Abstract
Gene therapy vectors based on the adeno-associated virus (AAV) are extremely efficient for gene transfer into post-mitotic cells of heart, muscle, brain, and retina. The reason for their exquisite tropism for these cells has long remained elusive. Here, we show that upon terminal differentiation, cardiac and skeletal myocytes downregulate proteins of the DNA damage response (DDR) and that this markedly induces permissivity to AAV transduction. We observed that expression of members of the MRN complex (Mre11, Rad50, Nbs1), which bind the incoming AAV genomes, faded in cardiomyocytes at ~2 weeks after birth, as well as upon myoblast differentiation in vitro; in both cases, withdrawal of the cells from the cell cycle coincided with increased AAV permissivity. Treatment of proliferating cells with short-interfering RNAs (siRNAs) against the MRN proteins, or with microRNA-24, which is normally upregulated upon terminal differentiation and negatively controls the Nbs1 levels, significantly increased permissivity to AAV transduction. Consistently, delivery of these small RNAs to the juvenile liver concomitant with AAV markedly improved in vivo hepatocyte transduction. Collectively, these findings support the conclusion that cellular DDR proteins inhibit AAV transduction and that terminal cell differentiation relieves this restriction.
Introduction
Viral vectors based on the adeno-associated virus (AAV) have become progressively very efficient tools for in vivo gene transfer in both the laboratory and in the clinic. Their growing popularity is due largely to the relative genetic simplicity, low risk of insertional mutagenesis, low immunogenicity, and capacity to drive transgene expression in a very persistent manner.1 Most notably, while largely ineffective in several cultured, replicating primary cells and cell lines, the various AAV vector serotypes can transduce at very high efficiency noncycling tissues in vivo, including heart (cardiomyocytes), skeletal muscle (skeletal myofibers), brain (neurons), retina (ganglionar cells, pigment epithelium and photoreceptors) and, possibly, to a lesser extent, liver (hepatocytes) and pancreas (both β-cells and acinar cells).1 The molecular reasons as to why the virus is particularly efficient in these cell types are still largely enigmatic. Of note, most of the AAV targets in vivo are post-mitotic cells, namely cells that are terminally differentiated and have permanently exited the cell cycle. Since the various AAV serotypes interact with widely expressed receptors,2 restriction to AAV transduction in nonpermissive cells most likely occurs after virion internalization.
Similar to its wild type counterpart, recombinant AAV displays a linear single-stranded DNA genome with identical terminal hairpin palindromes (inverted terminal repeats). Work performed by different laboratories has shown that one of the major, rate-limiting determinants of AAV permissivity relates to the way in which cellular DNA damage response (DDR) proteins process viral genomes once these reach the nucleus. In particular, multiple evidence indicates that both the error-prone, nonhomologous end-joining and the homologous recombination (HR) DNA repair pathways are involved in different ways in the processing of AAV genomes. These mechanisms appear to regulate the conversion of single-stranded DNA into transcription-competent, double-stranded (ds) DNA,3,4,5,6,7 the opening of inverted terminal repeats and circularization,8,9 the joining of several genomes to form linear concatamers that are eventually also circularized into large and stable episomal molecular species,7,10,11,12,13 or, finally, the expression of the viral transgenes.14
In particular, work from our laboratory, and that of others,' has demonstrated that, once internalized into the nucleus of cycling cells, AAV genomes physically interact with members of the MRN (Mre11, Rad50, Nbs1) complex.15,16 In eukaryotic cells, MRN controls the DDR by sensing DNA damage and governing the activation of the ataxia-telangiectasia mutated kinase;17 besides being a key component in the homology-directed repair, MRN also participates in the repair through classical and alternative NHEJ pathways.18,19 Multiple evidence indicates that the interaction between AAV genomes and cellular DDR proteins restricts AAV transduction in several cultured cell types. This conclusion is supported by the observations that cells that are genetically defective in ataxia-telangiectasia mutated or Nbs1 are naturally more permissive to transduction,6,7,10 that different DDR proteins physically bind the transducing AAV genomes,6,10,15 and that short-interfering RNAs (siRNAs) targeting Nbs1 or MDC115 or Rad50 (unpublished observations) increase transduction efficiency.
Precisely how this information, obtained mostly in cultured cells, relates to the exquisite permissivity to AAV transduction of post-mitotic cells in vivo still remains to be understood and represents the major focus of the work presented in this manuscript. The negative role of the DDR proteins suggests that a most favourable cellular environment for viral transduction would entail a radically reduced expression of these proteins. Supporting this possibility, genetic studies have demonstrated that the essential functions of the MRN complex, as the major sensor of double stranded breaks, are associated with DNA replication and that HR is not essential in nondividing cells.20 More specifically, in post-mitotic cells, ds DNA breaks are radically reduced, concomitant with the cessation of DNA replication, and the DDR proceeds differently from replicating cells, with a significant increase in the NHEJ pathway at the expense of an attenuation of the HR machinery.21
The downregulation of HR proteins in post-mitotic cells coincides with that of several factors required for G1-S progression, as well as G1- and S-phase checkpoints. Recent evidence indicates that the overall decrease in the expression of these factors is strictly correlated to terminal differentiation and that microRNAs (miRNAs) are integral components of this regulatory circuitry. In particular, differentiation of muscle precursors to multinucleated myofibers involves the function of several, different miRNAs. In particular, miR-24 was shown as an essential miRNA for the modulation of transforming growth factor-β-inhibited myogenesis.22 The same miRNA, however, exerts a much broader effect on a variety of other cell types. In particular, its overexpression downregulates several hundred genes, many of which are involved in cell-cycle regulation and DNA repair, including those coding for transcription factors c-myc and E2F2, dihydrofolate reductase, histone γ-H2AX, the cyclin-dependent kinases Cdc2 and CDK6, members of the pre-replication complex proteins, and others.23,24,25 Overexpression of miR-24 in different cell lines inhibits cell proliferation and results in cell cycle arrest;26 conversely, its inhibition in HeLa cells markedly increases cell growth.27 Based on this information, a reasonable possibility is that this miRNA, either directly or indirectly, through the modulation of the cell cycle, might also regulate AAV permissivity by inducing the downregulation of the DDR proteins that restrict AAV transduction.
Indeed, here we show that permissivity to AAV transduction in both cultured primary cardiomyocytes and skeletal myoblasts correlates strictly with the progress of their terminal differentiation. We also show that this process coincides with the reduced expression of members of the MRN complex, which instead restrict AAV transduction of cycling cells. siRNA-mediated downregulation of MRN proteins, or treatment with miR-24, which negatively regulates the Nbs1 levels, induces AAV permissivity in both cultured cells and in the juvenile liver in vivo.
