Integral membrane proteins (IMPs) are crucial cellular components, mediating the transfer of material and signals between the environment and the cytoplasm, or between different cellular compartments. Structural and functional analysis of IMPs is important; more than half of current pharmaceutical agents target proteins in this class. [1] IMP characterization is often challenging, and sometimes impossible, because of difficulties associated with handling these macromolecules.[2] IMPs in the native state display large hydrophobic surfaces, which are not compatible with an aqueous environment; therefore, detergents are required to extract IMPs from the lipid bilayer and to maintain the native state of the protein in solution.[3] Nonionic detergents, such as dodecyl-β-D-maltoside (DDM) and octyl-β-D-glucoside (OG), are generally preferred for these applications. Despite the comparatively mild nature of DDM, OG and related detergents, many membrane proteins denature and/or aggregate upon solubilization with these agents.[4]
Diverse strategies have been pursued to develop new tools for solubilization of IMPs from membranes and for maintenance of these proteins in a native-like state in aqueous solution. Techniques that are effective for solubilization are not always optimal for stabilization, and vice versa. These efforts have included exploration of novel amphiphilic molecules that depart from traditional detergent architectures. [5] Specifically tailored amphiphiles that facilitate IMP crystallization are particularly noteworthy.[5l,m,6] Amphiphilic polymers ('amphipols')[7] and discoidal lipid bilayers stabilized by an amphiphilic protein scaffold ('nanodiscs')[8] represent highly innovative approaches for stabilizing IMPs in native-like states in aqueous solution. It is not clear, however, whether either of these approaches can support growth of high-quality crystals for diffraction analysis. Furthermore, neither amphipols nor nanodiscs were designed to extract IMPs from biological membranes. Despite considerable progress in the development of new compounds and strategies for membrane protein solubilization and stabilization, new tools are needed, because many IMPs are currently refractory. Given the great variation in structure and physical properties among membrane proteins, it is very unlikely that a single amphiphile or amphiphile family will be optimal for every system, or even most systems, and exploration of new amphiphilic agents is therefore important for membrane protein biochemistry. Herein we report a class of structurally novel amphiphiles that display favorable behavior, relative to traditional detergents such as DDM, toward a diverse set of membrane proteins.
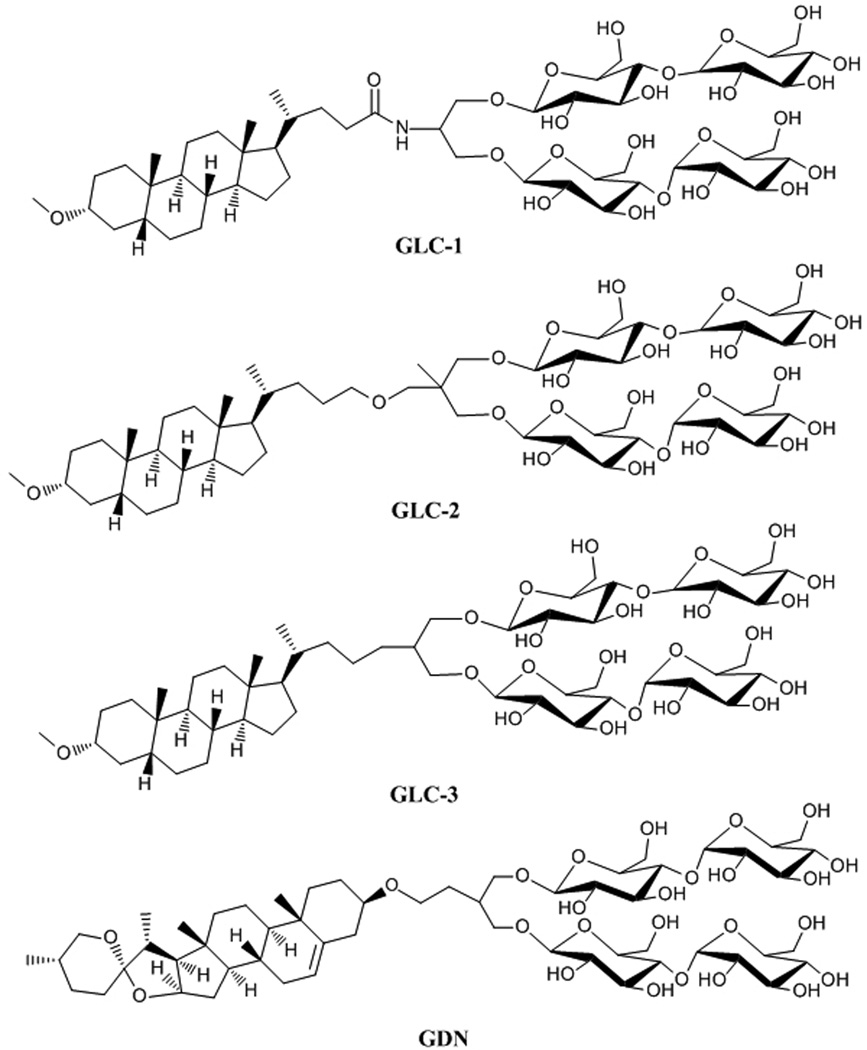
The new amphiphiles (Figure 1) all contain a rigid, steroid-based lipophilic group and a di-maltose hydrophilic group. Three of the new compounds are derived from lithocholic acid and are therefore designated #glyco-lithocholate# amphiphiles (GLC-1, GLC-2 and GLC-3); the fourth is derived from diosgenin and designated #glyco-diosgenin# (GDN). Many previously reported amphiphiles based on steroidal skeletons have been derivatives of cholic acid or deoxycholic acid, including members of the widely-used CHAPS family, cholate-based facial amphiphiles, and tandem-facial amphiphiles. [5a,h,k] In these cases the rigid steroidal units are facially