Abstract
The primary role of Actin-Depolymerizing Factors (ADFs) is to sever filamentous actin, generating pointed ends, which in turn are incorporated into newly formed filaments, thus supporting stochastic actin dynamics. Arabidopsis ADF4 was recently shown to be required for the activation of resistance in Arabidopsis following infection with the phytopathogenic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst) expressing the effector protein AvrPphB. Herein, we demonstrate that the expression of RPS5, the cognate resistance protein of AvrPphB, was dramatically reduced in the adf4 mutant, suggesting a link between actin cytoskeletal dynamics and the transcriptional regulation of R-protein activation. By examining the PTI (PAMP Triggered Immunity) response in the adf4 mutant when challenged with Pst expressing AvrPphB, we observed a significant reduction in the expression of the PTI-specific target gene FRK1 (Flg22-Induced Receptor Kinase 1). These data are in agreement with recent observations demonstrating a requirement for RPS5 in PTI-signaling in the presence of AvrPphB. Furthermore, MAPK (Mitogen-Activated Protein Kinase)-signaling was significantly reduced in the adf4 mutant, while no such reduction was observed in the rps5-1 point mutation under similar conditions. Isoelectric focusing confirmed phosphorylation of ADF4 at serine-6, and additional in planta analyses of ADF4's role in immune signaling demonstrates that nuclear localization is phosphorylation independent, while localization to the actin cytoskeleton is linked to ADF4 phosphorylation. Taken together, these data suggest a novel role for ADF4 in controlling gene-for-gene resistance activation, as well as MAPK-signaling, via the coordinated regulation of actin cytoskeletal dynamics and R-gene transcription.
Author Summary
The activation and regulation of the plant immune system requires the coordinated function of numerous pre-formed and inducible cellular responses. Following pathogen perception, plants not only activate specific defense-associated signaling, such as resistance (R) genes, but also redirect basic cellular machinery to support innate immune signaling. Within each of these processes, the actin cytoskeleton has been demonstrated to play a significant role in structural-based defense signaling in plants in response to pathogen infection. Most notably, the actin cytoskeleton of plants has been shown to play a role in structural-based defense signaling following fungal pathogen infection. Recent work from our laboratory has demonstrated that the actin cytoskeleton of Arabidopsis mediates defense signaling following perception of the phytopathogenic bacterium Pseudomonas syringae. Using a combination of genetic and cell biology-based approaches, we found that ADF4, a regulator of actin cytoskeletal dynamics, is required for the specific activation of R-gene-mediated signaling. By analyzing the activation of signaling following pathogen perception, we have identified substantial crosstalk between recognition of pathogen virulence factors (e.g., effector proteins) and the regulation of R-gene transcription. In total, our work highlights the intimate relationship between basic cellular processes and the perception and activation of defense signaling following pathogen infection.
Introduction
The actin cytoskeleton is an essential, dynamic component of eukaryotic cells, involved in numerous processes including growth and development, cellular organization and organelle movement, and abiotic and biotic stress signaling [1]. Underpinning these processes in plants is a tightly regulated genetic and biochemical mechanism driven by the function of more than 70 actin-binding proteins (ABPs), which through their coordinated activity, regulates the balance of free globular (G)-actin versus filamentous (F)-actin, of which nearly 95% is unpolymerized in plants [2], [3]. As a consequence of this large pool of free G-actin, the potential exists for explosive rates of polymerization following elicitation by a broad range of external stimuli, including pathogen infection [1]. Among the numerous ABPs in plants responsible for modulating the balance of G- to F-actin, one subclass, Actin-Depolymerizing Factors (ADFs), both sever and disassemble F-actin. In addition to its primary role in modulating host cytoskeletal architecture, a role for ADFs in defense signaling following pathogen infection is emerging [4], [5], [6].
The initiation of innate immune signaling in plants relies on multiple pre-formed and inducible processes to surveil, respond, and activate defense signaling following pathogen perception [7], [8]. In total, these responses can be cataloged based on two primary nodes of defense signaling: pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) [7]. In the case of PTI, perception and activation is typically mediated by extracellular plasma membrane-localized pattern recognition receptors (PRRs), which are responsible for the recognition of conserved pathogen motifs (i.e., PAMPs; e.g., flagellin, LPS, chitin). Recognition of PAMPs by PRRs initiates downstream signaling, including the activation of the Mitogen-Activated Protein Kinase (MAPK) signaling cascade, the generation of reactive oxygen species, and transcription of pathogen-responsive genes [9]. Arguably the best-characterized example of PTI signaling in plants is the activation of signaling associated with FLS2 (Flagellin Sensitive-2), a receptor-like kinase containing a serine/threonine kinase, which recognizes flagellin as well as the 22-amino acid peptide flg22 via the extracellular leucine rich repeat (LRR) domain [10], [11]. Activation of FLS2 by flg22 results in the association of FLS2 with BAK1 (BRI1-associated receptor kinase), as well as the phosphorylation of both FLS2 and BAK1 [12]. FLS2 ligand binding and association with BAK1 has been shown to activate the MAPK signaling pathway resulting in dual phosphorylation of conserved tyrosine and threonine residues of Arabidopsis (Arabidopsis thaliana) MAP kinases MPK3/6 [13], which in turn leads to transcription of PTI-related genes including FRK1 (Flg22-induced receptor kinase 1; [14]). The expression of FRK1, however, is believed to be both MAPK dependent and independent [14].
As a counter to the activation of PTI, many plant pathogens deploy secreted effector proteins, which induce a host response (e.g., ETI) - an enhanced PTI-like response, as well as a more robust, programmed cell death-like, response known as the hypersensitive response (HR) that is initiated via the direct or indirect recognition of pathogen effectors by host resistance (R) proteins [7]. As expected, numerous virulence targets of pathogen effectors identified thus far are components of PTI signaling pathways – with the hypothesis being that targeting PTI-components can lead to increased virulence of the pathogen [9], [15]. Among the best-characterized signaling pathways leading to the activation of ETI, as well as a mechanistic example of the functional overlap between PTI and ETI, is the recognition of the bacterial effector protein AvrPphB by the Arabidopsis resistance protein RPS5 (resistance to Pseudomonas syringae 5) [7]. RPS5 is a member of the coiled-coil (CC) nucleotide-binding-site (NBS) LRR R-gene family, required for recognition of Pseudomonas syringae pv. tomato DC3000 (Pst) expressing the cysteine protease effector protein AvrPphB [16], [17]. RPS5-mediated resistance signaling is dependent upon AvrPphB cleavage of the receptor-like cytoplasmic kinase (RLCK) AvrPphB-Susceptible 1 (PBS1), which in turn results in the activation of ETI [18]. Recently, it has been suggested that the virulence target of AvrPphB may in fact be another RLCK, the PTI component BIK1 (Botrytis-induced kinase; [15]). This hypothesis is based on the observation that not only does AvrPphB cleave BIK1, as well as other RLCKs, including PBL1 (PBS1-like 1), but also that cleavage in the absence of RPS5 results in a significant reduction in PTI responses. It should be noted, that while the bik1/pbl1 double mutant does have significant reductions in many PTI responses, bik1/pbl1 does not exhibit reduced MPK3/6 phosphorylation upon flg22 stimulation [15], [19].
In the current study, we report the identification of a reduction in the expression and accumulation of RPS5 mRNA in the absence of ADF4. In total, our data demonstrate that this reduction results in the down-regulation of PTI-signaling in the presence of the bacterial effector AvrPphB. Additionally, we demonstrate this reduction in PTI-signaling is due in part to an ADF4-dependent abrogation of the MPK3/6 branch of the MAPK pathway. From the standpoint of cellular dynamics and the activation of ETI, expression of RPS5 was restored in an ADF4 phosphorylation-dependent manner, demonstrating a link between ADF4 phosphorylation, activity (e.g., F-actin binding), RPS5 mRNA accumulation and subsequent resistance signaling. In addition to elucidating the signaling cascade from perception through MAPK activation, we identified a link between reduced actin cytoskeleton co-localization of ADF4 and the activation of RPS5-mediated resistance in a phosphorylation-dependent manner. In total, the work presented herein represents the first identification of link between the actin cytoskeleton, the dynamic control of ADF4, and the regulation of a resistance gene transcription.