Results
Efficiency of myocardial transduction with AAV vectors depends on cardiomyocyte withdrawal from the cell cycle
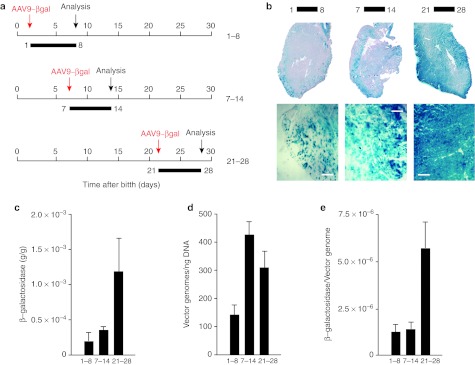
The adult heart is one of the tissues most permissive to transduction with various AAV serotypes, including AAV1, AAV2, AAV6, AAV8, and AAV9.2 During embryonic and fetal development, cardiomyocytes actively proliferate, while they permanently exit the cell cycle after birth. We recently observed that, at birth, over 30% of rat cardiomyocytes still incorporate the thymidine analogue bromodeoxyuridine, a marker of active proliferation, while this percentage drops to <3% in adult hearts.28 We, therefore, wished to assess whether the in vivo, proliferative potential of cardiomyocytes might also determine different permissivity to AAV transduction. We intraperitoneally injected animals at day 1, day 7, and day 21 after birth with AAV9-LacZ at a dose of 2.25 × 1010 vector genomes (vg)/g of body weight (n = 6 per group; Figure 1a). In all groups, 7 days postinjection animals were sacrificed, hearts collected, and vg copies and transgene expression assessed. Analysis of heart sections showed a markedly higher heart transduction in the animals injected at day 21 compared to those injected at days 1 or 7 (Figure 1b for histological analysis and Figure 1c for β-galactosidase quantification; approximately sixfold difference between day 1 and 21). Analysis of vg copies at the different time points revealed that animals injected at days 7 or 21 had a higher number of AAV copies in the heart compared to those injected at day 1 (Figure 1d); however, the highest transgene expression per DNA copy was observed in the animals injected at day 21 (Figure 1e).
Figure 1.
Efficiency of myocardial transduction with adeno-associated virus (AAV) vectors at different times after birth. (a) Schematic representation of the experimental workflow. AAV9-LacZ vectors were administered intraperitoneal (i.p.) in 1-, 7-, and 21-day-old mice; in all cases, analysis was performed 7 days after injection. (b) Histochemical staining for β-galactosidase expression—whole heart sections (upper panels) and enlargements (lower panels; Bar: 100 µm)—after AAV administration for the indicated time intervals (days). (c) Quantification of β-galactosidase expression in total heart tissue (n = 6 per group; mean values ± SD) after AAV administration for the indicated time intervals (days). (d) AAV vector DNA copy numbers per ng of genomic DNA (n = 6 per group; mean values ± SD) after AAV administration for the indicated time intervals (days). (e) Ratio between the amount of expressed β-galactosidase and AAV genome copy numbers (n = 6 per group; mean values ± SD) after AAV administration for the indicated time intervals (days).
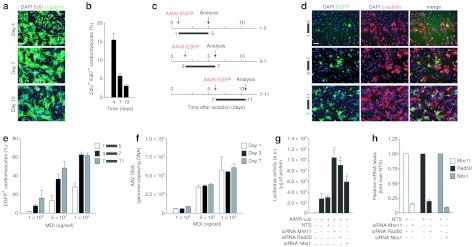
Next, we wanted to assess whether the markedly different permissivity to AAV transduction of cardiomyocytes of different age might be reproduced ex vivo. Purified neonatal rat cardiomyocytes maintain the capacity to undergo a few rounds of cell division in cell culture, as can be concluded by their capacity to incorporate the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) and to express the proliferation marker Ki67; this is rapidly lost in subsequent days, when they differentiate terminally, enlarge their cytoplasm and organize their sarcomere.28 The percentage of proliferating EdU and Ki67 double-positive cardiomyocytes, isolated immediately after birth, dropped from 15.2% at day 4 after plating to 2.6% at day 10 (Figure 2a for representative images and Figure 2b for quantification). We transduced these cells with an AAV6 vector-expressing enhanced green fluorescent protein (EGFP) at day 1, 3, and 7 after isolation and using three different multiplicities of infection (MOIs) (1 × 103, 5 × 103, and 1 × 104 vg/cardiomyocyte); in all cases, analysis of GFP fluorescence was performed after 4 days from transduction (Figure 2c). Permissivity of α-actinin-positive cardiomyocytes to AAV transduction was clearly correlated with the time from plating (Figure 2d). At an MOI of 5 × 103 vg/cell, the percentage of transduced cardiomyocytes rose from 13.3 ± 5.1% GFP+ cells to 37.6 ± 4.0% and 48.6 ± 7.3% when infection was performed at days 1, 3, and 7, respectively (Figure 2e). Notably, however, these differences in transduction efficiency did not denote a different capacity to internalize the vector; at 4 hours after transduction, the same amount of vector DNA was found inside the cells in all three groups of samples, as determined by quantitative PCR (~3.2 × 103 copies per ng of DNA at the m.o.i of 5 × 103 vg/cell; Figure 2f). Analogous considerations also apply to the other MOIs considered.
Figure 2.
Permissivity of cultured neonatal cardiomyocytes to adeno-associated virus (AAV) transduction. (a) Representative images of cardiomyocytes cultures at 4, 7, and 10 days after plating. Cells were stained with anti-α-sarcomeric actinin antibody (green) to reveal cardiomyocytes and ethynyl-2′-deoxyuridine (EdU) to assess DNA synthesis. Bar: 100 µm. (b) Percentage of proliferating (EdU+ and Ki67+) cardiomyocytes at day 4, 7, and 10 after plating (mean values ± SD of five independent experiments). (c) Schematic representation of experimental workflow. Neonatal rat cardiomyocytes were transduced with AAV6-EGFP at days 1, 3, and 7 after plating and analyzed 4 days after transduction. (d) Representative images of cardiomyocytes transduced with AAV6-EGFP at the indicated time intervals. Cells were fixed and immunostained with anti-α-sarcomeric actinin antibody (red) to reveal cardiomyocytes. Cell nuclei were stained with Hoechst 33342. (e) Percentage of transduced (GFP+) cardiomyocytes after AAV6-EGFP transduction for the indicated time intervals (mean values ± SD of three independent experiments). (f) Amount of AAV vector DNA genomes per ng of genomic DNA, determined by real-time quantitative PCR, at 4 hours postinfection, after transduction for day 1, 3, and 7 (mean values ± SD of three independent experiments). (g) Effects of knockdown of Mre11, Rad50, and Nbs1 using RNA interference (RNAi). Cardiomyocytes, isolated at day 1 after birth, were treated with the indicated short-interfering RNAs (siRNAs) and later transduced with AAV6-Luciferase. The levels of luciferase expression were tested 48 hours after transduction (mean values ± SD of three independent experiments)/NTS, nontargeting siRNA control. *Statistical significance (P < 0.05). (h) Levels of Mre11, Rad50, and Nbs1 mRNAs at 72 hours after transfection of the corresponding siRNAs, measured by real-time PCR in the same samples as in panel g. The results are shown relative to those measured in cardiomyocytes treated with the NTS control. EGFP, enhanced green fluorescent protein.