amphiphilic:[9] one side is hydrophilic, displaying either hydroxyl groups or carbohydrate units. In contrast, the hydrophobic units in the GLC and GDN amphiphiles introduced here are hydrophobic on both faces, and the hydrophilic moiety is appended to the periphery of the rigid hydrophobic unit. A cholesterol-based amphiphile, 'Chobimalt,'[5n] was recently described. This compound bears a linear tetrasaccharide that is structurally different from and less synthetically accessible than the di-maltose unit of the GLC and GDN amphiphiles. The capabilities of Chobimalt have been assessed with only a single IMP so far, the human kappa opioid receptor type 1 (hKOR1), and the results were less promising than the findings we report for the new GLC and GDN amphiphiles. hKOR1 could not be extracted in an active form from a biological membrane with chobimalt alone; however, use of chobimalt as an additive to stabilize DDM-solubilized hKOR1 was successful.
Figure 1.
Chemical structures of new amphiphiles (GLC-1, GLC-2, GLC-3 and GDN).
The studies described below involve multiple membrane proteins from various structural classes, in order to assess the potential of the GLC and GDN amphiphiles for broad utility. Initial experiments involved a conservative experimental design, in which IMPs were first extracted from the membrane with a conventional detergent and then assessed for stability after introduction of one of the new amphiphiles as the predominant solubilizing agent. Favorable outcomes of these initial studies led us to evaluate the new agents for membrane-extraction capabilities; GDN proved to be particularly effective in this context.
The four new compounds were easily prepared on a multi-gram scale, as is necessary if they are to serve as research tools (see supporting information). All four are highly soluble and display fairly low critical micelle concentrations (CMC; determined by fluorescent dye solubilization[10]; Table 1). These values are somewhat smaller than the CMC of DDM, which indicates a strong tendency of the new agents to self-assemble. Table 1 provides the hydrodynamic radius (Rh of the micelles formed by each amphiphile, as determined by dynamic light scattering (DLS). The micelles formed by GLC amphiphiles are slightly smaller than those formed by DDM, while the micelles formed by GDN are slightly larger.
Table 1.
Critical Micelle Concentration (CMC) of GLC/GDN amphiphiles and hydrodynamic radii (Rh) of their micelles (Mean ± SD, n = 5).
| MW[a] | CMC (µM) |
CMC (wt %) |
Rh (nm)[b] | |
|---|---|---|---|---|
| GLC-1 | 1112.3 | ~52 | ~0.0060 | 3.22 ± 0.03 |
| GLC-2 | 1127.3 | ~8.0 | ~0.00090 | 3.32 ± 0.04 |
| GLC-3 | 1083.3 | ~7.1 | ~0.00077 | 3.27 ± 0.08 |
| GDN | 1165.3 | ~18 | ~0.0021 | 3.86 ± 0.05 |
| DDM | 510.1 | ~170 | ~0.0087 | 3.42 ± 0.03 |
Molecular weight of detergents.
Hydrodynamic radius of micelles measured by dynamic light scattering.
Non-traditional amphiphiles: Conferring aqueous solubility on membrane proteins generally requires the use of a detergent or other amphiphilic agent. Here we introduce a new class of amphiphiles, each of which is based on a steroidal lipophilic group. These agents have been evaluated with several membrane proteins. The results show that the new amphiphiles confer enhanced stability to a variety of membrane proteins in solution relative to popular conventional detergents, such as dodecylmaltoside (DDM).
