras gene mutation status |
|
|
|
| Mutation vs. wild-type |
0.59 (0.25-.099) |
0.0459 |
|
|
B-Raf gene mutation status |
|
|
|
| Mutation vs. wild-type |
NS |
> 0.05 |
|
|
Pre-treatment CEA level (ng/ml) |
|
|
|
| ≤ 5 vs. > 5 | 0.52(0.33–0.83) | 0.0065 |
NS, not significant.
Predictive roles of K-Ras and B-Raf mutations on time to progression in CRC patients treated with irinotecan-based first-line palliative chemotherapy on the basis of multivariate analysis
These results are summarized in Table 3. Multivariate analysis identified the following independent favorable predictive factors in patients with disseminated CRC treated with irinotecan-based first-line palliative chemotherapy: Wild-type K-Ras gene (HR 0.59; p = 0.0459) and normal pretreatment CEA levels (HR 0.52; p = 0.0065).
However, this analysis did not reveal any significant differences between patients with and without resection of metastases, with different histological types of neoplasms and B-Raf gene mutation status.
Predictive roles of K-Ras and B-Raf mutations on time to progression in CRC patients treated with oxaliplatin-based first-line palliative chemotherapy on the basis of univariate analysis
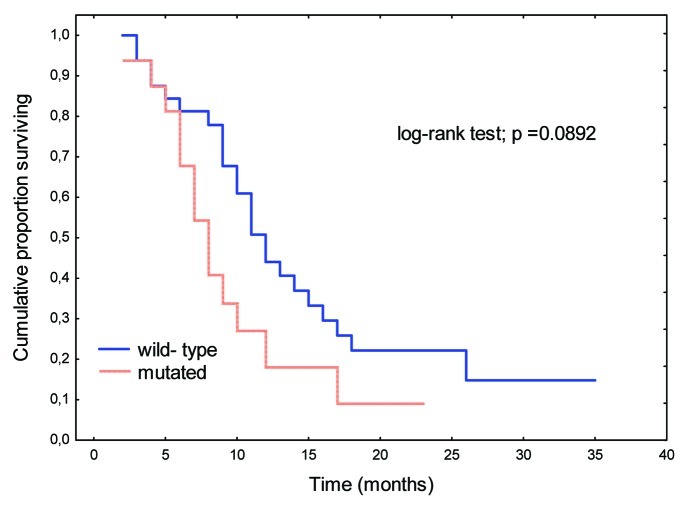
These results are summarized in Table 4. Univariate analysis of time to progression in patients treated with oxaliplatin-based first-line chemotherapy regimens reveals that increased CEA levels and resection of metastases exerted significant influences on median time to progression. Patients with increased pretreatment CEA levels had a time to progression of 8.0 mo compared with 13.0 mo in patients with normal CEA levels (p = 0.0084). Patients without resection of metastases had a time to progression of 9.0 mo relative to 16.0 mo in patients who underwent resection (p = 0.0226). Patients with tubular tumors showed a time to progression of 9.0 mo compared with 13.0 mo in those with other histological types (p = 0.0462). Patients with K-Ras gene mutations did not show a significant difference in time to progression when treated with oxaliplatin chemotherapy, when compared with those with the wild-type K-Ras gene (Fig. 4). The significance of B-Raf gene status, WHO performance status, and Karnofsky performance status could not be assessed.
Table 4. Univariate and multivariate analysis of time to progression (log-rank test) for oxaliplatin-based chemotherapy.