Results
ADF4 is required for RPS5 expression
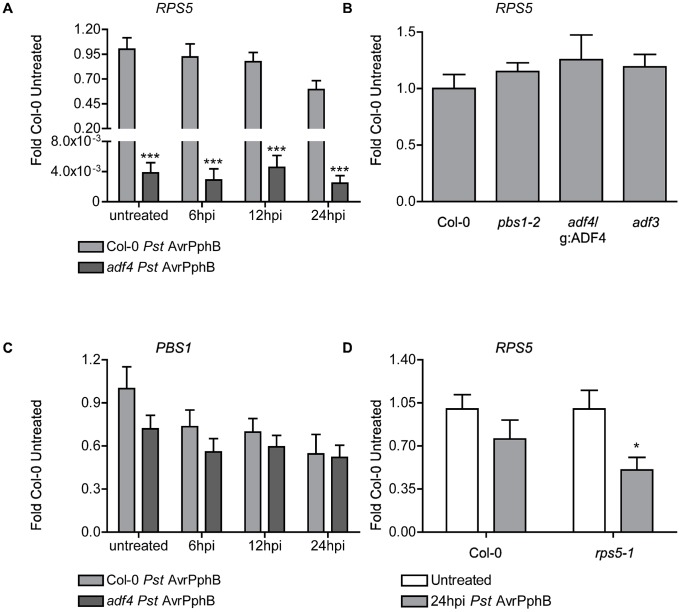
Previous work has shown that Arabidopsis Actin-Depolymerizing Factor-4 (ADF4) is required for resistance to Pst AvrPphB, however, the biochemical and genetic mechanism(s) associated with activation were largely undefined [4]. To elucidate the signaling cascade leading from the recognition of AvrPphB to the activation of resistance, we first investigated the expression of the resistance (R) gene (i.e., RPS5) required for the recognition of AvrPphB. As shown in Figure 1A, we found a significant reduction (∼250-fold) in the accumulation of RPS5 mRNA in the adf4 mutant compared to wild-type Col-0. It was further determined that there is no significant alteration in the expression of ADF4 in Col-0 during the course of infection with Pst AvrPphB (Figure S1). To address the possibility of positional effects in the adf4 T-DNA SALK line, Tian et al. [4] demonstrated that complementation of the adf4 mutant with native promoter-driven ADF4 restored resistance to Pst AvrPphB. Similarly, these lines also showed a restoration in mRNA expression of RPS5 (Figure 1B). The expression of RPS5 in a second ADF mutant, adf3, was not altered (Figure 1B), confirming that the loss of resistance is specific to ADF4, as previously reported [4]. To confirm that the loss of RPS5-mediated resistance in the adf4 mutant is specific to RPS5, we transformed the adf4 mutant with a RPS5-sYFP (adf4/35S:RPS5-sYFP; [20]) to uncouple RPS5 expression from native regulation. As shown in Figure S2, RPS5 mRNA (Figure S2A) and HR-induced cell death following AvrPphB recognition (Figure S2B) was restored. Taken together, this data demonstrates a direct and specific requirement of ADF4 for RPS5-mediated resistance.
Figure 1. ADF4 is required for RPS5 mRNA accumulation and resistance to Pseudomonas syringae expressing the cysteine protease effector AvrPphB.
Time-course of mRNA accumulation of (A) RPS5 and (C) PBS1 in Col-0 and adf4 mutant plants following dip inoculation with Pst AvrPphB. (B) Expression levels of RPS5 in Col-0, pbs1, adf4/g:ADF4, and adf3. (D) RPS5 mRNA accumulation in Col-0 and rps5-1, comparing each to their basal untreated levels at 24 hpi with Pst AvrPphB. Error bars represent mean ± SEM from two technical replicates of two independent biological repeats (n = 4). Statistical significance was determined using two-way ANOVA as compared to Col-0, with Bonferroni post test, where *p<0.05 and ***p<0.001. hpi = hours post inoculation.
To determine the specificity of the ADF4-RPS5 genetic interaction, we investigated if the mRNA expression of additional Arabidopsis R-genes are altered in the adf4 mutant. To this end, we examined the expression of RPS2 [21], RPM1 [22], RPS4 [23] and RPS6 [24]. As an additional measure, we monitored the mRNA accumulation of NDR1 (non race-specific disease resistance-1; [25], [26], [27]), a required component of most CC-NB-LRR defense signaling pathways in Arabidopsis, including RPS5. As shown in Figure S3, we did not observe a reduction in the resting levels of these mRNAs in the adf4 mutant. To confirm that increased susceptibility and the loss of the HR in the adf4 mutant is due to altered expression of RPS5 (i.e., mRNA reduction) and not a reduction in the expression of the AvrPphB cleavage target PBS1 [16], [17], [28], [29], [30], the expression of PBS1 mRNA was also measured. As shown in Figure 1C, we did not detect a significant difference between PBS1 expression in the adf4 mutant and Col-0. Additionally, there was no alteration of RPS5 mRNA expression in the functional PBS1 mutant, pbs1-2 ([30]; Figure 1B).
Our data present a role for ADF4 in the expression of RPS5, but not for the expression of PBS1, suggesting the loss of ETI in the adf4 mutant may be a direct result of reduced RPS5 expression (Figure 1A, Figure 1C). However, whether a role for AvrPphB in the down-regulation of RPS5 expression exists is unknown. In order to address this question, we measured the expression of RPS5 in both Col-0 and the RPS5 point-mutant, rps5-1; the rationale being that if AvrPphB negatively regulates the expression of RPS5, its expression should be reduced in the absence of the activation of ETI. In support of this hypothesis, as shown in Figure 1D, we observed a significant reduction in RPS5 expression in rps5-1 at 24 hpi following inoculation with Pst AvrPphB.
The virulence activity of AvrPphB blocks MAPK signaling in adf4
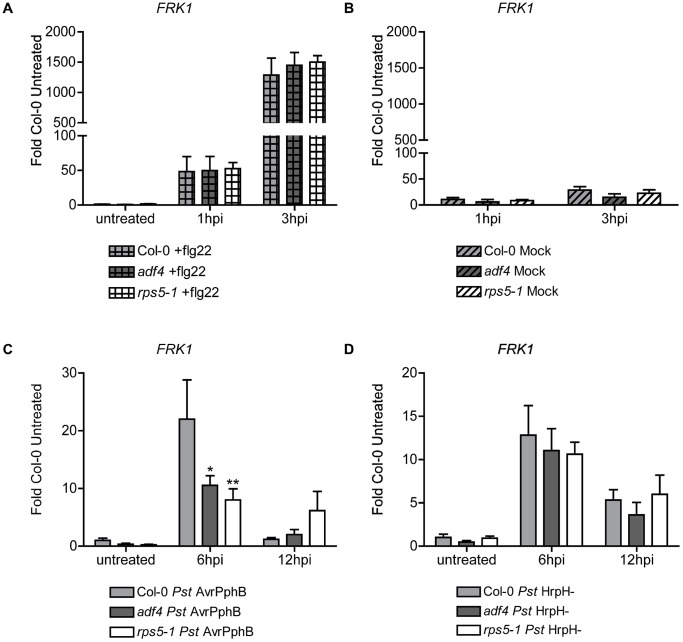
Based on our observations above, we hypothesize that absence of RPS5-derived ETI in adf4 is most likely due to the reduced expression of RPS5. Based on this, and given the significant overlap in signaling of ETI and PTI, particularly with regard to AvrPphB activity [9], [15], [31], we asked if PTI signaling is affected in the adf4 mutant. To address this question, we first monitored the activation of FRK1 expression, a transcriptional marker for FLS2 activation [14], in wild-type (WT) Col-0, adf4 and rps5-1. As shown in Figure 2A, when Col-0, adf4 and rps5-1 plants were treated with flg22, no significant changes in FRK1 mRNA expression were observed, and mock infiltration did little to activate FRK1 (Figure 2A, Figure 2B). As a second, complementary analysis of the fidelity of PTI-based signaling responses in the adf4 mutant, we also monitored root growth inhibition in the presence of flg22 (Chinchilla 2007, same as in the methods section). As shown in Figure S4, we did not observe a significant difference in root growth in adf4 in the presence of flg22 as compared to Col-0. In total, these data demonstrate that flg22-induced PTI-signaling is functional in both the rps5-1 and adf4 mutants. As an additional measure to ensure that the technique employed in Figure 2A and B did not have an adverse effects on RPS5 mRNA expression in either Col-0 or adf4, RPS5 mRNA was monitored following hand-infiltration with either flg22 or mock (i.e., buffer alone). As shown in Figure S5A, we observed that flg22-induced expressional changes of RPS5 mRNA was similar to that of mock, thus assuring the observed activation of FRK1 in Col-0 and adf4 (Figure 2A) can be attributed specifically to flg22, and is independent of the infiltration technique (Figure 2B), or changes in RPS5 expression (Figure S5A).
Figure 2. Flg22-induced receptor kinase 1 expression in the adf4 mutant is reduced when the effector protein AvrPphB is expressed in planta.
Relative expression levels of FRK1 mRNA in Col-0, adf4, and rps5-1 plants when treated with (A) 10 µM flg22, (B) mock inoculated with MgCl2 by hand infiltration (C) Pst AvrPphB, or (D) the hrpH − (Pst hrpH −). Error bars represent mean ± SEM from two technical replicates of two independent biological repeats (n = 4). Statistical significance was determined using two-way ANOVA, as compared to Col-0, with Bonferroni post test where *p<0.05 and **p<0.005. hpi = hours post-inoculation.