Both the in vivo and the ex vivo results indicate a marked discrepancy between the number of genome copies internalized by the cardiomyocytes and the efficiency at which these were actually expressed. This discrepancy is highly consistent with the possibility that viral transduction is blocked at a step post-virus entry in replicating cardiomyocytes.
Our previous findings indicate that, in poorly permissive cells, AAV genomes are found within the nucleus in close proximity to the sites where cellular DDR proteins and, in particular, members of the MRN complex accumulate.15 We, and others, have also shown that siRNA-mediated downregulation of these proteins15 or Adenovirus E1B55k-mediated degradation of Mre11,29 markedly induce AAV transduction in poorly permissive cell lines. We wondered, therefore, whether knockdown of MRN might also modify permissivity of neonatal cardiomyocytes. For this purpose, we reverse transfected, at the time of plating, primary rat cardiomyocytes, which are still replicating, with siRNAs against Mre11, Rad50, or Nbs1, or with a nontargeting siRNA (NTS) control, followed by transduction with an AAV6 vector-expressing luciferase. Analysis of luciferase expression after 48 hours indicated that all three siRNAs determined a significant increase of cell permissivity to AAV transduction (Figure 2g). Quantification of the mRNA levels of the three MRN proteins confirmed that the siRNA treatments were effective in all three cases (>85% knockdown; Figure 2h).
Taken together, these experiments indicate that, in replicating neonatal cardiomyocytes, MRN proteins negatively regulate AAV transduction.
Efficiency of in vivo AAV transduction correlates with the levels of MRN proteins
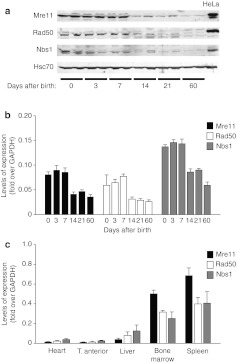
Next, we wondered whether the different permissivity of cardiac myocytes in vivo to AAV transduction at various times after birth correlated with the endogenous levels of expression of MRN proteins. To address this possibility, we assessed the levels of Mre11, Rad50, and Nbs1 in mouse heart from the early postnatal days to the adult age. We observed that these proteins were expressed abundantly in the neonatal heart, but that their levels abruptly decreased between day 7 and 14 (Figure 3a), a time point coincident with the withdrawal of cardiomyocytes from the cell cycle.28 This decrease in protein level was accompanied by a corresponding decrease in the respective mRNA levels, as evaluated by quantitative PCR from the same tissue samples (Figure 3b).
Figure 3.
Levels of Mre11/Rad50/Nbs1 (MRN) proteins in the heart at different times after birth and in other organs. (a) Western blot analysis of Mre11, Rad50, and Nbs1 proteins from whole mouse heart samples obtained at different times after birth—days 0 (neonatal), 3, 7, 14, 21, 60 (adult). Representative samples from two mice are shown for each time point. Hsc-70 was used as loading control. (b) Gene expression analysis of proteins of the MRN complex in the mouse heart at different age as determined by real-time PCR. Values (n = 5 per group; mean ± SD) are normalized over glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (c) Gene expression analysis of proteins of the MRN complex in different tissues of 2-month-old mice (adults), evaluated by real-time PCR. Values (n = 5 per group; mean ± SD) are normalized over GAPDH.
Next, we wanted to understand whether the amount of cellular MRN might more generally correlate with the different tissue permissivity to AAV transduction. We found that the expression levels of Mre11, Rad50, and Nbs1 differed markedly in the various mouse tissues, with their mRNAs being abundantly expressed in adult bone marrow and spleen, reduced in liver and very low in skeletal muscle and heart (Figure 3c). Of interest, we also observed that the levels of all three MRN proteins significantly decreased in the adult (age of 2 months) compared to the neonatal liver (Supplementary Figure S1a).
The rapid downregulation of the expression of MRN proteins in the heart at days 7–14 after birth and the generally low levels of these proteins in adult post-mitotic tissues correlate very well with the known permissivity of these tissues to AAV transduction. These results are thus consistent with the notion that MRN proteins restrict vector transduction in vivo.
Terminal differentiation of myoblasts coincides with downregulation of MRN proteins and induction of permissivity to AAV transduction
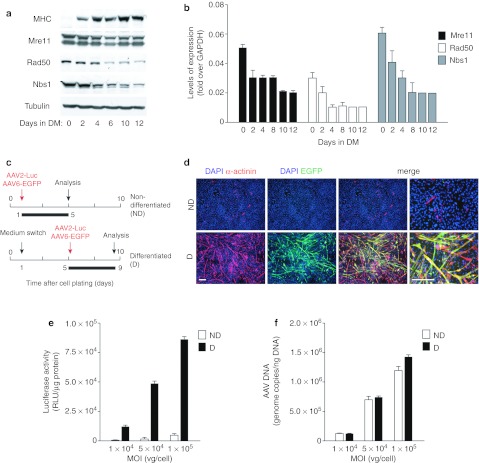
C2C12 murine myoblasts are a well-characterized model of skeletal muscle cell differentiation.30 When cultured with high concentrations of growth factors (nondifferentiating (ND) medium), these homogeneous cells proliferate as mononucleated progenitor cells. In contrast, when cultured with low concentrations of growth factors (differentiating (D) medium), they start expressing muscle differentiation proteins and fuse to form terminally differentiated, multinucleated myotubes. Consequently, we wished to verify whether the differentiation state of these cells might modify the levels of MRN proteins and permissivity to AAV transduction. The levels of Rad50 and Nbs1 proteins progressively decreased from day 4 in differentiation medium, parallel to the increase in the differentiation marker myosin heavy chain (Figure 4a). Mre11 protein levels did not appreciably change during the time of this study, however, the mRNA levels of all three MRN members were decreased after the switch to the differentiation medium (Figure 4b).
Figure 4.