| Univariate analysis | |||
|---|---|---|---|
| Clinical parameter | n | Median time to progression (months) | p value |
|
Age |
|
|
|
| < 75 y |
47 |
10 |
0.9252 |
| ≥ 75 y |
2 |
- |
|
|
Gender |
|
|
|
| Male |
25 |
11 |
0.6149 |
| Female |
24 |
9.7 |
|
|
Primary tumor localization |
|
|
|
| Sigmoid colon |
24 |
11.6 |
0.2375 |
| Colon/Rectum |
25 |
9 |
|
|
WHO performance status |
|
|
|
| · 0–1 |
49 |
10 |
0.3185 |
| · 2–3 |
0 |
- |
|
|
Histological type |
|
|
|
| Tubular |
22 |
13 |
0.0462 |
| Others |
27 |
9 |
|
|
Pretreatment CEA level (ng/ml) |
|
|
|
| · ≤ 5 |
25 |
13 |
0.0084 |
| · > 5 |
21 |
8 |
|
|
Resection of metastases |
|
|
|
| Yes |
18 |
16 |
0.0226 |
| No | 31 | 9 | |
| Multivariate analysis | |||
|---|---|---|---|
|
Clinical parameter |
HR (95% CI) |
p value |
|
| Histological type |
NS |
> 0.05 |
|
| Tubular vs. others |
|
|
|
| Resection of metastases vs. no resection of metastases |
0.43 (0.21–0.90) |
0.0249 |
|
| K-Ras gene mutation |
0.49 (0.24–0.99) |
0.0451 |
|
| mutation vs. wild-type |
|
|
|
| Pretreatment CEA level (ng/ml) |
NS |
> 0.05 |
|
| ≤ 5 vs. > 5 | |||
NS, not significant.
Figure 4. Time to progression according to K-Ras gene mutation status.
Predictive roles of K-Ras and B-Raf mutations on time to progression in CRC patients treated with oxaliplatin-based first-line palliative chemotherapy on the basis of multivariate analysis
These results are summarized in Table 4. Multivariate analysis identified resection of metastases (HR 0.43; p = 0.0249) and wild-type K-Ras gene (HR 0.49; p = 0.0451) as independent favorable predictive factors in patients with disseminated CRC who were treated with oxaliplatin-based first-line palliative chemotherapy regimens.
However, no statistically significant effects of CEA levels and types of neoplasm could be seen. The significance of B-Raf gene status, WHO performance status, and Karnofsky performance status could not be assessed due to the small number of patients.
Discussion
Cancer treatment is increasingly based on targeted therapy, i.e., morphological identification of tumor histology, tumor staging and identification of target pathways and molecules. New insights into signaling processes gone astray in carcinogenesis broaden the scope of molecular diagnosis in cancer. Identification and validation of new prognostic and prognostic markers allow physicians to offer patient-targeted therapy from a broader range of options.
Presently known biomarkers for CRC include the genetic instability status of the tumor, KRAS mutation status as a negative predictive marker for the overall rate of response to anti-EGFR treatment in patients with metastatic cancer, and BRAF mutation as an unfavorable prognostic marker.18
The introduction of molecularly targeted drugs for the treatment of advanced CRC is based on emerging data on the molecular mechanisms responsible for its origin and development. Disturbances in the RAS/RAF/MEK/ERK signaling pathway are the most frequent and perhaps the most important observed defects, with activating mutations in the K-Ras and B-Raf genes playing key roles.
The aims of this study were to evaluate the incidence of B-Raf and K-Ras gene mutation in patients with CRC regardless of disease stage, and to determine the prognostic significance of these mutations on time to progression in response to treatment with palliative chemotherapy. The role of select clinical and pathological variables as potential prognostic factors was also examined.
Our analysis revealed K-Ras gene mutations in our patient population with an incidence of 32.6% with most K-Ras mutations located in codon 12 (27.8%) compared with codon 13 (3.7%), similar to previously reported data.8,19 We estimate the incidence of B-Raf gene mutations at 6.2%, occurring predominantly in exon 15. Further, our analysis shows that B-Raf mutations in exon 15 (V599E) account for nearly 90% of all mutations. These results are similar to previously published data.16,17,20
Interestingly, women present with a higher rate of K-Ras gene mutations relative to men. A higher incidence of B-Raf mutations was seen in patients with low-grade neoplasms, primary tumor location outside the sigmoid colon, and neoplasms other than tubular.
In our analysis, no significant influence on survival was seen in patients with mutations either in the K-Ras or B-Raf genes relative to the general population. However, patients with K-Ras mutations in codon 12 showed significantly decreased survival rates compared with those with mutations in codon 13.