Recent work from Zhang et al. [15] suggests that FRK1 mRNA accumulation is reduced in the rps5-1 mutant following flg22 treatment of protoplasts expressing AvrPphB. This raises the question of the relationship between the activation of PTI-signaling in parallel with the activation of ETI. To investigate the downstream signaling response(s) associated with the activation of RPS5-mediated resistance, we measured the expression of FRK1 mRNA accumulation in Col-0, adf4, and rps5-1 when inoculated with Pst AvrPphB. As shown in Figure 2C, we observed a significant decrease in FRK1 mRNA expression in both the adf4 and rps5-1 mutants, as compared to Col-0, at 6 hpi with Pst AvrPphB. Coupled with the results of Zhang et al. [15], this would suggest that the adf4 mutant has a decreased level of RPS5. In support of this, we did not detect a significant difference between FRK1 expression in the adf4 and rps5-1 mutants when inoculated with flg22 (Figure 2A), demonstrating that the mutants had equivalent signaling potential following to FLS2 activation, and that ultimately, the reduction in FRK1 expression is a direct result of a loss in ETI, most likely due to a reduction in RPS5 mRNA expression and accumulation (Figure 1A).
It is possible that our observations described above could be an indirect result of cross-talk of PTI response signaling pathways in adf4 and rps5-1 in the presence of Pst. To test this, FRK1 mRNA expression in Col-0, adf4 and rps5-1 following inoculation with the type three secretion system (T3SS) mutant Pst hrpH − was assessed to differentiate PTI from ETI in the ADF4-RPS5 signaling node. As shown in Figure 2D, we detected no difference in FRK1 mRNA expression between Col-0, adf4 or rps5-1. Additionally, RPS5 mRNA expression following Pst hrpH − inoculation (Figure S5B) and elf18-induced PTI-signaling in Col-0 and adf4 (Figure S6) further supports these observations. When challenged with Pst expressing the catalytically inactive AvrPphB-C98S isoform [16], [18], both WT Col-0 and the adf4 mutant showed increased expression levels of FRK1 mRNA, in agreement with previously published data [15] (Figure S7A). A loss of induction of the HR in Col-0, adf4 and rps5-1 when challenged by Pst AvrPphB-C98S variant [18] confirms the catalytic inactivity of AvrPphB-C98S (Figure S7B).
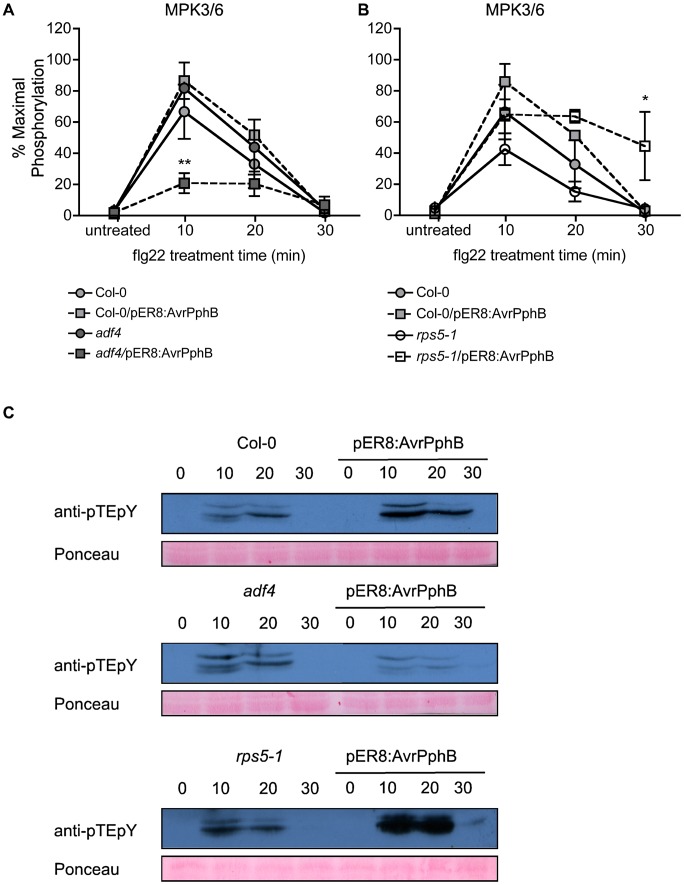
At this point, we reasoned that altered FRK1 expression in both the rps5-1 and adf4 mutants is due to a specific block in the MAPK signal cascade, most likely a function of the virulence activity of AvrPphB in the absence of ETI. To examine MAPK activation in the presence of both flg22 and AvrPphB, in the absence of pathogen, Col-0, adf4 and rps5-1 plants were transformed with an estradiol-inducible AvrPphB construct (i.e., Col-0/pER8:AvrPphB, adf4/pER8:AvrPphB and rps5-1/pER8:AvrPphB) to enable us to monitor the interplay between flg22 perception (i.e., PTI) and AvrPphB (i.e., ETI). As shown in Figure 3A and Figure 3C, when phosphorylation of both MPK3 and MPK6 was measured in response to flg22, a significant reduction in adf4/pER8:AvrPphB was observed as compared to Col-0 at 10 minutes; this reduction was not observed in adf4, and Col-0/pER8:AvrPphB. Interestingly, no significant reduction of MPK3 and MPK6 was observed in the rps5-1/pER8:AvrPphB 10 minutes after flg22 treatment (Figure 3B and Figure 3C). This observation suggests a potential combinatory role for ADF4 in both the expression of RPS5 (Figure 1A), resulting in reduced PTI-signaling (Figure 2C), as well as in the proper regulation of MAPK-signaling in the presence of AvrPphB (Figure 3A and Figure 3C). Estradiol induction of AvrPphB is shown in Figure S8.
Figure 3. Mitogen Activated Protein Kinase (MAPK) phosphorylation is reduced in the adf4 mutant in the presence of AvrPphB.
(A) Percent maximal phosphorylation of the MPK3/6 TEY motif in Col-0 and the adf4 mutant, +/− AvrPphB, followed by 1 µM flg22 treatment. (B) Percent maximal phosphorylation of the MPK3/6 TEY motif in Col-0 and the rps5-1 mutant, +/− AvrPphB, followed by 1 µM flg22 treatment. AvrPphB expression was induced at 48 h pre-treatment with 100 µM estradiol in Col-0, adf4 and rps5-1 mutant plants containing an estradiol-inducible AvrPphB transgene (pER8:AvrPphB). Statistical significance was determined using two-way ANOVA as compared to Col-0 untreated, with Bonferroni post test, where *p<0.05, **p<0.005, n = 3. (C) Western blot analysis of MPK3/6 TEY phosphorylation.
Phosphorylated ADF4 is required for RPS5 expression and subsequent activation of resistance
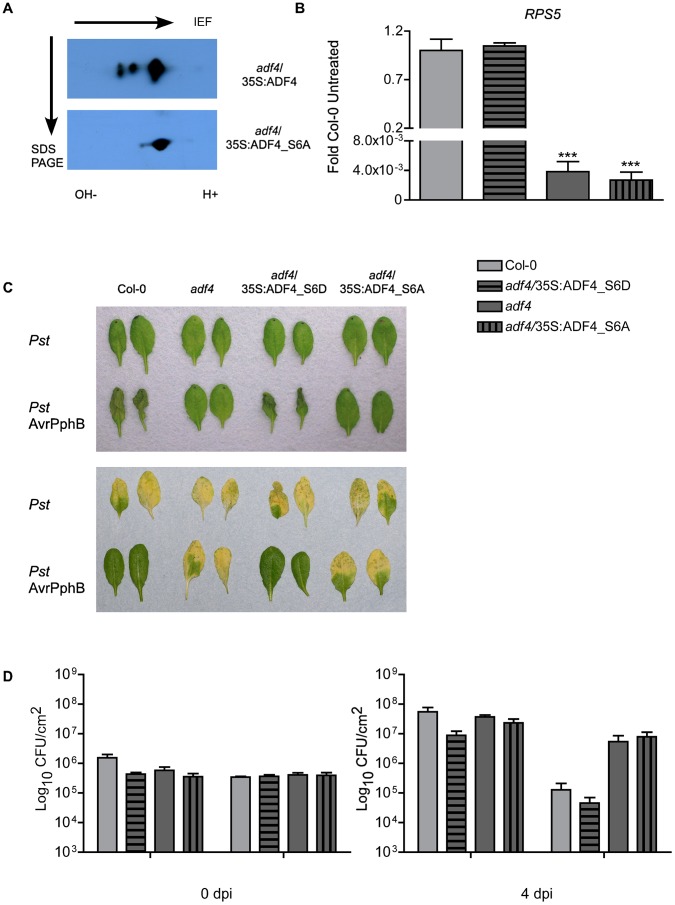
ADF4 mediated actin depolymerization is regulated in large part by the phosphorylation status of ADF. Indeed, previous work has demonstrated that mammalian cofilin/ADF activity is regulated by phosphorylation at serine-3, and that de/phosphorylation at this residue is responsible for the regulating the activation of actin depolymerization [32]. In plants, a direct correlation between the phosphorylation status of ADF and its function has not been demonstrated; however, ADF4 function is presumed to be regulated in a manner similar to that of mammalian cofilin [32], [33], [34]. Herein, we demonstrate for the first time that Arabidopsis ADF4 is indeed phosphorylated at serine-6, and that the phosphorylation status directly correlates with its activity and function of actin cytoskeletal dynamics. ADF4 and the phospho-null ADF4_S6A (i.e., serine-6 to alanine) plant lines were generated by expressing T7:ADF4 and T7:ADF4_S6A in the adf4 mutant under the control of a constitutive promoter (adf4/35S:ADF4 and adf4/35S:ADF4_S6A). As shown in Figure 4A, after 2D isoelectric focusing (IEF) and SDS PAGE, native ADF4 shows a differential IEF profile than the phospho-null ADF4_S6A. In order to determine if phosphorylation of ADF4 affects RPS5 expression, an additional phosphorylation isoform line was generated: a phospho-mimic isotype, reflecting a serine to aspartic acid change at amino acid position 6 (i.e., S6D) expressed in the adf4 mutant background (adf4/35S:ADF4_S6D). As shown in Figure 4B, the phosphomimetic isoform, adf4/35S:ADF4_S6D, restored RPS5 mRNA expression, while the phospho-null isoform, adf4/35S:ADF4_S6A, did not. A second independent transgenic Arabidopsis line expressing the ADF4 phosphorylation mutants were generated and tested for RPS5 expression to ensure that altered mRNA expression was not due to a positional transgene insertion effect (Figure S9A).