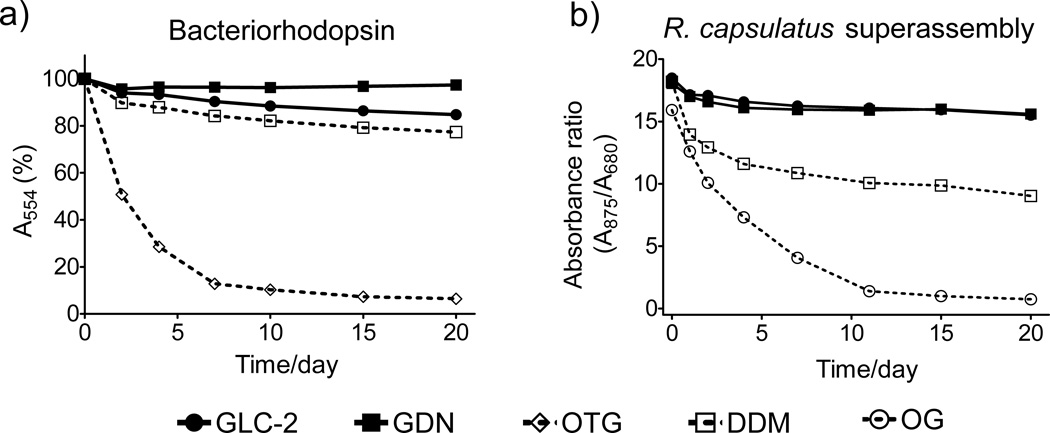
In order to assess the potential utility of new amphiphiles as tools for IMP manipulation, multiple protein systems must be examined. We used DDM as a benchmark for conventional detergent performance in each case, because DDM is probably the most commonly employed amphiphilic agent in membrane protein research. We focused initially on bacteriorhodopsin (bR), which has been commonly used for evaluation of novel amphiphiles because stability can be assessed conveniently via spectrophotometry.[5b,c] bR was extracted from the native purple membrane with 2.0 wt % octyl-β-D-thioglucoside (OTG),[11] and following ultracentrifugation to remove insoluble debris, the bR solution was diluted with amphiphile solutions to give 0.2 wt % OTG + 1.6 wt % new agent or DDM. The absorbance of the solutions at 554 nm was measured periodically over 20 days. Figure 2a shows that two new agents, GLC-2 and GDN, are more effective than conventional detergents OTG and DDM at maintaining the native structure (see Figure S1 for results with other agents). GDN was the best of the new agents, showing negligible loss in protein integrity after 20 days. When we conducted the assay at a lower amphiphile concentration, 0.2 wt % OTG + 0.8 wt % new agent or DDM, similar results were obtained (Figure S1).
Figure 2.
Stability of (a) bR and (b) R. capsulatus LHI-RC superassembly at RT as a function of time. Agents were tested at 0.2 wt % OTG + 1.6 wt % amphiphile for bR and at CMC + 0.04 wt % for the R. capuslatus superassembly.
We turned next to a more challenging system, the photosynthetic superassembly from Rhodobacter capsulatus,[12] which contains the light harvesting I (LHI) complex and the reaction center (RC) complex. This superassembly contains > 30 protein molecules; integrity can be assessed based on the 875 nm/680 nm absorbance ratio.[5i] The superassembly was extracted from the native membrane with 1.0 wt % DDM and purified with DDM at its CMC (0.009 wt %). This preparation was diluted with solutions containing new agents, so that residual DDM (0.0004 wt %) was far below its CMC. The final concentration of each agent was CMC + 0.04 wt %. Figure 2b shows that the LHI-RC superassembly is substantially more stable over 20 days when solubilized by GLC-2 or GDN relative to solubilization with DDM or OG. Similar results were obtained with different detergent concentrations (Figure S2).