Previous studies have shown that mutations of the K-Ras gene in patients with metastatic CRC are a predictive marker of poor response to anti-EGFR therapy alone or in combination with chemotherapy, relative to patients with WT tumors.21-28 However, Richman et al.23 could not establish any prognostic significance of K-Ras and B-Raf mutations in patients with disseminated CRC and treated only with chemotherapy.
Our study did not establish a prognostic role for B-Raf mutation status in CRC patients in contrast to the results obtained by Tol et al.,29 who observed significantly shorter OS in patients with these mutations. Their retrospective analysis was conducted in a relatively small group of patients with stage IV disease being treated with anti-EGFR therapy, which may have significantly affected survival rates in patients with WT tumors. Similarly, a very high incidence of mutations may account for the differences between our data and results from a previous study in which B-Raf mutations had a prognostic significance in patients with stage II or III CRC.30 In the present analysis, a subgroup of patients with disseminated CRC (M+) and wild-type B-Raf genes tended toward longer survival rates relative to those with B-Raf mutations, although this difference was not statistically significant.
In this study, univariate analysis of the role of clinical and pathological variables revealed a positive, statistically significant influence of the following factors on overall patient survival: female gender, primary tumor localization in sigmoid colon, CEA level within normal limits, good performance status (WHO: 0–1 or Karnofsky Performance Status Scale 81–100%) and lack of metastases in regional lymph nodes. Multivariate analysis identified primary tumor localization in sigmoid colon, lack of metastases in regional lymph nodes, CEA level within normal limits and good performance status according to WHO criteria (0–1) as favorable independent prognostic factors.
Lagautriere et al.31 conducted a retrospective analysis of CRC patients being treated surgically to determine prognostic factors, and identified age, preoperative CEA level, performance status, ileus, and clinical and pathological staging as influencing OS. On the other hand, clinical parameters such as gender, primary tumor localization, and pathological staging had no influence. The retrospective nature of their analysis and differences in inclusion criteria between studies may account for observed differences. Other studies did not show an effect of age on patient survival,32,33 although the prognostic significance of primary tumor localization relative to other localizations has been observed.34
The predictive significance of molecular factors in response to treatment is a fundamental problem in oncology. Available data concerning possible influence of molecular parameters on chemotherapy treatment is strictly limited. Therefore, we performed an analysis of the influence of K-Ras and B-Raf mutations on time to progression in CRC patients being treated with palliative first-line chemotherapy based on irinotecan and oxaliplatin.
Multivariate analysis revealed a predictive significance for K-Ras mutations with respect to time to progression in patients treated with chemotherapy based on irinotecan and oxaliplatin as first-line chemotherapy. However, there was no predictive significance for B-Raf gene mutation status in patients treated with irinotecan or oxaliplatin (evaluation not performed due to a small n). Both univariate and multivariate analyses of time to progression in patients treated with irinotecan showed that pretreatment CEA level was a predictive factor. Resection of metastases was found to be a statistically significant predictive factor by univariate, but not by multivariate analysis. Additionally, univariate analysis revealed that pretreatment CEA level and histopathological type of neoplasm also influence time to progression. However, these factors were not identified by multivariate analysis.
To sum up, K-Ras mutation status, pretreatment CEA level and resection of metastases appear to be predictive of time to progression in CRC patients treated with chemotherapy regimens based on irinotecan and oxaliplatin in first-line therapy. Our results regarding CEA level and resection of metastases are similar to those published by Fong et al.35 and confirms the predictive significance of K-Ras and B-Raf gene status in patients treated with targeted anti-EGFR therapy.23-26,35-38
There is not much evidence for the predictive significance of K-Ras and B-Raf gene mutation status in patients treated solely with chemotherapy, including that based on irinotecan and oxaliplatin.23 Further, most studies examined the effects of treatment with single-agent anti-EGFR therapy or its combination with chemotherapy. Similarly, predictive significance could not be established for K-Ras gene mutations in patients with colorectal cancer, non-small cell lung carcinoma and other solid tumors treated with conventional chemotherapy.38
Our results suggest that determination of K-Ras and B-Raf mutation status in patients qualified for anti-EGFR therapy alone or in combination with chemotherapy can greatly assist in predicting the success or failure of these treatments. Moreover, K-Ras mutation status should be determined in patients qualified for chemotherapy based on irinotecan or oxaliplatin. The role of B-Raf mutation status remains unclear.