Figure 4. Phosphorylation of ADF4 is required for RPS5 mRNA expression.
(A) Western blot of isoelectric focusing (IEF) and SDS PAGE analysis of wild type ADF4 (upper) and phospho-null ADF4_S6A (lower). Arrows indicate direction of IEF and SDS PAGE. (B) The relative expression levels of RPS5 were determined by qRT-PCR. (C) HR phenotypes at 22 hours after bacterial infiltration (upper), disease phenotypes at 4 dpi (lower). (D) Enumeration of bacterial growth at 0 and 4 dpi. HR and bacterial population experiments were repeated at least 3 times. Error bars, representing mean ± SEM, were calculated from two (A; n = 4) or three (D; n = 9) technical replicates of two independent biological repeats. Statistical significance was determined using two-way ANOVA, comparing adf4 to Col-0, with Bonferroni post test, where *p<0.05; ***p<0.001. hpi = hours post inoculation; dpi = days post inoculation.
To confirm that the ADF4 phosphomimetic constructs were functional in their ability to restore resistance in the adf4 mutant, the induction of HR and disease phenotypes, as well as bacterial growth were assessed to determine the relationship between ADF4 phosphorylation and resistance activation through AvrPphB-RPS5. As shown in Figure 4C and 4D, inoculation of adf4 mutant plants expressing the phosphomimetic (ADF4_S6D) with Pst AvrPphB restored the WT Col-0 resistance phenotype, both in terms of HR (Figure 4C, top panel), disease symptoms (Figure 4C, lower panel), and bacterial growth at 4 dpi (Figure 4D). Conversely, inoculation of the phospho-null-expressing plants (i.e., adf4/35S:ADF4_S6A) with Pst AvrPphB resulted in the absence of HR (Figure 4C, top panel), the development of disease symptoms (Figure 4C, lower panel), and an increase growth of the pathogen (Figure 4D), similar to that observed in the adf4 mutant. As a control, to correlate transgene expression levels with our observations, the relative expression levels of both ADF4_S6A and ADF4_S6D were assessed by western blot to confirm that the observed restoration of RPS5 with the phosphomimetic isoform was in fact due to the phosphorylation status and not an artifact of expression (Figure S9B). In total, our data confirms a restoration in resistance, as well as supports the hypothesis that phosphorylated ADF4 is required for resistance to Pst AvrPphB. Similarly, and in agreement our phosphorylation data, expression of FRK1 following Pst AvrPphB inoculation in the adf4/35S:ADF4_S6D mutant was similar to that observed in Col-0, whereas the adf4/35S:ADF4_S6A plants had an FRK1 expression pattern similar to the adf4 mutant (Figure S10).
Phosphorylation of ADF4 reduces its co-localization with F-actin, but does not influence nuclear targeting
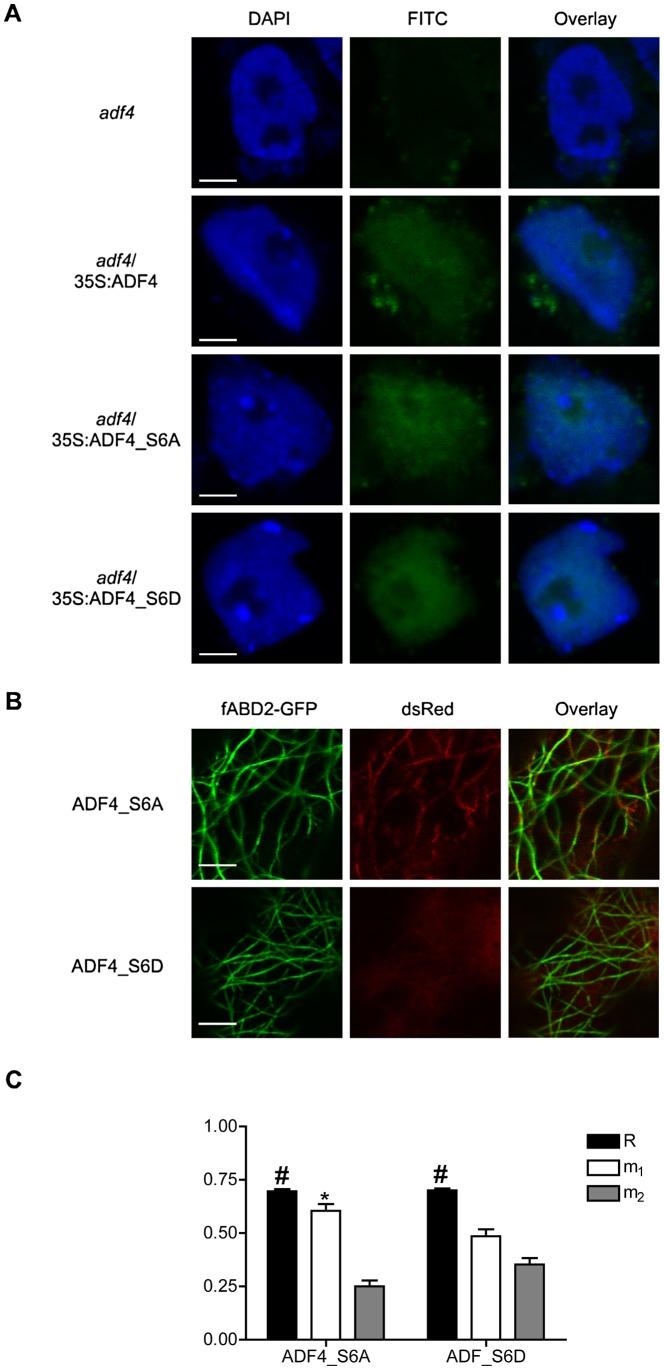
As shown above, phosphorylated ADF4 is required for the accumulation of RPS5 mRNA, as well as for resistance signaling in response to Pst AvrPphB (Figure 4). Previous work has demonstrated the potential for nuclear localization of ADFs, supportive of a role for actin and ADFs in regulating gene transcription [35], [36], [37]. To this end, we sought to determine if translocation of ADF4 into the nucleus is dependent upon the phosphorylation status of ADF4. As shown in Figure 5A, we found that ADF4, ADF4_S6A and ADF4_S6D are all present in the nucleus. This data would suggest that perturbation of RPS5 expression in the adf4/35S:ADF4_S6A plants is not due to an inability of phospho-null ADF4 to enter the nucleus. However, the phospho-null ADF_S6A (ds-Red_ADF4) does show an increased co-localization with the actin cytoskeleton (filamentous Actin Binding Domain 2-GFP; fABD2-GFP), as well as the formation of filamentous like structures in the ADF4_S6A panel (Figure 5B). Conversely, phosphomimetic ADF4_S6D is more diffuse within the cytosol and has reduced co-localization with the actin cytoskeleton (Figure 5B).
Figure 5. Confocal microscopy demonstrates phosphorylation of ADF4 affects cytoskeletal localization, but not nuclear localization.
(A) Laser-scanning confocal microscopy of adf4, adf4/35S:ADF4, adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D isolated nuclei; DAPI stained nuclei (blue), immunochemistry FITC (green), and overlay. Bar = 2 µm. (B) Images of transiently expressed fABD2-GFP (green), dsRed- ADF4 _S6A/_S6D (red), and overlay in Nicotiana benthamiana taken by laser-scanning confocal microscopy. Bar = 5 µm. (C) Graphical representation of the overlay coefficient according to Manders (R) and the co-localization coefficients m1 and m2. Error bars, representing mean ± SEM, were calculated from two biological repeats (n = 40). Overlap coefficient (R) is considered to be co-localized when #R = 0.6 to 1.0, and co-localization coefficients indicate co-localization when *m1>0.5 and *m2>0.5.