Permissivity of C2C12 cells to adeno-associated virus (AAV) transduction markedly increases during cell differentiation. (a) Western blot of Mre11, Rad50, and Nbs1 proteins during differentiation of C2C12 cells. C2C12 myoblasts were incubated in differentiation medium (DM) for 0, 2, 4, 6, 10, and 12 days to induce differentiation and formation of myotubes. Myosin heavy chain (MHC) was used as a differentiation marker and β-tubulin as a loading control. (b) Gene expression analysis of Mre11, Rad50, and Nbs1 by real-time PCR during the C2C12 differentiation process. The graphs represent the change in gene expression over GAPDH (mean values ± SD of three independent experiments). (c) Schematic representation of the experimental setup for analysis of AAV transduction of C2C12 cells. Cultures of nondifferentiated (ND) or differentiated cells (D, 4 days in differentiation medium) were transduced with AAV vectors expressing enhanced green fluorescent protein (EGFP) or luciferase at different multiplicities of infection (MOIs); analysis was performed 4 days after transduction. (d) Representative images of nondifferentiated (ND) and differentiated (D) C2C12 cells transduced with AAV6-EGFP at an MOI of 1 × 105 vg. Four days after transduction cells were fixed and immunostained with anti-α-sarcomeric actinin antibody (red) to reveal formation of differentiated myotubes. Cell nuclei were stained with DAPI. The enlarged merged figures are shown in the rightmost panels. Bars: 200 µm. (e) Quantification of luciferase activity per µg of total protein in nondifferentiated (ND) and differentiated (D) C2C12 cultures transduced with AAV2-Luciferase at the MOI of 1 × 104, 5 × 104, and 1 × 105 vg. Graphs represent mean values ± SD of three independent experiments. (f) Amount of AAV vector genomes internalized by ND and D C2C12 cells at 4 hours postinfection, determined by real-time quantitative PCR (mean values ± SD of three independent experiments).
To assess AAV transduction efficiency, myoblasts and myotubes were transduced with an AAV6-EGFP vector for 4 days in ND or D medium, respectively (Figure 4c). Transduction was not apparent for myoblasts, while a very large number of polynucleated, α-actinin-positive myotubes were immediately evident in the cell cultures that had been maintained in differentiating conditions (Figure 4d). The same experiments were also repeated using an AAV2-Luciferase vector, to obtain a precise quantification of transduction efficiency. At all the three investigated multiplicities of infection, the luciferase activity per µg of cell lysate was >20-fold higher in myotubes than in myoblasts (Figure 4e). Notably, however, also in this case, myoblasts and myotubes internalized comparable amounts of vg, as evaluated by quantitative PCR performed at 4 hours after transduction (Figure 4f). This again suggests that, in myotubes, AAV transduction is restricted at a time point after vector internalization.
Expression of the cell-cycle regulator miR-24 increases along myoblast differentiation and during heart development
The molecular mechanisms behind the downregulation of MRN and other DDR proteins in post-mitotic cells are still not well understood, but are, most likely, part of a broader modification of gene expression along cellular differentiation. One of the miRNAs that regulate post-mitotic differentiation of several cell types, including myoblasts22 is miR-24. This miRNA is clustered closely with miR-23 and miR-27 at two genomic loci known as the miR-24-1 and -2 gene clusters.31 All three miRNAs were reported to be highly expressed in post-mitotic tissues such as skeletal muscle, heart, and brain.22 Of note, miR-24 was also found to be upregulated during cardiac hypertrophy and to induce hypertrophic growth when overexpressed in primary cardiomyocytes.32
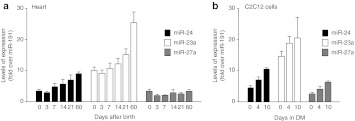
Based on these observations, we wanted to understand whether miR-24 might, directly or indirectly, be involved in the regulation of AAV permissivity. We first analyzed the levels of miR-24, 23a, and 27a in the mouse heart from early neonatal days to adult age. The expression of miR-24 and 23a increased 2.5-fold from day 0 to 60 after birth, while no change was apparent in the expression of miR-27a (Figure 5a). Moreover, miR-24 and the other miRNAs of the same cluster were also markedly upregulated in a time-dependent manner during the process of C2C12 myoblast differentiation in vitro (Figure 5b).
Figure 5.
Levels of expression of microRNAs 24, 23a, and 27a. (a) Expression of miR-24, miR-23a, and miR-27a in mouse heart at different times after birth (days 0 (neonatal), 3, 7, 14, 21, 60 (adult)) as determined by real-time PCR. Graphs represent the change in gene expression over miR-191, shown as mean values ± SD; n = 5 per group. (b) Expression of miR-24, miR-23a, and miR-27a during the differentiation of C2C12 cells (0, 4, and 10 days in differentiation medium (DM)) as determined by real-time PCR. Graphs represent mean values ± SD of three independent samples, normalized to miR-191.
miR-24 negatively regulates Nbs1 levels and enhances permissivity to AAV
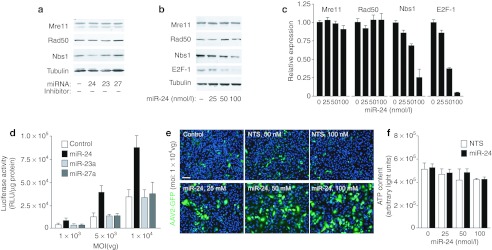
Based on the observation that the downregulation of MRN parallels the miR-24 cluster increase, we wondered whether overexpression of miR-24, miR-23a, or miR-27 precursors might induce downregulation of Nbs1, Mre11, or Rad50. As shown in Figure 6a, we found that treatment of HeLa cells with 50 nmol/l miR-24 precursor determined a decrease in the levels of Nbs1 protein, in addition to that of E2F-1, as already described.23 Reduction in the levels of both Nbs1 and E2F-1 were clearly dose-dependent, as shown for representative immunoblots in Figure 6b and quantified in Figure 6c (4.1-fold reduction of Nbs1 with 100 nmol/l miR-24 precursor). Through 3′UTR reporter experiments we were unable to detect direct targeting of miR-24 to the Nbs1 3′ UTR (data not shown); however, another direct miR-24 target, c-myc, is known to positively regulate expression of Nbs1 and its downmodulation upon miR-24 transfection might thus explain the observed decrease of Nbs1.23
Figure 6.
Expression of miR-24 precursor leads to downregulation of Nbs1 and enhances permissivity to adeno-associated virus (AAV) in vitro. (a) Mre11, Rad50, and Nbs1 protein levels in HeLa cells 72 hours after transfection with precursors of miR-24, 23a, and 27a, at a final concentration of 50 nmol/l. β-Tubulin was used as a control for protein levels. (b) Mre11, Rad50, and Nbs1 protein levels in HeLa cells 72 hours after transfection with 25, 50, and 100 nmol/l miR-24 precursor. E2F-1 was used as control of miR-24 efficacy, since the E2F family members are known targets of miR-24.23 (c) Quantification of immunoblots showing the relative expression of Mre11, Rad50, Nbs1, and E2F-1 in HeLa cells treated with the indicated amounts of miR-24 precursor for 72 hours. The results show mean values ± SD of three independent experiments. (d) Luciferase activity of lysates from HeLa cells transfected with miR-24, miR-23a, or miR-27a precursors (100 nmol/l) or control cells (white bars) after transduction with AAV2-Luciferase at three different multiplicities of infection (MOIs) (1 × 103, 5 × 103, and 1 × 104 vg per cell). The graph shows mean values ± SD of three independent experiments. (e) Representative images of HeLa cells transfected with miR-24 precursor (25, 50, 100 nmol/l) or nontargeting short-interfering RNA (siRNA) (NTS; 50 and 100 nmol/l) for 48 hours and then transduced with AAV2-EGFP (MOI 1 × 104). Images were taken 24 hours after AAV transduction. Control cells represent nontransfected cells transduced with AAV2-EGFP. Bar: 100 µm. (f) Analysis of ATP production, as a measurement of cell viability of HeLa cells transfected with miR-24 precursor at the indicated concentrations. The experimental conditions were as in panel e (mean ± SD of three independent experiments). EGFP, enhanced green fluorescent protein.