Patients and Methods
Ethical approval for research
The research approved by the appropriate local ethical committees (reference numbers: WIM-50/2008 and WIM-45/2009).
Patients
273 consecutive patients (median age 65 y, range 25–85 y) with CRC who were treated between 2006 and 2010 at the Oncology Department of the Military Institute of the Heath Services, Warsaw, were included in this study (See Table 1 for an overview of patient characteristics).
Inclusion criteria consisted of a confirmed histopathological diagnosis of colorectal cancer, availability of adequate primary tumor material, and a lack of effect of chemotherapy or radiotherapy on the tumor.
Surgically removed primary tumor tissue specimens were fixed in formalin and converted into paraffin blocks for further analysis.
Tumor specimens and histological examination
Primary tumor tissue collected from colorectal cancer patients was fixed in 10% neutral buffered formalin for 24 h and converted into paraffin blocks. Serial 5 µm-thick sections of each paraffin block corresponding to representative areas of the tumors were stained with hematoxylin/eosin (H&E) and the presence of tumor tissue verified by an experienced pathologist.
DNA isolation
DNA from paraffin-embedded tissue was prepared from 10–30 µm sections after macrodissection, to ensure they contained at least 80% tumor cells. Tissue samples were extracted with xylene and ethanol to remove paraffin and placed in 1% SDS/proteinase K (10 mg/ml) at 56°C overnight. DNA was isolated using the NucliSens easyMAG platform (bioMérieux) for automated nucleic acid extraction.
K-Ras and B-Raf mutation analysis
Mutation analysis at codons 12 and 13 of the K-Ras gene, and exons 11 and 15 of the B-Raf gene was performed by direct sequencing of amplified PCR products. Genomic DNA was amplified by PCR using the following primers: FS 5′- TCA TTA TTT TTA TTA TAA GGC CTG CTG - 3′, RS 5′-CAA GAT TTA CCT CTA TTG TTG GAT CA-3′ (for codons 12 and 13 in exon 2 of K-Ras), BF11 5′-TCCCTCTCAGGCATAAGGTAA-3′, BR11 5′-TTATTGATGCGAACAGTGAATAT-3′ (for a glycine-rich loop region in exon 11 of the B-Raf gene), B2F 5′-TCATAATGCTTGCTCTGATAGGA–3′, B1R 5′-TAACTCAGCAGCATCTCAGG–3′ (for activation domain in exon 15 of the B-Raf gene). PCRs were performed in a total volume of 10 µl containing 2 µl of extracted genomic DNA, 1X PCR buffer, 1.5 mmol/L MgCl2, 0.2 µmol/L of each primer, 0.1 mmol/L dNTPs and 1U of Taq DNA polymerase (EURx Ltd., Gdansk, Poland).
PCR conditions were as follows: 95°C for 10 min and 40 cycles of 95°C for 20 sec, 56°C for 30 sec (K-Ras and B-Raf in exon 11), 57°C for 30 sec (B-Raf in exon 15), 72°C for 30 sec, and finally 5 min at 72°C. Amplification products were purified using the DNA Gel-Out Kit (DNA GDANSK). Automated sequencing was performed using the Big Dye Terminator Cycle Sequencing kit version 3.1 (Applied Biosystems).
Sequencing reactions were purified using the ExTerminator Kit (DNA GDANSK), and analyzed on an ABI PRISM 377 DNA sequencer (Applied Biosystems). A wild-type control DNA sample (without K-Ras and B-Raf mutations) and a known mutation sample were also included in the experiment. The presence of a mutation was confirmed by sequencing at least two independent PCR products.
Enriched PCR-RFLP analysis for K-Ras codon 12 mutations detection
Detection of K-Ras mutations in codon 12 was performed by enriched non-radioactive single-step PCR-restriction fragment length polymorphism (RFLP) as described previously (Banerjee et al., 1997), with some modifications.