To confirm our observations of a phosphorylation-specific alternation in the co-localization of our ADF4 isoforms (i.e., S6A versus S6D) with the actin cytoskeleton, we next performed a red-green analysis on the collected images, calculating the overlap coefficients, according to Manders (R). In short, this analysis will determine the actual overlap of the red/green signals in our collected images [38], providing an in vivo quantification of the co-localization of ADF4 with the actin cytoskeleton. As shown in Figure 5C, both ADF4_S6A and ADF4_S6D were found to have a significant R-value, 0.697±0.009 and 0.701±0.009 respectively, with significant differences in co-localization of ADF4_S6A and ADF4_S6D based on co-localization coefficients m1 and m2. For a red-green pairing, such as was preformed in our analysis, m1 refers to the fraction of red pixels co-localized with green pixels, while m2 is the fraction of green pixels co-localized with red pixels. The m1 values for ADF4_S6A and ADF4_S6D are 0.604±0.032 and 0.485±0.033 respectively, while the m2 values are 0.250±0.028 and 0.353±0.030 (Figure 5C). The co-localization coefficients suggest a significant co-localization of ADF_S6A with fABD2, but not for ADF4_S6D. In total, these observations are in agreement with previous reports of phosphorylated cofilin having reduced binding to both G- and F-actin [39].
Discussion
Understanding the mechanism(s) of pathogen effector recognition, as well as elucidating the putative virulence function(s) of these secreted proteins, provides the foundation for our understanding of innate immune signaling in plants [8]. Using a combination of cell biology, biochemical, and genetics-based approaches, we show that ADF4 is required for the specific activation of RPS5-mediated resistance. In both plants and animals, the actin cytoskeletal network plays a broad role in numerous cellular processes, including cell organization, growth, development and response to external stimuli, including pathogen infection. Herein, we propose a mechanism through which the expression of the R-gene RPS5 is under the control of the actin binding protein ADF4, in a phosphorylation dependent manner, independent of nuclear localization, which subsequently affects co-localization with actin, suggesting a possible cytoskeletal role in gene transcription (Figure 6).
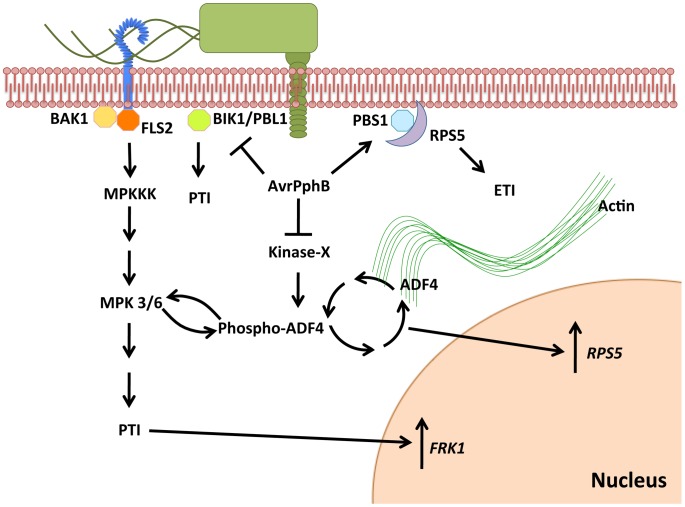
Figure 6. Proposed model illustrating the virulence and avirulence function of the bacterial cysteine protease AvrPphB through an ADF4-dependent mechanism.
Following delivery of AvrPphB into the plant cells by Pst via the T3SS, AvrPphB targets multiple innate immune signaling pathways, including: 1) PTI, via the cleavage of BIK1 kinase; 2) ETI, via the cleavage of the kinase PBS1, a guardee of the resistance protein RPS5. We propose a potential role for AvrPphB in the modulation of actin cytoskeletal dynamics via the targeting of an unknown kinase responsible for the phosphorylation of ADF4 that ultimately results in reduced expression of RPS5, as well as specific down-regulation of MAP kinase signaling. ADF4 translocation into the nucleus is independent of phosphorylation status, however, F-actin co-localization and RPS5 gene expression are dependent upon the phosphorylation of ADF4.
In animal cells, a complex signaling network involving Rho-GTPase activation, actin cytoskeletal dynamics, and the interplay between pathogen virulence has been extensively characterized [1]. In plants, however, the elucidation of the genetic link between pathogen virulence and the regulation of actin cytoskeletal dynamics has only recently been described [4], [5]. In plant-pathogen interactions, the effects of modulation to the host actin cytoskeleton have been best characterized using a combination of pharmacological and cell biology-based approaches to monitor focal orientation of F-actin filaments to the site of infection during fungal pathogenesis [6], [40], [41], [42], [43]. As a first step towards elucidating the mechanism of activation of RPS5-meditated resistance, we examined the expression levels of Arabidopsis genes associated with resistance to Pst AvrPphB. We observed a marked reduction in mRNA levels of the R-gene RPS5, while the protein kinase PBS1 was not affected (Figure 1B, Figure 1C). Additionally, the mRNA levels of R-genes unrelated to the recognition of AvrPphB were not affected in the adf4 mutant (Figure S2B). From these data, we conclude that ADF4 is specifically required for the expression of RPS5 and subsequent resistance to Pst AvrPphB.
The initiation of resistance signaling in plants following pathogen infection engages a multitude of processes, including PRR activation [12], MAPK signaling [14] and transcriptional reprogramming [44]. In the current study, our observation of a reduction in PTI-signaling in the adf4 mutant supports our hypothesis that RPS5 mRNA levels correlate with reduced levels of RPS5 protein. In support of this, we observed a reduction in FRK1 transcript accumulation in the presence of AvrPphB in both the adf4 and rps5-1 mutants. This observation is in agreement with recent reports, including a study demonstrating a physical interaction between FLS2 and RPS5, which would suggest that PTI and ETI signaling is more interdependent than previously hypothesized [45]. Subsequent analysis of upstream MAPK components partially attributed diminished FRK1 mRNA levels to a reduced activation of MPK3/6. Herein, we did not detect a significant reduction in flg22-induced phosphorylation of MPK3/6 in either Col-0/pER8:AvrPphB or rps5-1/pER8:AvrPphB; however, in adf4/pER8:AvrPphB plants, a significant reduction in MPK3/6 phosphorylation following flg22 treatment was observed (Figure 3). MAPK signaling is often primarily associated with PTI (i.e. flagellin activation of the FLS2 receptor); however, many reports have demonstrated the necessity of these components for ETI. For example, in tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum) the requirement of MAPK signaling-components for AvrPto- and N-mediated ETI has been well documented [46], [47], [48]. Our data would suggest that in the case of AvrPphB, R-Avr activation does not specifically induce MPK3/6 within 48 hours of estradiol-induced expression of AvrPphB (Figure 3B). Furthermore, the absence of perturbation to MPK3/6 in the rps5-1/pER8:AvrPphB suggest that while it appears recognition is important for aspects of PTI-signaling i.e. FRK1 mRNA expression (Figure 2C), MAPK-signaling specifically is independent of the need for recognition (Figure 3B).
One possible explanation for reduced MAPK-signaling in the absence of ADF4 reflects the virulence activity of AvrPphB. Indeed, recent work has demonstrated a physical interaction between BIK1 and the FLS2 receptor upon ligand activation – an association that is required for the activation of PTI-signaling [15]. As a mechanism linking with the virulence activity of AvrPphB with both PTI and ETI, cleavage of BIK1 by AvrPphB results in reduced PTI-signaling in the absence of recognition (i.e. the rps5-1 mutant). Our observation of a reduction in MPK3/6 phosphorylation in adf4, but not Col-0 nor rps5-1, would suggest an additional role for ADF4 in regulation of MAPK-signaling, while the reduced FRK1 in adf4 and rps5-1 as compared to Col-0, supports the aforementioned potential virulence activity of AvrPphB, as well as a possible role for recognition (i.e. ETI) in the protection/recovery of the targeted PTI-signaling pathway. Although the mechanism(s) utilized by Arabidopsis to preserve the integrity of the MAPK- and PTI-signaling pathway are not yet fully understood, it is possible that ETI-induced SA accumulation, which has been demonstrated to prime and enhance accumulation of MPK3/6, can be partially responsible for the recovery of MAPK signaling in Col-0 [49]. Another possible contribution to the reduction in PTI-signaling associated with loss of ETI is the aforementioned direct association of FLS2 with RPS5 [45].