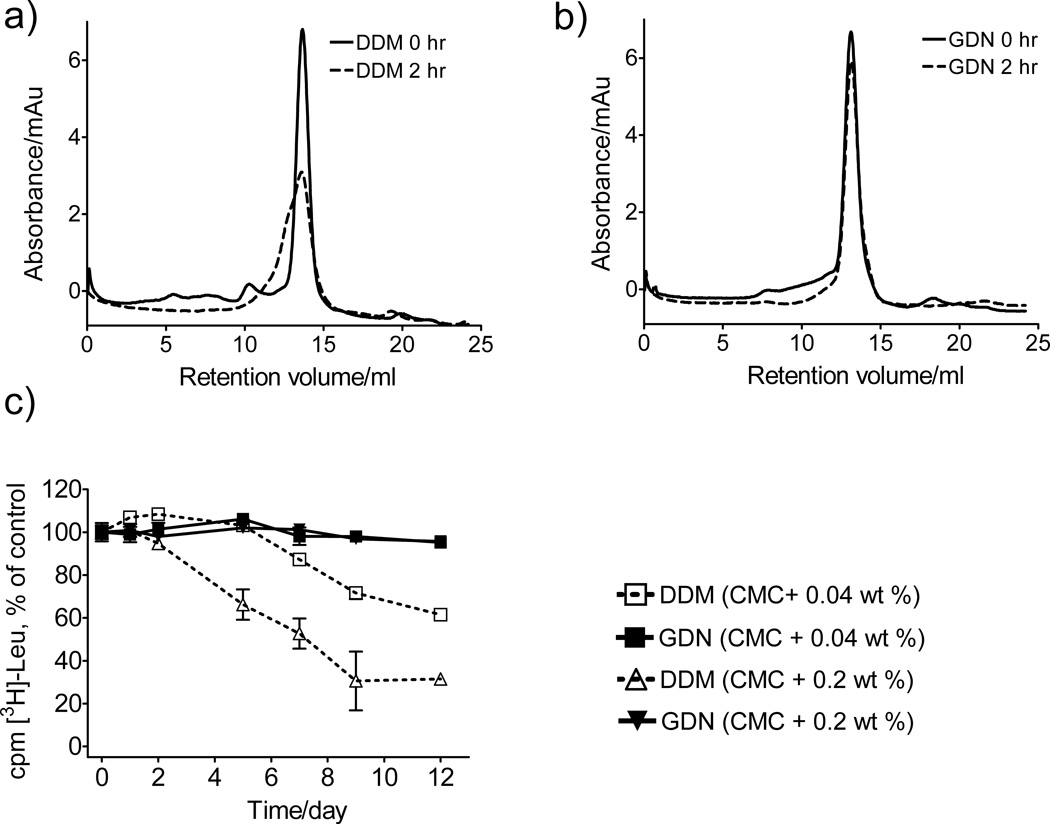
The promising behavior manifested by GDN in terms of the stability of bR and the R. capsulatus superassembly prompted us to examine this amphiphile with the murine cytidine-5’-monophosphate-sialic acid transporter (CMP-Sia).[13] The protein was initially extracted from S. cerevisiae membranes with 1% DDM and isolated in buffer containing 0.03% DDM. The final purified protein (6 mg/ml) was diluted 1:100 into solutions containing DDM or GDN at 0.042 wt % (which corresponds to CMC + 0.033 wt % for DDM and CMC + 0.040 wt % for GDN). The DDM- and GDN-solubilized CMP-Sia were analyzed by gel filtration before and after incubation for 2 hr at 30°C (Figure 3a,b). The results show GDN to be superior to DDM: CMP-Sia solubilized with DDM displays ~50% integrity after the 2 hr period, while GDN-solubilized protein retains >90% integrity. The favorable effect of GDN on CMP-Sia stability was further supported by N-[4-(7-diethylamino-4-methyl-3-coumarinyl) phenyl]maleimide (CPM) assay results when we evaluated the detergents at CMC + 0.04 wt % (Figure S3a).[14] Two other membrane proteins were examined with the CPM assay, the rhomboid intramembrane serine protease GlpG[15] and succinate:quinone oxidoreductase (SQR),[16] both of which were expressed in Escherichia coli. In both cases, the results suggest that the new GLC/GDN amphiphiles are superior to DDM at maintaining native structure (Figure S3b,c).
Figure 3.
(a, b) Gel filtration analysis for CMP-Sia and (c) time course activity of LeuT (scintillation proximity assay (SPA), based on [3H]-Leu binding). Gel filtration analysis was performed at a detergent concentration of 0.042 wt %, before or after incubation of solubilized CMP-Sia at 30 °C for 2 hr. SPA was conducted with detergents at CMC + 0.04 wt % or CMC + 0.2 wt % with LeuT stored at RT. Results are expressed as % activity relative to the day 0 measurements (mean ± s.e.m., n = 2).
The new amphiphiles were evaluated for the ability to maintain the leucine transporter (LeuT) from Aquifex aeolicus in a functional state.[17] The transporter was initially extracted with DDM and then diluted with amphiphile-containing solutions to generate amphiphile concentration of CMC + 0.04 wt % or CMC + 0.2 wt %. At both concentrations, GDN was very effective at maintaining LeuT activity, as indicated by binding of radiolabeled leucine, with preservation of >95 % of initial activity after 12 days (Figure 3c). In contrast, DDM-solubilized LeuT lost activity over this period. The GLC amphiphiles, too, were superior to DDM in terms of maintaining LeuT activity, although they did not match GDN (Figure S4).