First-round PCR primers K1 5′-ACT GAA TAT AAA CTT GTG GTA GTT GGA CCT-3′ and DD5P 5′-TCA TGA AAA TGG TCA GAG AA-3′ were designed to create a restriction site for the restriction endonuclease BstOI (Promega) within the amplified product. The upstream primer K1 is immediately upstream of K-Ras codon 12 and introduces a G to C substitution at the first position of codon 11, creating a BstOI restriction site (5′-CCTGG-3′) in the amplified fragment. This site overlaps with the first 2 nucleotides of codon 12 and is lost when a codon 12 mutation is present. As a result, the restriction endonuclease BstOI recognizes the sequence 5′-CCTGG-3′ in K-Ras codon 12 wild-type PCR products and digests them, without affecting mutant PCR products.
Second-round PCR primers K1 and K2 5′-TCA AAG AAT GGT CCT GGA CC-3′ created another restriction site in the final segment of the PCR product, which served as an internal control for the restriction digestion. PCR products containing codon 12 mutations were mainly amplified in the second round, because wild-type products were digested in the previous step. These products will contain only one restriction site for BstOI near their 3′-end. Any non-digested PCR products containing wild-type codon 12 sequence that are amplified during the second PCR round will contain two BstOI restriction sites—one identical to that in the mutant molecules and the second overlapping with the codon 12 sequence (introduced by the K1 primer).
The products of the second PCR amplification were also digested with BstOI. The digestion products were electrophoresed on a 3% agarose gel and stained with ethidium bromide. Non-restricted PCR products were 157 bp, wild-type products were 113 bp and mutant codon 12 products were 142 bp in size. A normal control DNA sample (without the K-Ras codon 12 mutation) and a known mutation sample were included in all experiments. The results of PCR detection were verified by direct DNA sequencing.
Statistical analysis
The Chi-square test was used to investigate the differences between the 2 treatment groups with respect to baseline characteristics and response rates. Time to disease progression and overall survival (OS) were summarized as Kaplan-Meier estimates. The log-rank test was used in the Kaplan–Meier survival analyses to assess the effect of variables on time to disease progression and OS.
Multivariate analyses of time to disease progression and OS were performed by Cox proportional-hazard regression using the forward stepwise method; all variables found to be significant in the univariate analysis were included in the multivariate analysis. Statistical calculations were performed using STATISTICA for Windows Version 7.0 software.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
The authors thank the Proper Medical Writing (infrared group s.c.) for the technical and language assistance in the preparation of this paper.
Contributors: Study concepts, R.S., L.B., R.Ch.; Study design, R.S., L.B., R.Ch.; Data acquisition, M.R., M.S., M.C.; Quality control of data and algorithms, L.Ch., W.K., C.Sz.; Data analysis and interpretation, R.S., L.B., R.Ch.; Statistical analysis, L.B.; Manuscript preparation, R.S., L.B., J.K., M.R., M.S.; Manuscript editing, R.S, R.Ch., J.K.; Manuscript review, J.N., C.Sz.; All authors read and approved the final manuscript.