To test whether miR-24 overexpression and consequent Nbs1 downregulation might result in higher cellular permissivity to AAV transduction, we infected HeLa cells with AAV2-Luciferase at three different MOIs at 48 hours after transfection with 100 nmol/l miR-24, miR-23a, or miR-27 precursors. As shown in Figure 6d, overexpression of the miR-24 precursor significantly increased AAV transduction at all three, investigated vector concentrations (showing a 3.6-fold increase at the MOI 5 × 103 vg/cells). Precursors of miR-23a and 27a had no effect either on MRN complex expression or on AAV2 transduction.
Finally, a marked increase in the number of fluorescent cells compared to nontransfected cells or cells transfected with control, NTS was also observed when using an AAV2-EGFP vector, followed by quantitative analysis of cell fluorescence (with a fivefold increase at 100 nmol/l miR-24 precursor; Figure 6e). Analysis of ATP content by a chemiluminescence-based assay in these cells revealed that, at the concentrations used, cell treatment with miR-24 did not induced significant cellular toxicity (Figure 6f).
MRN knockdown or treatment with miR-24 precursor increase liver permissivity to AAV transduction in vivo
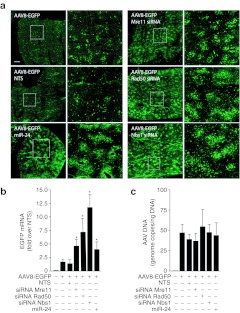
Next, we wanted to assess whether transfection of the siRNAs against Mre11, Rad50, and Nbs1, or of the miR-24 precursor, might also be effective in improving AAV transduction in vivo. We choose to perform our experiments by portal vein injection in 1-month-old mice, since there is evidence that AAV transduction of the liver becomes progressively more efficient from the neonatal age onward.33 Consistent with this notion, we also noticed that, in contrast to the above described decrease in the levels of MRN mRNAs in the adult liver, on the opposite, those of miR-24 were twice as higher in the adult compared to the neonate (Supplementary Figure S1b). One-month old mice (n = 6 per group) were injected, into the portal vein, with cationic lipid formulations containing siRNAs against Mre11, Rad50, or Nbs1, the miR-24 precursor or a NTS control (150 pmol); these formulations were mixed together with an AAV8 vector in which the EGFP gene was under the control of the ApoE/hAAT promoter, which does not undergo transcriptional silencing over time in the liver.34 The animals were sacrificed at day 10 after treatment to analyse the extent of transduction. Analysis of EGFP fluorescence indicated significant increase of liver transduction in the animals that received the three siRNAs against MRN members and, to a lesser extent, in those treated with miR-24 (representative pictures shown in Figure 7a). Quantification of the levels of the EGFP mRNA revealed an average 4.9-fold, 7.4-fold, and 11.9-fold induction for Mre11, Rad50, and Nbs1 respectively, and of 4.0-fold for miR-24, compared to the nontargeting RNA control (Figure 7b). We also analysed the number of vg in the total DNA extracted from two lobes of the transduced livers, and detected no significant differences in the animals treated with the various small RNAs (Figure 7c). This observation is again in agreement with the conclusion that the marked increase in transduction efficiency observed upon anti-MRN siRNA or miR-24 delivery is exerted after AAV genome internalization into the cells.
Figure 7.
In vivo delivery of short-interfering RNAs against Mre11/Rad50/Nbs1 (MRN) components and of miR-24 precursor increases adeno-associated virus (AAV) transduction of juvenile mice livers. (a) Representative images of liver sections at day 10 after portal vein injection of AAV8-ApoE/hAAT-EGFP together with siRNAs against Mre11, Rad50, and Nbs1 or the miR-24 precursor, as indicated. For each treatment, the panel on the right side shows a magnification of the squared area shown on the left panel. Bar: 1 mm. (b) Quantification of the levels of the enhanced green fluorescent protein (EGFP) mRNA in the liver samples as in panel a. Real-time PCR quantifications were normalized over the amounts of cellular GAPDH and expressed as fold-values over samples treated with the nontargeting siRNA (NTS; mean values ± SD; n = 6 per group). The asterisks denote statistical significance (P < 0.05). (c) Quantification of the number of AAV vector genomes in total DNA samples from the liver of animals treated as in panel a (mean values ± SD; n = 6 per group).
Discussion
In this work, we show that permissivity to AAV transduction strictly coincides with the process of terminal differentiation of skeletal and cardiac myocytes. In vivo, cardiomyocytes enter a permanent postmitotic state within a couple of weeks from birth and this markedly increases the efficacy of whole heart transduction by systemically administered AAV vectors. In cultured cells, induction of differentiation of both skeletal and cardiac myoblasts parallels a remarkable increase in permissivity of these cells to AAV infection. Both in vivo and in vitro, terminal cell differentiation is accompanied by a drastic decrease in the levels of proteins of the MRN complex, a finding which is in agreement with previous reports on the relatively lower levels of these factors in adult animals compared to the embryo35,36 and the dispensability of MRN for maintenance of genomic integrity in terminally differentiated cells.20 Members of the MRN complex are known to efficiently recognize DNA damage in response to external agents or ensuing during DNA replication,17 and to direct repair primarily through the HR pathway,37 which takes place during the S-phase of the cell cycle.38 Postmitotic cells, which have permanently exited the cell cycle, downregulate expression of most S-phase required factors, including proteins required for DNA replication and HR, whereas DNA repair mainly occurs by nonhomologous end joining.21,38
The correlative evidence that the down regulation of MRN proteins is paralleled by the increased cell permissivity to AAV transduction is in agreement with the conclusion that in cycling cells, including most cultured cell lines, these proteins restrict AAV transduction. This is supported by a series of previous findings obtained both by us and by other laboratories, including the observations that: (i) in cultured cells, AAV DNA genomes accumulate in the nucleus into specific foci (AAV foci), which overlap with DNA repair foci formed by Mre11, Rad50, Nbs1, and the MDC-1 proteins;15 (ii) in poorly permissive cells, Mre11 and other cellular DDR proteins physically interact with AAV ssDNA, as shown by chromatin immunoprecipitation experiments;6,15 (iii) cell treatment with agents determining genome-wide DNA damage increase AAV permissivity while decreasing Mre11 binding to the AAV genomes and the percentage of DDR foci colocalizing with AAV foci;15 (iv) cell infection with adenovirus or the specific expression of adenovirus E4ORF6/E1B55k induces degradation of Mre11 and increases AAV transduction;29 (v) cells defective for Nbs1 and ataxia-telangiectasia mutated are naturally permissive to AAV in the absence of any treatment.6,7,10,15 The observations, reported in this manuscript, that treatment of rat neonatal cardiomyocytes with siRNAs against Mre11, Rad50, and Nbs1 increases AAV transduction and that siRNAs against the same proteins, once injected in vivo, markedly improve transduction of the mouse juvenile liver further support the conclusion that MRN restricts AAV transduction.