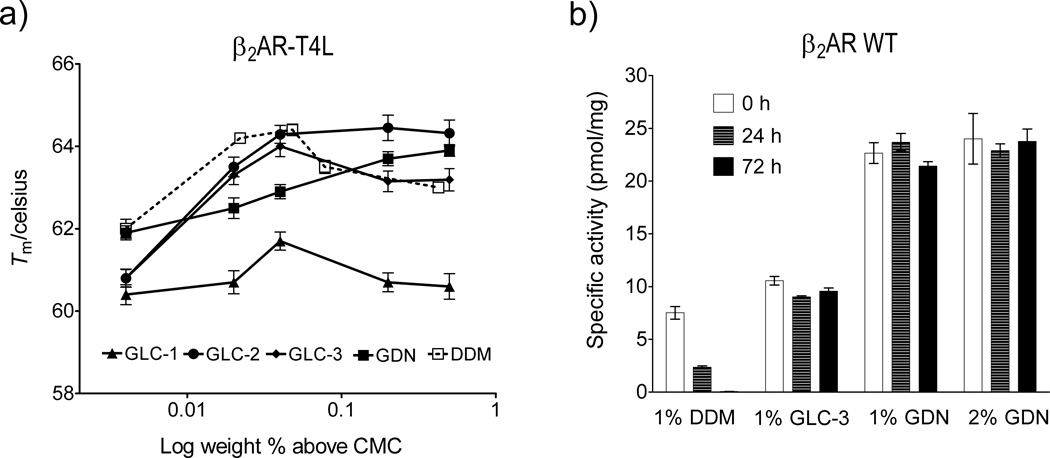
To assess the new amphiphiles with a GPCR, we turned to a human β2 adrenergic receptor-T4-lysozyme fusion protein (β2AR-T4L).[18] Stability was assessed via optical absorption measurements of β2AR-T4L bound to the inverse agonist carazolol. β2AR-T4L was initially solubilized and purified with DDM, and this detergent was then exchanged for the agent to be evaluated. The fluorescence emission maximum of carazolol occurs at 356 nm in aqueous solution, but emission is shifted to 341 nm in the receptor-bound state. The 341:356 nm peak intensity ratio was used to monitor the relative amounts of intact and denatured β2AR-T4L, with Tm defined as the temperature at which the 341:356 nm peak intensity ratio is half-way between that of fully native receptor and the fully denatured receptor. Figure 4a shows how Tm varies as a function of amphiphile concentration. At relatively low concentrations (< CMC + 0.05 wt %), DDM was superior to the new amphiphiles. However, GLC-2 and GDN became superior to DDM at higher concentrations.
Figure 4.
(a) Melting temperatures (Tm) of β2AR-T4L, and (b) β2AR WT activity as a function of time, for proteins solubilized with new amphiphiles or DDM. Tm values for β2AR-T4L are plotted in terms of wt % of the amphiphile. β2AR WT was extracted with 1 wt % or 2 wt % amphiphile, and activity was measured periodically by radioligand-binding assay using the antagonist [3H]-dihydroalprenolol. The solubilized β2 AR WT samples were stored at 4 °C.
In the examples discussed so far, conventional detergents such as DDM were used to extract IMPs from the membrane, and then in most cases the solution of detergent-solubilized protein was diluted with amphiphile-containing solutions to evaluate the new agents. With this approach it is possible that the small amount of residual conventional detergent could affect protein stability. To exclude this possibility, GLC-3 and GDN were used to extract wild type β2AR (β2AR WT)[19] from the membrane. Receptor activity was measured via a binding assay involving the antagonist [3H]-dihydroalprenolol. The DDM-solubilized receptor showed low initial activity and rapidly decomposed (Figure 4b). GLC-3-solubilized receptor showed initial activity similar to that of DDM-solubilized receptor, but in this case activity was maintained over 72 hr. GDN-solubilized β2AR WT showed remarkable behavior: high initial activity (> 3-fold increase relative to that seen with DDM) that did not vary over 72 hr.
We turned to a δ-opioid receptor-T4L fusion (δOR-T4L), another GPCR, to compare GDN with the recently reported amphiphile MNG-3[5l], which has proven to be essential for crystallization of several other GPCR constructs.[6] Consistent with prior observations, MNG-3-solubilized δOR-T4L showed higher activity than DDM-solubilized δOR-T4L (Figure S5). Remarkably, GDN-solubilized δOR-T4L displayed even higher activity. Thus, GDN seems to be very promising for further GPCR solubilization efforts.