Sources of support: Support from the Military Institute of The Health Services, Warsaw, Poland, Grant No. WIM-50/2008 and WIM-45/2009.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/21813
References
- 1.The GLOBOCAN Project http://globocan.iarc.fr/
- 2.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–54. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 3.Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17:1263–93. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- 4.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle. 2009;8:1168–75. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czajkowski R, Placek W, Tadrowski T, et al. Geny BRAF, NRAS, HRAS w liniach komórkowych czerniaka ludzkiego. Przegl Dermatol. 2009;96:265–70. [Google Scholar]
- 6.Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol. 2006;33:392–406. doi: 10.1053/j.seminoncol.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 8.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 9.Nishimura S, Sekiya T. Human cancer and cellular oncogenes. Biochem J. 1987;243:313–27. doi: 10.1042/bj2430313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–32. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–5. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 12.Haluska F, Pemberton T, Ibrahim N, Kalinsky K. The RTK/RAS/BRAF/PI3K pathways in melanoma: biology, small molecule inhibitors, and potential applications. Semin Oncol. 2007;34:546–54. doi: 10.1053/j.seminoncol.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Nandan MO, Yang VW. An update on the biology of RAS/RAF mutations in colorectal cancer. Curr Colorectal Cancer Rep. 2011;7:113–20. doi: 10.1007/s11888-011-0086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan W, Liu J. Epithelial ovarian cancer: focus on genetics and animal models. Cell Cycle. 2009;8:731–5. doi: 10.4161/cc.8.5.7848. [DOI] [PubMed] [Google Scholar]
- 15.Khanafshar E, Lloyd RV. The spectrum of papillary thyroid carcinoma variants. Adv Anat Pathol. 2011;18:90–7. doi: 10.1097/PAP.0b013e3182026da6. [DOI] [PubMed] [Google Scholar]
- 16.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 17.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 18.Cervera P, Fléjou JF. Changing pathology with changing drugs: tumors of the gastrointestinal tract. Pathobiology. 2011;78:76–89. doi: 10.1159/000315535. [DOI] [PubMed] [Google Scholar]
- 19.Chang YS, Yeh KT, Chang TJ, Chai C, Lu HC, Hsu NC, et al. Fast simultaneous detection of K-RAS mutations in colorectal cancer. BMC Cancer. 2009;9:179. doi: 10.1186/1471-2407-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 21.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 22.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 23.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 24.Van Cutsem E, Rougier P, Köhne CH. A meta-analysis of the CRYSTAL and OPUS studies combining cetuximab with chemotherapy (CT) as 1st-line treatment for patients (pts) with metastatic colorectal cancer (mCRC): Results according to KRAS and BRAF mutation status. ECCO-ESMO 2009; Abstr.No: 6.077. [Google Scholar]
- 25.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–8. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 26.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 27.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697–705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 28.Petrelli F, Borgonovo K, Cabiddu M, Ghilardi M, Barni S. Cetuximab and panitumumab in KRAS wild-type colorectal cancer: a meta-analysis. Int J Colorectal Dis. 2011;26:823–33. doi: 10.1007/s00384-011-1149-0. [DOI] [PubMed] [Google Scholar]
- 29.Tol J, Dijkstra JR, Klomp M, Teerenstra S, Dommerholt M, Vink-Börger ME, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997–2009. doi: 10.1016/j.ejca.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 31.Lagautrière F, Valvano L, Chaazl M, Benchimol D, Bernard JL, Bourgeon A, et al. [Prognostic factors in colorectal adenocarcinoma] Ann Ital Chir. 1998;69:491–6, discussion 496-7. [PubMed] [Google Scholar]
- 32.Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, et al. Irinotecan Study Group Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. N Engl J Med. 2000;343:905–14. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 33.Folprecht G, Seymour MT, Saltz L, Douillard JY, Hecker H, Stephens RJ, et al. Irinotecan/fluorouracil combination in first-line therapy of older and younger patients with metastatic colorectal cancer: combined analysis of 2,691 patients in randomized controlled trials. J Clin Oncol. 2008;26:1443–51. doi: 10.1200/JCO.2007.14.0509. [DOI] [PubMed] [Google Scholar]
- 34.Wray CM, Ziogas A, Hinojosa MW, Le H, Stamos MJ, Zell JA. Tumor subsite location within the colon is prognostic for survival after colon cancer diagnosis. Dis Colon Rectum. 2009;52:1359–66. doi: 10.1007/DCR.0b013e3181a7b7de. [DOI] [PubMed] [Google Scholar]
- 35.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–18, discussion 318-21. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 37.Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, et al. Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28:4706–13. doi: 10.1200/JCO.2009.27.6055. [DOI] [PubMed] [Google Scholar]
- 38.Loriot Y, Mordant P, Deutsch E, Olaussen KA, Soria JC. Are RAS mutations predictive markers of resistance to standard chemotherapy? Nat Rev Clin Oncol. 2009;6:528–34. doi: 10.1038/nrclinonc.2009.106. [DOI] [PubMed] [Google Scholar]