In plants, ADF localization is intimately associated with actin reorganization [50]. At present, a full understanding of how translocation of ADFs into the nucleus occurs has not been defined [51]; moreover, the precise function within the nucleus is unclear [36]. The current hypothesis is the translocation of ADFs, as well as other ABPs, into the nucleus may serve a chaperone function [39]. In support of this, actin, as well as several actin-binding proteins (including ADFs), has recently been shown to be present in the nuclei of Arabidopsis [36]. This data support the hypothesis that in addition to actin, ABPs and actin-related proteins (ARPs) may have specific functions within the nucleus, including chromatin assembly and remodeling, as well as participation in various steps of RNA transcription and processing [36], [52]. It is quite possible that ADF4 either facilitates nuclear translocation of specific actin isoforms required for processes related to the expression of RPS5, or, ADF4 itself is required for gene expression (i.e., transcription), as has been demonstrated to be the cased for other ARPs. Mechanistically, however, it is unclear how ADF proteins are translocated into the nucleus. Plant ADFs do not have a conserved nuclear localization signal sequence, as is found in the vertebrate ADFs/cofilins; however, plant ADFs do have two regions with basic amino acids which are similar to domains in other plant proteins that function together as a nuclear localization signal (NLS) [53]. To date, the function of these domains has not been explored. Our data, as well as a recent study by Kandasamy et al. [36], suggests that these two regions of basic amino acids may be both sufficient for translocation to the nucleus, which is not affected by the phosphorylation status of ADF4 at serine-6 (Figure 5).
In the current study, we demonstrate that ADF4 phosphorylation influences both actin cytoskeletal localization, and ultimately, RPS5 mRNA expression (Figure 4, Figure 5). In total, our data provide prima facie evidence for an actin-based regulatory mechanism controlling R-gene expression, and further support the emerging hypothesis that there are critical physiological roles for phosphorylated ADFs in plants [39]. Phosphorylation of cofilin, the predominant ADF found in animal cells, is regulated in part through the action of LIM kinase [54], and results in a reduced affinity of cofilin for F-actin. To this end, ADF phosphorylation has commonly been viewed as an inactivation mechanism, however, recent data suggest that this is not the case [39]. In plant-pathogen interactions, numerous defense-associated processes are regulated by kinase phosphorylation [15], [18], [55], [56]. Conversely, the regulatory mechanisms controlling the phosphorylation, and subsequent regulation of actin dynamics, have not been well established, nor has the crosstalk between ADF regulation and innate immune signaling been fully defined. One obvious disconnect in the link between innate immune signaling and kinase activity in plants and animals is that plants do not have a kinase family homologous to mammalian LIM kinases [54], and thus, ADF phosphorylation is likely mediated by the activity of additional kinase(s), such as calcium dependent protein kinases [33]. One interesting hypothesis in support of the work described herein is that the kinase responsible for the phosphorylation of ADF4 may be a virulence target of AvrPphB. This hypothesis is supported in part by Figure 1D, in which RPS5 expression is significantly reduced in the rps5-1 point mutant following inoculation with Pst AvrPphB. Additionally, the observed requirement of ADF4 for MAPK-signaling in the presence of AvrPphB (Figure 3A) lends support for the idea of ADF4, or the kinases required for its regulation as potential virulence targets. In this regard, such a mechanism would further solidify a link between the virulence function and activity of AvrPphB and the role of the actin cytoskeleton in controlling RPS5 transcription and disease signaling.
Materials and Methods
Plant growth, transformation, and bacterial growth assays
Arabidopsis plants were grown in a BioChambers walk-in growth chamber (model FLX-37; Winnipeg, Manitoba, Canada) at 20°C under a 12-hour light/12-hour dark cycle, with 60% relative humidity and a light intensity of 100 µmol photons m−2s−1. Transformation of Arabidopsis, as well as selection of transformants, was performed as described by Clough and Bent [57].
Pseudomonas syringae pv. tomato DC3000 (Pst) strains were grown as previously described [4]. Four-week-old plants were used for bacterial inoculations. For growth assays and qRT-PCR analyses, whole plants were dip inoculated into bacterial suspensions of 3×108 colony-forming units (cfu) mL−1 in 10 mM MgCl2 containing 0.1% Silwet L-77. Three 0.7 cm diameter leaf disks were collected from three plants for bacterial growth assays, as previously described [4]. The hypersensitive response (HR) was analyzed by hand infiltrating bacterial suspension in 10 mM MgCl2 at 5×107 cfu mL−1 and scoring leaves for tissue collapse 20 to 24 hours post inoculation.
flg22 infiltration was performed at a concentration of 1–10 µM in 10 mM MgCl2, as previously described [27]. Col-0 and adf4 plants were grown upright on plates containing MS media for 10 days±10 nM flg22 in a GC8-2H growth chamber (Environmental Growth Chambers LTD., Winnipeg, Manitoba, Canada) at 20°C under a 12-hour light/12-hour dark cycle, with 60% relative humidity and a light intensity of 120 µmol photons m−2s−1. Analysis of flg22 inhibition of root growth was performed as previously described [12].
Plasmid construction
The native promoter driven pMD1-g:ADF4 (g:ADF4) was constructed as described in Tian et al. [4]. Primer sequences 5′-GCGGTCGACATGGCTAATGCTGCGTCAGGAATGG-3′ (forward ADF4), 5′-GCGGTCGACATGGCTAATGCTGCGGCAGGAATGG-3′ (forward ADF4_S6A), 5′-GCGGTCGACATGGCTAATGCTGCGGACGGAATGG-3′ (forward ADF4_S6D) and 5′- GCGGTCGACATGGCTAATGCTGCGTCAGGAATGG -3′ (reverse for all 3) were used to add SalI restriction enzyme sites (underlined) for cloning ADF4 and its phospho-mutants into pMD1:35S:T7 [27].
Nuclei isolation and immunocytochemistry
Nuclei isolations were conducted as described in Kandasamy et al. [36]. Approximately 1 g of 2- to 3-week old adf4/35S:ADF4, _S6A, and _S6D Arabidopsis seedlings, grown upright on MS medium plates were used for each nuclear extraction. The isolated nuclei were fixed on chrome alum slides, permeabilized, and incubated with primary antibody T7-monoclonal (EMD Chemicals, Gibbstown, NJ, USA), secondary anti-mouse IgG-FITC (Sigma-Aldrich) and DAPI (Sigma-Aldrich) before imaging [36].
Laser-scanning confocal microscopy and co-localization analysis
Isolated nuclei and transiently expressed dsRed-ADF4 constructs, and fABD2-GFP generated using Agrobacterium tumefaciens-mediated transient expression in Nicotiana benthamiana, were imaged using laser confocal scanning microscopy using a 60×/1.42 PlanApo N objective on an Olympus FV1000 (Olympus America Inc, Center Valley, PA), as described in Tian et al. [58]. Co-localization was preformed utilizing FluoView FV1000 (System Analysis Software, Olympus). An area of each image was selected for analysis containing <50% fABD2-GFP occupancy in order to examine true co-localization and not artificial co-localization due to over abundance of fABD2-GFP. Thresholds were set manually to account for background, and overlap coefficient according to Manders (R), and co-localization coefficients m1 and m2 were generated by the FV1000-ASW. Co-localization coefficient equations used can be found in Table S1.
RNA isolation and qRT-PCR analysis
Total RNA was extracted from leaves using the PrepEase Plant RNA Spin kit (USB Affymetrix, Santa Clara, CA, USA). First-strand cDNA was synthesized from 1 µg total RNA using the First-Strand cDNA Synthesis kit (USB Affymetrix). Primers used for quantitative real-time PCR (qRT-PCR) are listed in Table S2. qRT-PCR was performed using the Mastercycler ep Realplex system (Eppendorf AG, Hamburg, Germany), as previously described [27], using the Hot Start SYBR Master mix 2× (USB Affymetrix). Ubiquitin (UBQ10) was used as an endogenous control for amplification. Fold Col-0 was determined using the following equation: (relative expression)/(relative expression of Col-0 untreated), where “relative expression” = 2(−ΔCt), where ΔCt = Ctgene of interest−CtUBQ10.
Statistical analysis
All data were analyzed using GRAPHPAD PRISM Software (San Diego, California, USA). Values are represented as mean ±SEM. All statistical analysis was performed using two-way ANOVA, followed by the Bonferroni post-test as compared to Col-0. In Figure 2C, a two-way ANOVA, followed by the Bonferroni post-test was performed in order to determine if there is a significant difference between rps5-1 and adf4. In Figure S1, an unpaired student t-test with a 95% confidence interval was performed to determine if change over time was significant. P values≤0.05 are considered significant, where *p<0.05; **p<0.01 and ***p<0.005.
Immunoblot analysis
Western blot analysis of phosho-MPK3/6 was performed using 40 µg total protein, utilizing anti-pTEpY (Cell Signaling Technology, Danvers, MA, USA), while analysis of adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D was preformed using 20 µg total protein, utilizing anti-T7-HRP (EMD Chemicals, Gibbstown, NJ, USA), as previously described [26].