By what mechanism is this restriction exerted? A most likely possibility is that, by virtue of its ss DNA nature and the presence of hairpins at its extremities, the AAV genome is recognized as a form of “damaged” DNA by the cellular DDR machinery. The observation that 3′ overhangs (such as the origin of the AAV DNA) and hairpins are preferred substrates for Mre11 binding39 and that AAV nuclear foci contain the activated form of Nbs1, phosphorylated at S-343,15 support this possibility. We speculate that binding of MRN itself might block single- to ds DNA conversion of the AAV genomes, a step which has long been recognized as one of the most limiting during AAV infection.3,4 In addition, in the process of genomic DNA repair, Mre11 acts as an exonuclease, generating 3′ overhangs that become substrates for HR.39 Although not formally demonstrated here, it might be envisaged that the resection of ss AAV genomes is deleterious to AAV-mediated transduction of protein-coding transgenes. Besides prevention of single-stranded to ds-DNA conversion or degradation of internalized AAV DNA, other possibilities to explain inhibition of AAV processing exist. Among these, recent work in cultured cells has shown that the interaction of DDR proteins might induce transcriptional silencing of the AAV genomes, similar to that which occurs for damaged cellular chromatin.14 Whatever the mechanism of inhibition, it is worth noting that MRN binding to the viral genomes and subsequent recruitment of the cellular HR machinery might well explain why AAV vectors carrying portions of cellular genomic DNA promote HR with their cellular homologue sequences at an efficiency that is 2–3 orders of magnitude higher than plasmid DNA.40
Multiple evidence indicates that various cellular barriers to AAV transduction exist and that some of these can be overcome by different AAV serotypes.2 Restriction of AAV transduction by DDR proteins, however, most likely acts at a step involving genome processing and is thus common to all serotypes. As a matter of fact, in this work, we have exploited the vector serotypes known to be among the most efficient for transduction of the various investigated tissues, including AAV9 for cardiac transduction after systemic administration,41 AAV6 for transduction of isolated cardiomyocytes,42 AAV2 and AAV6 for C2C12 myocytes (L. Zentilin and M. Giacca, unpublished results), and AAV8 for transduction of the liver.43,44 Restriction of AAV transduction by MRN, however, appeared to apply in all cases, consistent with the conclusion that it affects a step subsequent to virion internalization.
When considered collectively, the results obtained favour a model by which terminal cell differentiation coincides with reduced MRN expression and increased AAV permissivity. Terminal differentiation of both cardiomyocytes and myoblasts is accompanied by an increase in the levels of miR-24. In addition to this miRNA, we found that the other two miRNAs from the miR-24-2 cluster (miR-23a and miR-27a) were also induced during myotube formation, whereas only miR-23a during differentiation of cardiomyocytes, consistent with the notion that, while all being encoded within a short genomic region, the different miRNAs of this cluster can be transcriptionally regulated in an independent manner.31 The work of other laboratories has shown that, in muscle and heart, not only is miR-24 strongly induced during myogenesis, but it is also maintained at high levels in terminally differentiated adult tissues.22 In our experiments, delivery of a miR-24 precursor RNA induced downregulation of Nbs1 in a dose-dependent manner, while leaving the levels of Mre11 and Rad50 unaffected. This is most likely the indirect consequence of downregulation of the c-myc transcription factor, which is targeted by miR-2423 and in turn negatively regulates Nbs1 expression.45 In addition, E2F-1, an indirect miR-24 target, has been shown to directly bind MRN through specific interaction with Nbs1.46 Downregulation of Nbs1 by miR-24 would concur with the induction of cell permissivity to AAV transduction, again reinforcing the conclusion that the levels of MRN correlate with restriction to AAV transduction.
Based on the effect of miR-24 precursor delivery to cultured cells, we wished to explore the effect of this molecule in vivo. A number of investigators have reported that AAV vectors are efficient in permanently transducing the adult liver,47 and this information has been successfully brought into the clinic for gene therapy of different, inherited disorders.1 A few of these studies have also reported that, in contrast to that of the adult, the neonate and juvenile liver are much less prone to persistent transduction by the same vectors.33,48 Since the adult liver contains mainly nonproliferating hepatocytes, with a turnover of 180–400 days49 while neonatal hepatocytes proliferate to sustain organ growth, the difference between the efficiency of AAV transduction in adult and neonatal liver has been largely attributed to the loss of episomal AAV genomes, in the latter case, upon cell proliferation.48 An alternative possibility is that the neonatal hepatocytes restrict AAV transduction due to expression of inhibitory DDR proteins. This possibility is very well supported by the observation that the levels of Mre11, Nbs1, and Rad50 are decreased in adults versus neonatal mice, while, conversely, the levels of miR-24 are increased. Even more significant, in our experiments the simultaneous delivery of an AAV8 vector together with siRNAs against the three MRN proteins or an RNA corresponding to the miR-24 precursor significantly augmented the extent of AAV transduction of the juvenile liver. Whether these treatments might be routinely used to improve AAV transduction in vivo of poorly permissive tissues is an interestingly possibility that we wish to further pursue.
Materials and Methods
Cell cultures. C2C12 mouse myoblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. To induce cell differentiation into myotubes, the growth medium was switched to Dulbecco's modified Eagle's medium with 1% horse serum when cells reached 80% confluence. HeLa cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics. Neonatal rat ventricular myocytes were obtained as previously described50 and detailed in the Supplementary Materials and Methods.