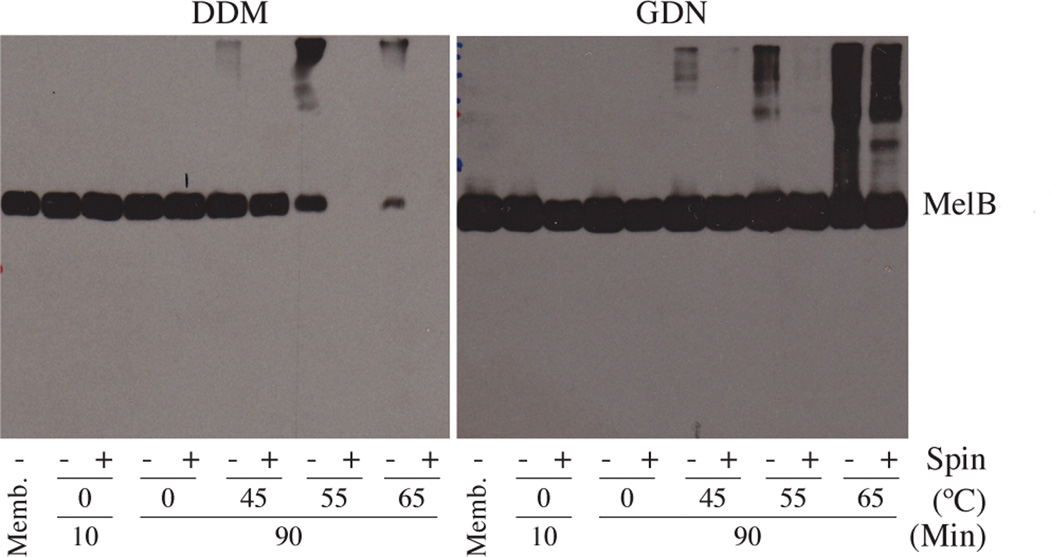
Because GDN displayed particularly favorable behavior in the preceding studies, this agent was further characterized with Salmonella typhimurium melibiose permease (MelB) expressed in E. coli.[20] We used 1.5 wt % DDM or GDN to extract MelB from E. coli membranes at 0°C for 10 min or 90 min and then removed aggregated material via ultracentrifugation. The amount of MelB in solution was determined by SDS-PAGE with immunoblot detection (Figure 5). DDM could quantitatively extract MelB under these conditions; GDN was not quite as efficient in extraction, although a substantial yield of MelB was obtained. We assessed the effect of DDM and GDN on MelB thermostability by solubilizing the protein at the elevated temperatures for 90 min. DDM gave a high yield of soluble MelB at 45°C, but at 55°C no soluble protein was obtained; presumably MelB denatured and aggregated at the higher temperature in the presence of DDM. In contrast, GDN provided large amounts of soluble protein at 55°C and even at 65°C. Interestingly, GDN could quantitatively extract the protein at elevated temperatures. This result raises the possibility that GDN may be more useful for extracting membrane proteins at high temperatures relative to low temperatures (e.g., 4°C or 25 °C). When we used MelB of E. coli, similar results were obtained (Figure S6).
Figure 5.
SDS-12% PAGE and Western blot analysis of MelB. Samples were analyzed by SDS-PAGE analysis, and MelB was detected using anti-histidine tag antibody. Each sample contained 10 µg protein. For extracts generated with each detergent or amphiphile at each temperature, one sample was subjected to ultracentrifugation (+), and a comparison sample was not). (As a control, an untreated membrane sample ('Memb.'; no ultracentrifugation) was included in each gel.
The favorable MelB extraction performance of GDN led us to examine this amphiphile for extraction of other IMPs. Comparable results were obtained when the LHI-RC superassembly was extracted from R. capsulatus membranes with either 2 wt % GDN or 1 wt % DDM (GDN molecular weight is more than twice that of DDM) (Figure S7). For β2AR WT extraction from insect cell membranes, 1 or 2 wt % GDN was more effective than was 1 wt % DDM; only a very small amount of β2AR WT was detected with 1 wt % OG (Figure S7). DDM and GDN were used to extract a CMP-Sia fusion protein bearing green fluorescent protein (GFP) at the C-terminus, after expression in Saccharomyces cerevisiae. The amount of solubilized protein was estimated by total fluorescence. GDN (2 wt %), DDM (1 wt %) and OG (1 wt %) gave ~70%, ~80% and ~50% extraction yields, respectively. Overall, results with several systems show that GDN is generally very effective at extracting embedded proteins from biological membranes.