2D IEF was preformed on 500 mg of total lysate from adf4/35S:ADF4 and adf4/35S:ADF4_S6A. The lysates were precipitated using chloroform∶methanol (1∶4) and reconstituted in Urea buffer (7 M Urea, 2 M Thiourea, 2% CHAPS, 2% ASA-14, 50 mM DTT, 0.2% Biolyte ampholytes and 0.1% bromophenol blue). Isoelectric focusing was conducted according to manufacturing guidelines at the proteomics core at Michigan State University Research Technology Support Facility (Bio-Rad). Immunoblot analysis was preformed as above.
Supporting Information
ADF4 expression does not change during the course of infection with Pseudomonas syringae expressing AvrPphB. The expression levels of ADF4 in Col-0, over time, when inoculated with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB). Error bars represent mean ± SEM from two technical replicates of two independent biological replicates (n = 4). hpi = hours post inoculation. An unpaired student t-test with a 95% confidence interval was performed to determine if change over time was significant, where p>0.05 is considered not significant.
(TIF)
Expression of 35S:RPS5-sYFP in adf4 recovers the Hypersensitive Response. (A) RPS5 expression in two adf4 mutant-complemented lines expressing 35S:RPS5-sYFP, adf4/35S:RPS5-sYFP-4 and adf4/35S:RPS5-sYFP-12. (B) Hypersensitive Response (HR) in adf4/35S:RPS5-sYFP-4 and adf4/35S:RPS5-sYFP-12 when challenged with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB; left) and untreated (right).
(TIF)
The adf4 mutant does not have altered expression of other resistance genes. The mRNA expression levels of RPS2, RPM1, RPS4, RPS6 and NDR1 in Col-0 and adf4. Error bars represent mean ± SEM from two technical replicates of two independent biological replicates (n = 4). hpi = hours post inoculation.
(TIF)
adf4 mutants are sensitive to fl22 in root length assay. (A) Graphical representation of root lengths of Col-0 and adf4 grown 10 days in the presence (+flg22) or absence (−flg22) of 10 nM flg22. Error bars represent mean ± SEM from two independent biological replicates (n = 32–46). Statistical significance was determined using two-way ANOVA, with Bonferroni post test, where ***p<0.001. (B) Col-0 and adf4 seedlings grown for 10 days±10 nM flg22.
(TIF)
Expression of RPS5 mRNA is not affected by treatment with flg22, or by inoculation with the hrpH − mutant of Pseudomonas syringae . Real-time PCR analysis of RPS5 mRNA accumulation in Col-0 and adf4 following (A) flg22 treatment, mock inoculation or (B) dip-inoculation with the hrpH − mutant of Pseudomonas syringae (Pst hrpH −). Expression was determined by qRT-PCR, utilizing amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, were calculated from two technical replicates of two independent biological repeats (n = 4). Statistical significance was determined using two-way ANOVA as compared to Col-0, with Bonferroni post test, where ***p<0.001. hpi = hours post inoculation.
(TIF)
Both Col-0 and adf4 have induced FRK1 expression when treated with elf18. Relative expression levels of FRK1 in Col-0 and adf4 mutant plants, hand infiltrated with elf18. All expression values were determined by qRT-PCR, with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of one biological repeat (n = 2). hpi = hours post inoculation.
(TIF)
Increased FRK1 expression in Col-0 and adf4 when challenged by Pst AvrPphB-C98S, and HR phenotypes in Col-0, adf4 , and rps5-1 . (A) The expression levels of FRK1 in Col-0, adf4 and rps5-1 following dip-inoculation with Pseudomonas syringae expression the AvrPphB catalytic mutant C98S (Pst AvrPphB-C98S). All expression values were determined by qRT-PCR, with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of three biological replicates (n = 6). hpi = hours post inoculation. (B) HR phenotypes in Col-0, adf4 and rps5-1 when hand inoculated with Pst AvrPphB-C98S.
(TIF)
Estradiol-inducible expression of avrPphB in Col-0, adf4 and rps5-1 . Induction of avrPphB expression in Col-0, adf4 and rps5-1 plants containing the estradiol-inducible avrPphB construct pER8:AvrPphB following 48 h pre-treatment with 100 µM estradiol. Expression values were determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative two technical replicates of one biological repeat (n = 2).
(TIF)
RPS5 mRNA expression in additional adf4 /35S:ADF4_S6A and adf4 /35S:ADF4_S6D lines confirm observed RPS5 expression is not due to positional effects of the transgene nor disproportionate levels of protein levels of protein expression. (A) The expression level of RPS5 in a second set of adf4/35S:ADF4_S6A (adf4/35S:ADF4_S6A-2) and adf4/35S:ADF4_S6D (adf4/35S:ADF4_S6D-2) transgenic lines, as compared to the first line shown in Figure 4A. All expression values were determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of one biological repeat (n = 2). hpi = hours post inoculation. (B) Relative protein levels of ADF4_S6A and ADF4_S6D in adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D as determined by western blot when probed with anti-T7-HRP. Ponceau blot is shown to demonstrate equal loading.
(TIF)
FRK1 expression in adf4 /35S:ADF4_S6A and adf4 /35S:ADF4_S6D lines confirm link between RPS5 expression and FRK1 in the presence of Pseudomonas syringae expressing AvrPphB. Relative expression levels of FRK1 mRNA following dip-inoculation with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB) in adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of two independent biological replicates (n = 4). Statistical significance was determined using two-way ANOVA as compared to Col-0, with Bonferroni post test, where *p<0.05. hpi = hours post inoculation.
(TIF)
qRT-PCR primers used in this study.
(DOCX)
Mathematical equations used for co-localization overlap coefficient determination.
(DOCX)
Acknowledgments
We would like to thank Doug Whitten (MSU RTSF) for assisting with the 2D IEF, Melinda Frame and Elizabeth Savory for confocal microscopy assistance, Caitlin Thireault for assisting with screening of adf4/35S:RPS5-sYFP lines, and Alyssa Burkhardt and Jeff Chang for critical reading of the manuscript. Special thanks to Roger Innes for providing the AvrPphB C98S catalytic mutant and the RPS5-sYFP construct.
Funding Statement
This work was supported by a Early CAREER award from the National Science Foundation (IOS_0641319) and a National Science Foundation Arabidopsis 2010 grant (IOS_1021044) to BD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Day B, Henty JL, Porter KJ, Staiger CJ (2011) The pathogen-actin connection: A platform for defense signaling in plants. Annu Rev Phytopathol 49: 483–506. [DOI] [PubMed] [Google Scholar]
- 2. Gibbon BC, Kovar DR, Staiger CJ (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Snowman BN, Kovar DR, Shevchenko G, Franklin-Tong VE, Staiger CJ (2002) Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, et al. (2009) Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol 150: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clément M, Ketelaar T, Rodiuc N, Banora MY, Smertenko A, et al. (2009) Actin-Depolymerizing Factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21: 2963–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, et al. (2007) Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144: 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- 8. Knepper C, Day B (2010) From perception to activation: The molecular-genetic and biochemical landscape of disease resistance signaling in plants. The Arabidopsis Book 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang J, Zhou J-M (2010) Plant immunity triggered by microbial molecular signatures. Mol Plant 3: 783–793. [DOI] [PubMed] [Google Scholar]
- 10. Gomez-Gomez L, Boller T (2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- 11. Gomez-Gomez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant J 18: 277–284. [DOI] [PubMed] [Google Scholar]
- 12. Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, et al. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Ann Rev Plant Biol 61: 621–649. [DOI] [PubMed] [Google Scholar]
- 14. Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- 15. Zhang J, Li W, Xiang T, Liu Z, Laluk K, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- 16. Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A 104: 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warren RF, Merritt PM, Holub E, Innes RW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152: 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, et al. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233. [DOI] [PubMed] [Google Scholar]
- 19. Feng F, Yang F, Rong W, Wu X, Zhang J, et al. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118. [DOI] [PubMed] [Google Scholar]
- 20. Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, et al. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- 23. Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR- NBS-LRR family of disease-resistance genes. Plant J 20: 265–277. [DOI] [PubMed] [Google Scholar]
- 24. Kim SH, Kwon SI, Saha D, Anyanwu NC, Gassmann W (2009) Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol 150: 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, et al. (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965. [DOI] [PubMed] [Google Scholar]
- 26. Knepper C, Savory E, Day B (2011) The role of NDR1 in pathogen perception and plant defense signaling. Plant Signal Behav 6: 1114–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knepper C, Savory EA, Day B (2011) Arabidopsis NDR1 is an Integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol 156: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao F, Vacratsis PO, Bao Z, Bowers KE, Fierke CA, et al. (2003) Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc Natl Acad Sci U S A 100: 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Innes R (2003) New effects of type III effectors. Mol Microbiol 50: 363–365. [DOI] [PubMed] [Google Scholar]