Production of AAV stocks. All the AAV vectors used in this study were generated by the AAV Vector Unit (AVU) at ICGEB Trieste (http://www.icgeb.org/avu-core-facility.html) according to established procedures, as detailed in the Supplementary Materials and Methods. All vectors are based on the AAV2 genome and the transgenes are driven by the cytomegalovirus immediated early promoter, with the exception of AAV-ApoE/hAAT-EGFP, in which EGPF is controlled by the ApoE/hAAT promoter.34
In vivo myocardial transduction. Animals were injected intraperitoneally with AAV9-LacZ at a dose of 2.25 × 1010 vg/g of body weight. An appropriate volume of viral vector solution (~30 µl in neonatal mice to ~100 µl in 21-day old mice) was injected slowly using a U-100 insulin syringe. Seven days postinjection animals were sacrificed, the hearts collected and divided into two parts. One half was slowly frozen using isopentane/liquid nitrogen and stored at –80 °C until processed for X-gal staining and determination of AAV vg copy number; the other half was frozen immediately in liquid nitrogen and stored at –80 °C until processed for quantification of β-galactosidase expression.
Ex vivo AAV transduction of cardiomyocytes. Cardiomyocytes were transduced with AAV6-EGFP at day 1, 3, and 7 after isolation. Cells were transduced at three different MOIs (1 × 103, 5 × 103, and 1 × 104 vg). Four days after transduction, cells were fixed and immunostained with anti-α-sarcomeric actinin antibody to reveal cardiomyocytes, as detailed in the Supplementary Materials and Methods.
Cardiomyocyte proliferation assay. Cardiomyocytes growing on BD Primaria 96-well plates were fixed at days 4, 7, or 10, following 20-hour incubation with EdU. Cells were immunostained with α-sarcomeric actinin antibody to identify cardiomyocytes and the cell proliferation marker Ki67; EdU incorporation was revealed using the Click-iT EdU Alexa Fluor555, according to the manufacturer's instructions; cell nuclei were stained with Hoechst 33342. Analysis of EdU and Ki67 double positive cardiomyocytes was performed using an ImageXpress Micro automated high-content screening microscope (Molecular Devices, Sunnyvale, CA) equipped with a 10× objective; analysis was performed using the MetaXpress software (Molecular Devices) as detailed in the Supplementary Materials and Methods.
AAV transduction of ND and D C2C12 cells. C2C12 cells were seeded onto 96-well plate at density of 5,000 cells/well and, after 24 hours, transduced with AAV6-EGFP or AAV2-Luciferase at MOIs of 1 × 104, 5 × 104, and 1 × 105 vg. Cells were passaged 2 days after plating when they reached confluence.
To induce differentiation of C2C12 into myotubes, the growth medium was switched to differentiation medium 24 hours after plating (96-well plate at density of 5,000 cells/well). Four days after the medium switch cells were transduced with AAV6-EGFP or AAV2-Luciferase.
Both ND and D cells were fixed with 4% paraformaldehyde 4 days after transduction. Immunostaining was performed using anti-α-sarcomeric actinin antibody to reveal formation of differentiated myotubes.
Luciferase activity was determined four days after transduction. Luminescence was measured using PerkinElmer EnVision 2104 Multilabel Reader (Perkin Elmer, Waltham, MA) using beetle-Juice reagents (PJK, Kleinblittersdorf, Germany). Luciferase activity was expressed as the ratio between relative light units and total protein (in µg) of the sample.
Analysis of β-galactosidase expression. For histochemical detection of β-galactosidase, frozen heart sections (3.5 µm) were fixed in phosphate-buffered saline containing 2% formaldehyde and 0.2% glutaraldehyde at room temperature for 15 minutes. After being washed with in phosphate-buffered saline containing 0.02% NP-40 three times, sections were incubated at 37 °C overnight in phosphate-buffered saline containing 5 mmol/l potassium ferricyanide, 5 mmol/l potassium ferrocyanide, 2 mmol/l MgCl2, 2 mg/ml X-Gal, and 0.02% NP-40.
After staining, sections were dehydrated by graded alcohol, cleared with Bio-Clear, mounted with EuKit mounting medium (Sigma -Aldrich, St Louis, MO), and protected with cover slides. Pictures were taken using an Olympus CX40 microscope (Olympus, Center Valley, PA).
The Beta-Glo Assay System (Promega) was used to quantify the β-galactosidase expression after AAV-LacZ transduction. Tissues were disrupted using electric homogenizer in Reporter lysis buffer (Promega). The grams of β-galactosidase were calculated based on a standard curve, obtained from serial dilutions of β-galactosidase enzyme (Sigma).
miRNA precursor transfection and AAV transduction of HeLa cells. We reverse transfected HeLa cells (96-well plate, 104 cells/well) with 25, 50, or 100 nmol/l miR-24 (Qiagen, Hilden, Germany) or siGENOME Non-Targeting siRNA (Dharmacon, Lafayette, CO) using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. After 48 hours, cells were transduced with AAV2-EGFP or AAV2-Luciferase at MOI of 103, 5 × 103, or 104. Transduction efficiency was determined 24 hours postinfection.
siRNA transfection of rat neonatal cardiomyocytes. Rat neonatal cardiomyocytes were reverse transfected using Lipofectamine RNAiMAX according to the manufacturer's instructions. siRNAs targeting Mre11, Rad50, and Nbs1 (SMARTpool) and NTS control were purchased from Dharmacon. Cardiomyocytes were transduced with AAV6-Luc at 72 hours after transfection; luciferase activity was determined 48 hours after transduction). To control the effect of siRNA knockdown on Mre11, Rad50, and Nbs1 expression, cells were reverse transfected in 35-mm dishes and harvested 72 hours after transfection. Expression was evaluated by quantitative real time-PCR, as detailed in the Supplementary Materials and Methods.
AAV transduction of the liver. Juvenile mice (1-month old) were injected into the portal vein with a mix of transfection reagent (Lipofectamine RNAiMAX) and siRNA against Mre11, Rad50, Nbs1, miR-24 precursor or a nontargeting control together with AAV8-ApoE/ hAAT-EGFP at a dose of 5 × 1010 vg; a volume of 50 µl was injected slowly using a U-100 insulin syringe. Animals were sacrificed 10 days after injection to evaluate AAV transduction. Livers were collected, carefully divided in lobes, and a piece of every lobe was processed for evaluation of GFP+ cells (by fluorescent microscopy) and quantification of GFP expression by quantitative PCR. For fluorescent microscopy, liver tissue was fixed in 4% paraformaldehyde overnight at 4 °C, cryoprotected in 30% sucrose overnight at 4 °C and then frozen at –20 °C. Liver sections (5 µm) were imaged using a ImageXpress Micro automated high-content screening microscope (Molecular Devices) equipped with a 4× objective.
SUPPLEMENTARY MATERIAL Figure S1. Levels of Mre11, Rad50, Nbs1mRNAs, and miR-24 in neonatal and adult liver samples. Materials and Methods.
Acknowledgments
The authors are grateful to Marina Dapas and Michela Zotti for superb technical support in AAV production, to Mauro Sturnega for help in animal experimentation and to Suzanne Kerbavcic for excellent editorial assistance. A.E. is recipient of a FEBS Long-term Fellowship; J.L. is recipient of a fellowship from the Croatian Science Foundation and the Central European Initiative Research Fellowship Programme, Marie Curie COFUND. This work was supported by grant GGP11068 from the Telethon Foundation, Italy and Advanced Grant 250124 from the European Research Council (ERC) to M.G.