The results reported here suggest that GDN could prove to be a useful tool for membrane protein research. Promising behavior has been observed for GLC amphiphiles as well, in some cases. It is particularly noteworthy that our tests have included membrane protein systems that vary in terms of structure and function. These studies have included systems that display only limited stability when solubilized with conventional detergents, such as the R. capsulatus photosynthetic superassembly, LeuT, MelB, two forms of β2AR, and δOR-T4L. DDM is probably the most popular conventional detergent for IMP manipulations, and we have shown that GDN consistently matches or exceeds DDM in terms of both extracting and stabilizing diverse membrane proteins.
We recently introduced the MNG amphiphile series,[5l] molecules that are structurally quite different from GDN; MNG amphiphiles have already proven their worth by enabling the acquisition of new GPCR crystal structures.[6] The present report includes a direct comparison new steroidal agent GDN with MNG-3, which suggests that GDN could be generally useful as a new tool for membrane protein solubilization and stabilization. Since the MNG and GDN molecular structures are very different, it is possible that these two types of amphiphile will manifest distinct (and perhaps complementary) advantages among the large set of membrane proteins that have yet to be tamed in the laboratory. The current work did not include comparative studies with amphipols or nanodiscs, both of which have proven to be excellent for stabilization of many membrane protein systems, because neither of these types of agent is likely to be useful for extraction of intrinsic membrane proteins from lipid bilayers.
Typical detergents such as DDM, OG and LDAO have simple alkyl chains as the lipophilic groups. In the presence of a membrane protein, these amphiphiles associate with one another to cover the hydrophobic surfaces of the protein, resulting in protein-detergent complexes (PDCs).[5m,21] The overall architectures of the amphiphiles introduced here are similar to those of classical detergents in that the new compounds are neither facially amphiphilic nor polymeric. Consequently, the new agents are anticipated to associate with membrane protein similarly to classical detergents. Since, however, the lipophilic groups of our new steroid-derived amphiphiles are rigid and flat, we anticipate that these molecules will display a stronger tendency to associate with complementary protein surfaces than do conventional detergents, and we propose that this tendency underlies the favorable solubilization and stabilization properties we have demonstrated here. Cholesterol and its derivatives are known to stabilize the oxytocin receptor and β2AR via direct protein-cholesterol interactions.[22] Because GDN, the GLCs and cholesterol contain similar steroidal moieties, the new amphiphiles could mimic these interactions of cholesterol with membrane proteins.
Important questions remain to be addressed regarding the precise roles of the steroidal units of the new amphiphiles. However, even before these issues are explored, the potential promise of the new amphiphiles as tools for membrane protein manipulation is evident from their success with the range of systems discussed above.
Experimental Section
Synthesis and characterization of amphiphiles, detergent screening and stabilization measurements: Details may be found in the Supporting Information.
Supplementary Material
Acknowledgements
This work was supported by NIH grant P01 GM75913 (S.H.G.), NS28471 (B.K.), the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F4-2007-201924, EDICT Consortium (B.B., K.G., U.G.), the Danish National Research Council (C.J.L., U.G.), the Lundbeck Foundation (S.G.F.R., C.J.L., U.G.), the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant number 2008-0061856 to P.S.C., K.H.C.), and NIH grants R01 GM95538 and R21 HL087895 (to L.G.). R.R. was funded by the Defence, Science and Technology Laboratories (DSTL), Porton Down, UK. We thank Philip Laible (Argonne National Laboratory) and, Elodie Point and Jean-Luc Popot (Université Paris-7) for supplying membrane preparations from R. capsulatus and purple membrane, respectively. We also thank Chiara Lee and David Drew (Imperial College London) and, Jonathan Ruprecht (Medical Research Council-Mitochondrial Biology Unit, Cambridge) and Gary Cecchini (University of California, San Francisco) for providing purified GlpG and SQR samples, respectively.
Footnotes
Supporting information for this article is available on the WWW under http://www.chemeurj.org/ or from the author.
Contributor Information
Prof. Pil Seok Chae, Email: pchae@hanyang.ac.kr.
Prof. Lan Guan, Email: lan.guan@ttuhsc.edu.
Prof. Claus J. Loland, Email: cllo@sund.ku.dk.
Prof. Bernadette Byrne, Email: b.byrne@imperial.ac.uk.
Prof. Brian K. Kobilka, Email: kobilka@stanford.edu.
Prof. Samuel H. Gellman, Email: gellman@wisc.edu.