- 30. Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26: 101–112. [DOI] [PubMed] [Google Scholar]
- 31. Lu D, Wu S, Gao X, Zhang Y, Shan L, et al. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, et al. (1998) Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393: 809–812. [DOI] [PubMed] [Google Scholar]
- 33. Allwood EG, Anthony RG, Smertenko AP, Reichelt S, Drøbak BK, et al. (2002) Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 14: 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shvetsov A, Berkane E, Chereau D, Dominguez R, Reisler E (2009) The actin-binding domain of cortactin is dynamic and unstructured and affects lateral and longitudinal contacts in F-actin. Cell Motil Cytoskeleton 66: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burgos-Rivera B, Ruzicka DR, Deal RB, McKinney EC, King-Reid L, et al. (2008) ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol Biol 68: 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kandasamy MK, McKinney EC, Meagher RB (2010) Differential sublocalization of actin variants within the nucleus. Cytoskeleton 67: 729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meagher RB, Kandasamy MK, Smith AP, McKinney EC (2010) Nuclear actin-related proteins at the core of epigenetic control. Plant Signal Behav 5 E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zinchuk V, Grossenbacher-Zinchuk O (2011) Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr Prot Cell Biol 4: Unit 419. [DOI] [PubMed] [Google Scholar]
- 39. Bamburg JR, Bernstein BW (2010) Roles of ADF/cofilin in actin polymerization and beyond. F1000 biology reports 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hardham AR, Jones DA, Takemoto D (2007) Cytoskeleton and cell wall function in penetration resistance. Curr Opin Plant Biol 10: 342–348. [DOI] [PubMed] [Google Scholar]
- 41. Hardham AR, Takemoto D, White RG (2008) Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol 8: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takemoto D, Hardham AR (2004) The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol 136: 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takemoto D, Jones DA, Hardham AR (2006) Re-organization of the cytoskeleton and endoplasmic reticulum in the Arabidopsis pen1-1 mutant inoculated with the non-adapted powdery mildew pathogen, Blumeria graminis f. sp. hordei. Mol Plant Pathol 7: 553–563. [DOI] [PubMed] [Google Scholar]
- 44. Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qi Y, Tsuda K, Glazebrook J, Katagiri F (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917. [DOI] [PubMed] [Google Scholar]
- 47. Jin H, Liu Y, Yang KY, Kim CY, Baker B, et al. (2003) Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 33: 719–731. [DOI] [PubMed] [Google Scholar]
- 48. Oh CS, Martin GB (2011) Tomato 14-3-3 protein TFT7 interacts with a MAP kinase kinase to regulate immunity-associated programmed cell death mediated by diverse disease resistance proteins. JBC 286: 14129–14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, et al. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jiang CJ, Weeds AG, Khan S, Hussey PJ (1997) F-actin and G-actin binding are uncoupled by mutation of conserved tyrosine residues in maize actin depolymerizing factor (ZmADF). Proc Natl Acad Sci U S A 94: 9973–9978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bamburg JR (1999) Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Ann Rev Cell Develop Biol 15: 185–230. [DOI] [PubMed] [Google Scholar]
- 52. Castano E, Philimonenko VV, Kahle M, Fukalova J, Kalendova A, et al. (2010) Actin complexes in the cell nucleus: new stones in an old field. Histochem Cell Biol 133: 607–626. [DOI] [PubMed] [Google Scholar]
- 53. Shieh MW, Wessler SR, Raikhel NV (1993) Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol 101: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bernard O (2007) Lim kinases, regulators of actin dynamics. Intl J Biochem Cell Biol 39: 1071–1076. [DOI] [PubMed] [Google Scholar]
- 55. Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, et al. (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu J, Elmore JM, Lin ZJ, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 58. Tian M, Win J, Savory E, Burkhardt A, Held M, et al. (2011) 454 Genome sequencing of Pseudoperonospora cubensis reveals effector proteins with a QXLR translocation motif. Mol Plant-Microbe Interact 24: 543–553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ADF4 expression does not change during the course of infection with Pseudomonas syringae expressing AvrPphB. The expression levels of ADF4 in Col-0, over time, when inoculated with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB). Error bars represent mean ± SEM from two technical replicates of two independent biological replicates (n = 4). hpi = hours post inoculation. An unpaired student t-test with a 95% confidence interval was performed to determine if change over time was significant, where p>0.05 is considered not significant.
(TIF)
Expression of 35S:RPS5-sYFP in adf4 recovers the Hypersensitive Response. (A) RPS5 expression in two adf4 mutant-complemented lines expressing 35S:RPS5-sYFP, adf4/35S:RPS5-sYFP-4 and adf4/35S:RPS5-sYFP-12. (B) Hypersensitive Response (HR) in adf4/35S:RPS5-sYFP-4 and adf4/35S:RPS5-sYFP-12 when challenged with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB; left) and untreated (right).
(TIF)
The adf4 mutant does not have altered expression of other resistance genes. The mRNA expression levels of RPS2, RPM1, RPS4, RPS6 and NDR1 in Col-0 and adf4. Error bars represent mean ± SEM from two technical replicates of two independent biological replicates (n = 4). hpi = hours post inoculation.
(TIF)
adf4 mutants are sensitive to fl22 in root length assay. (A) Graphical representation of root lengths of Col-0 and adf4 grown 10 days in the presence (+flg22) or absence (−flg22) of 10 nM flg22. Error bars represent mean ± SEM from two independent biological replicates (n = 32–46). Statistical significance was determined using two-way ANOVA, with Bonferroni post test, where ***p<0.001. (B) Col-0 and adf4 seedlings grown for 10 days±10 nM flg22.
(TIF)
Expression of RPS5 mRNA is not affected by treatment with flg22, or by inoculation with the hrpH − mutant of Pseudomonas syringae . Real-time PCR analysis of RPS5 mRNA accumulation in Col-0 and adf4 following (A) flg22 treatment, mock inoculation or (B) dip-inoculation with the hrpH − mutant of Pseudomonas syringae (Pst hrpH −). Expression was determined by qRT-PCR, utilizing amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, were calculated from two technical replicates of two independent biological repeats (n = 4). Statistical significance was determined using two-way ANOVA as compared to Col-0, with Bonferroni post test, where ***p<0.001. hpi = hours post inoculation.
(TIF)
Both Col-0 and adf4 have induced FRK1 expression when treated with elf18. Relative expression levels of FRK1 in Col-0 and adf4 mutant plants, hand infiltrated with elf18. All expression values were determined by qRT-PCR, with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of one biological repeat (n = 2). hpi = hours post inoculation.
(TIF)
Increased FRK1 expression in Col-0 and adf4 when challenged by Pst AvrPphB-C98S, and HR phenotypes in Col-0, adf4 , and rps5-1 . (A) The expression levels of FRK1 in Col-0, adf4 and rps5-1 following dip-inoculation with Pseudomonas syringae expression the AvrPphB catalytic mutant C98S (Pst AvrPphB-C98S). All expression values were determined by qRT-PCR, with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of three biological replicates (n = 6). hpi = hours post inoculation. (B) HR phenotypes in Col-0, adf4 and rps5-1 when hand inoculated with Pst AvrPphB-C98S.
(TIF)
Estradiol-inducible expression of avrPphB in Col-0, adf4 and rps5-1 . Induction of avrPphB expression in Col-0, adf4 and rps5-1 plants containing the estradiol-inducible avrPphB construct pER8:AvrPphB following 48 h pre-treatment with 100 µM estradiol. Expression values were determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative two technical replicates of one biological repeat (n = 2).
(TIF)
RPS5 mRNA expression in additional adf4 /35S:ADF4_S6A and adf4 /35S:ADF4_S6D lines confirm observed RPS5 expression is not due to positional effects of the transgene nor disproportionate levels of protein levels of protein expression. (A) The expression level of RPS5 in a second set of adf4/35S:ADF4_S6A (adf4/35S:ADF4_S6A-2) and adf4/35S:ADF4_S6D (adf4/35S:ADF4_S6D-2) transgenic lines, as compared to the first line shown in Figure 4A. All expression values were determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of one biological repeat (n = 2). hpi = hours post inoculation. (B) Relative protein levels of ADF4_S6A and ADF4_S6D in adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D as determined by western blot when probed with anti-T7-HRP. Ponceau blot is shown to demonstrate equal loading.
(TIF)
FRK1 expression in adf4 /35S:ADF4_S6A and adf4 /35S:ADF4_S6D lines confirm link between RPS5 expression and FRK1 in the presence of Pseudomonas syringae expressing AvrPphB. Relative expression levels of FRK1 mRNA following dip-inoculation with Pseudomonas syringae expressing AvrPphB (Pst AvrPphB) in adf4/35S:ADF4_S6A and adf4/35S:ADF4_S6D determined by quantitative real-time PCR (qRT-PCR), with amplification of UBQ10 as an endogenous control. Error bars, representing mean ± SEM, are representative of two technical replicates of two independent biological replicates (n = 4). Statistical significance was determined using two-way ANOVA as compared to Col-0, with Bonferroni post test, where *p<0.05. hpi = hours post inoculation.
(TIF)
qRT-PCR primers used in this study.
(DOCX)
Mathematical equations used for co-localization overlap coefficient determination.
(DOCX)