Supplementary Material
Levels of Mre11, Rad50, Nbs1mRNAs, and miR-24 in neonatal and adult liver samples.
REFERENCES
- Mingozzi F., and, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A., and, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Ferrari FK, Samulski T, Shenk T., and, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF., and, Wilson JM. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P., and, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Zentilin L, Marcello A., and, Giacca M. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J Virol. 2001;75:12279–12287. doi: 10.1128/JVI.75.24.12279-12287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlioglu S, Benson P., and, Engelhardt JF. Loss of ATM function enhances recombinant adeno-associated virus transduction and integration through pathways similar to UV irradiation. Virology. 2000;268:68–78. doi: 10.1006/viro.1999.0137. [DOI] [PubMed] [Google Scholar]
- Inagaki K, Ma C, Storm TA, Kay MA., and, Nakai H. The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol. 2007;81:11304–11321. doi: 10.1128/JVI.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma HI, Li J, Sun L, Zhang J., and, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Choi VW, McCarty DM., and, Samulski RJ. Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol. 2006;80:10346–10356. doi: 10.1128/JVI.00841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Lacaze N, Snyder RO, Gluzman R, Bohl D, Lagarde C., and, Danos O. Structure of adeno-associated virus vector DNA following transduction of the skeletal muscle. J Virol. 1999;73:1949–1955. doi: 10.1128/jvi.73.3.1949-1955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Li J., and, Samulski RJ. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaud-Budloo M, Le Guiner C, Nowrouzi A, Toromanoff A, Chérel Y, Chenuaud P.et al. (2008Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle J Virol 827875–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi MP., and, McCarty DM. Differential effects of DNA double-strand break repair pathways on single-strand and self-complementary adeno-associated virus vector genomes. J Virol. 2010;84:8673–8682. doi: 10.1128/JVI.00641-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervelli T, Palacios JA, Zentilin L, Mano M, Schwartz RA, Weitzman MD.et al. (2008Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins J Cell Sci 121Pt 3349–357. [DOI] [PubMed] [Google Scholar]
- Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M., and, Weitzman MD. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH., and, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P., and, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- Xie A, Kwok A., and, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman CA, De S., and, Petrini JH. Rad50 is dispensable for the maintenance and viability of postmitotic tissues. Mol Cell Biol. 2009;29:483–492. doi: 10.1128/MCB.01525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonatto M, Latella L., and, Puri PL. DNA damage and cellular differentiation: more questions than responses. J Cell Physiol. 2007;213:642–648. doi: 10.1002/jcp.21275. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G.et al. (2008Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation Nucleic Acids Res 362690–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E.et al. (2009miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements Mol Cell 35610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D., and, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R, Jr, Srikantan S.et al. (2008p16(INK4a) translation suppressed by miR-24 PLoS ONE 3e1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Song B, Mishra PJ, Wang Y, Humeniuk R, Banerjee D.et al. (2009MiR-24 tumor suppressor activity is regulated independent of p53 and through a target site polymorphism PLoS ONE 4e8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J., and, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collesi C, Zentilin L, Sinagra G., and, Giacca M. Notch1 signaling stimulates proliferation of immature cardiomyocytes. J Cell Biol. 2008;183:117–128. doi: 10.1083/jcb.200806091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M, Weitzman MD. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;18:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K., and, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y.et al. (2009Characterization of function and regulation of miR-24-1 and miR-31 Biochem Biophys Res Commun 380660–665. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD.et al. (2006A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure Proc Natl Acad Sci USA 10318255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SC, Dane AP, Spinoulas A, Logan GJ., and, Alexander IE. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol Ther. 2008;16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Huber RM, Bowling W, Pearline R, Kennedy SC, Flye MW.et al. (1996Liver-directed gene therapy: a retroviral vector with a complete LTR and the ApoE enhancer-alpha 1-antitrypsin promoter dramatically increases expression of human alpha 1-antitrypsin in vivo Hum Gene Ther 7637–645. [DOI] [PubMed] [Google Scholar]
- Wilda M, Demuth I, Concannon P, Sperling K., and, Hameister H. Expression pattern of the Nijmegen breakage syndrome gene, Nbs1, during murine development. Hum Mol Genet. 2000;9:1739–1744. doi: 10.1093/hmg/9.12.1739. [DOI] [PubMed] [Google Scholar]
- Kim KK, Daud AI, Wong SC, Pajak L, Tsai SC, Wang H.et al. (1996Mouse RAD50 has limited epitopic homology to p53 and is expressed in the adult myocardium J Biol Chem 27129255–29264. [DOI] [PubMed] [Google Scholar]
- Arnaudeau C, Lundin C., and, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Krüger I, Thompson LH., and, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M, Bree RT., and, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW., and, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki K, Fuess S, Storm TA, Gibson GA, Mctiernan CF, Kay MA.et al. (2006Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8 Mol Ther 1445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipo I, Fechner H, Pinkert S, Suckau L, Wang X, Weger S.et al. (2007Differential internalization and nuclear uncoating of self-complementary adeno-associated virus pseudotype vectors as determinants of cardiac cell transduction Gene Ther 141319–1329. [DOI] [PubMed] [Google Scholar]
- Vandendriessche T, Thorrez L, Acosta-Sanchez A, Petrus I, Wang L, Ma L.et al. (2007Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy J Thromb Haemost 516–24. [DOI] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G., and, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Chiang YC, Teng SC, Su YN, Hsieh FJ., and, Wu KJ. c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J Biol Chem. 2003;278:19286–19291. doi: 10.1074/jbc.M212043200. [DOI] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA.et al. (2001Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis Mol Cell Biol 216006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang H, Bell P, McCarter RJ, He J, Calcedo R.et al. (2010Systematic evaluation of AAV vectors for liver directed gene transfer in murine models Mol Ther 18118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flageul M, Aubert D, Pichard V, Nguyen TH, Nowrouzi A, Schmidt M.et al. (2009Transient expression of genes delivered to newborn rat liver using recombinant adeno-associated virus 2/8 vectors J Gene Med 11689–696. [DOI] [PubMed] [Google Scholar]
- Magami Y, Azuma T, Inokuchi H, Kokuno S, Moriyasu F, Kawai K.et al. (2002Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver Liver 22419–425. [DOI] [PubMed] [Google Scholar]
- Deng XF, Rokosh DG., and, Simpson PC. Autonomous and growth factor-induced hypertrophy in cultured neonatal mouse cardiac myocytes. Comparison with rat. Circ Res. 2000;87:781–788. doi: 10.1161/01.res.87.9.781. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Levels of Mre11, Rad50, Nbs1mRNAs, and miR-24 in neonatal and adult liver samples.