References
- 1.Liu J, Rost B. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.a) White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; b) Moller JV, le Maire J. J. Biol. Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 4.a) Garavito RM, Ferguson-Miller S. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; b) Bowie JU. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; c) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; d) Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Proc. Natl. Acad. Sci. USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Hjelmeland LM. Proc. Natl. Acad. Sci. USA. 1980;77:6368–6370. doi: 10.1073/pnas.77.11.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schafmeister CE, Miercke LJW, Stroud RM. Science. 1993;262:734–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]; c) McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew. Chem. 2000;112:774–777. Angew. Chem. Int. Ed. 2000, 39, 758-761. [PubMed] [Google Scholar]; d) McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Privé GG. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; e) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; f) Polidori A, Presset M, Lebaupain F, Ameduri B, Popot JL, Breyton C, Pucci B. Bioorg. Med. Chem. Lett. 2006;16:5827–5831. doi: 10.1016/j.bmcl.2006.08.070. [DOI] [PubMed] [Google Scholar]; g) Zhao, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc. Natl. Acad. Sci. USA. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Zhang Q, Ma X, Ward A, Hong W-X, Jaakola VP, Stevens RC, Finn MG, Chang G. Angew. Chem. 2007;119:7153–7155. doi: 10.1002/anie.200701556. Angew. Chem. Int. Ed. 2007, 46, 7023-7025. [DOI] [PubMed] [Google Scholar]; i) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Chae PS, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2010;132:1953–1959. doi: 10.1021/ja9085148. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]; l) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J�L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Hovers J, Potschies M, Polidori A, Pucci B, Raynal S, Bonneté F, Serrano-Vega MJ, Tate CG, Picot D, Pierre Y, Popot J�L, Nehmé R, Bidet M, Mus-Veteau I, Busskamp H, Jung K�H, Marx A, Timmins PA, Welte W. Mol. Membr. Biol. 2011;28:170–180. doi: 10.3109/09687688.2011.552440. [DOI] [PMC free article] [PubMed] [Google Scholar]; n) Howell SC, Mittal R, Huang L, Travis B, Breyer RM, Sanders CR. Biochemistry. 2010;49:9572–9583. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Rasmussen SGF, Choi H-J, Fung J-J, Casarosa P, Pardon E, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:236–240. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rosenbaum DM, Zhang C, Lyons J, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shwa DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunnahara RK, Kobilka BK. Nature. 2011;477:540–541. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka B. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Haga K, Kruse AC, Asada H, Kobayashi TY, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Popot JL. Annu. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]; c) Popot J-L, Althoff T, Bagnard D, Banères J-L, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Crémel G, Dahmane T, dela Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH, Kühlbrandt W, Le Bon C, Martinez KL, Picard M, Pucci B, Rappaport F, Sachs JN, Tribet C, van Heijenoort C, Wien F, Zito F, Zoonens M. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 8.Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y, Ho DM, Gottlieb CR, Kahne D, Bruck MA. J. Am. Chem. Soc. 1992;114:7319–7320. [Google Scholar]
- 10.Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 11.Bazzacco P, Sharma KS, Durand G, Giusti F, Ebel C, Popot J-L, Pucci B. Biomacromolecules. 2009;10:3317–3326. doi: 10.1021/bm900938w. [DOI] [PubMed] [Google Scholar]
- 12.Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]
- 13.Newstead S, Kim H, von Heijne G, Iwata S, Drew D. Proc. Natl. Acad. Sci. USA. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov A, Mileni M, Chien EY, Hanson MA, Stevens RC. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Urban S. Biochem. J. 2010;425:501–521. doi: 10.1042/BJ20090861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsefield R, Iwata S, Byrne B. Curr. Protein Pept. Sci. 2004;5:107–118. doi: 10.2174/1389203043486847. [DOI] [PubMed] [Google Scholar]
- 17.a) Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]; b) Quick M, Javitch JA. Proc. Natl. Acad. Sci. USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H�J, Yao X�J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 19.Kobilka BK. Trends Pharmacol. Sci. 2011;32:213–218. doi: 10.1016/j.tips.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a) Timmins P, Pebay-Peyroula E, Welte W. Biophys. Chem. 1994;53:27–36. doi: 10.1016/0301-4622(94)00073-5. [DOI] [PubMed] [Google Scholar]; b) Le Miare M, Champeil P, Moller JV. Biophys. Acta. 2000;(1508):86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 22.a) Gimpl G, Fahrenholz F. Biochim. Biophys. Acta. 2002;(1564):384–392. doi: 10.1016/s0005-2736(02)00475-3. [DOI] [PubMed] [Google Scholar]; b) Yao Z, Kobilka B. Anal. Biochem. 2005;343:344–346. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]; c) Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.