Abstract
Existing evidence suggests that endothelial lipase (EL) plays an important role in high-density-lipoprotein (HDL) metabolism. Because rabbits are a useful animal model for the study of human lipid metabolism and atherosclerosis, we characterized rabbit tissue EL expression and investigated its relationship with plasma HDL levels in normal and hyperlipidemic rabbits. First, we cloned the rabbit EL (rEL) cDNA and gene. Using Northern blotting, real-time RT-PCR, Western blotting, and in situ hybridization, we revealed that rEL mRNA was highly expressed in the cholesterol synthesis-related organs including the liver, testis, and adrenal along with its expression in the lung, kidney, bone marrow, and small intestine. Interestingly, Watanabe heritable hyperlipidemic (WHHL) rabbits, a model of human familial hypercholesterolemia, had lower plasma levels of HDLs than normal rabbits. The plasma HDL levels in WHHL rabbits were inversely associated with high levels of plasma rEL proteins and hepatic expression of rEL mRNA. Injection of rEL-specific antisense oligonucleotides into rabbits resulted in an elevation of plasma large-sized HDLs. Furthermore, we demonstrated that the rEL mRNA was expressed by both endothelial cells and macrophages in the lesions of aortic atherosclerosis of WHHL rabbits. EL is expressed in multiple tissues and may have many physiological and pathophysiological functions such as in the regulation of cholesterol metabolism and atherosclerosis. Our results suggest that EL is an important regulator of plasma HDL levels in rabbits.
Keywords: Endothelial lipase, HDL, rabbit, hypercholesterolemia, antisense
INTRODUCTION
Endothelial lipase (EL) which was discovered independentlyby two laboratories in 1999(1, 2), belongs to the triglyceride lipase family including lipoprotein lipase (LPL) and hepatic lipase (HL). Despite high sequence similarities, the members of this family have different relative preferences for lipid substrates. EL has high phospholipase activity but low triglyceride lipase activity, which is opposite to the low phospholipase activity and high triglyceride lipase activity of LPL. On the other hand, HL hydrolyzes triglycerides and phospholipids equally(3). Compelling evidence suggests that EL plays an important role in the HDL metabolism(3, 4). In EL knock-out (KO) mice, deletion of the EL gene resulted in increased plasma levels of HDL-cholesterol (HDL-C) whereas overexpression of human EL gene led to the reduction of HDL-C levels in EL transgenic mice(5). Moreover, clinical studies revealed that the plasma EL mass is inversely associated with plasma HDL-C levels and high level of EL proteins correlates with inflammation, metabolic syndrome, and coronary atherosclerosis(6). In addition to HDL metabolism, EL is involved in apoB-containing lipoprotein metabolism(7). These findings suggest that inhibition of EL would elevate the HDL-C levels. On the other hand, EL may directly or indirectly participate in the pathogenesis of atherosclerosis through catalytic and non-catalytic “bridging” functions although this hypothesis has not been definitively verified. EL inactivation was found to be anti-atherogenic in apo-E KO mice(8) but this was not confirmed by other studies(9) even though EL immuno-reactive proteins were found in human atherosclerotic lesions(10). Therefore, it remains to be clarified whether EL is anti-atherogenic or pro-atherogenic in mouse models. The major concern regarding whether inhibition of EL activity is potentially beneficial for the treatment of atherosclerosis was raised from the finding that deletion of the EL gene in KO mice also resulted in elevation of LDL in addition to HDL(5, 8).
Because there are many differences in lipid metabolism and cardiovascular system between humans and mice, it may not be adequate to use mouse models to verify EL-inhibition therapeutic effects for humans. Yasuda et al. suggested that there is a need to develop alternative non-murine animal models to elucidate EL functions in vivo(11). Rabbits are a useful animal model for the study of human lipid metabolism and atherosclerosis because they can easily develop hypercholesterolemia and atherosclerosis on a cholesterol diet and the lesions of atherosclerosis of both aorta and coronary arteries resemble to human atherosclerotic pathology(12, 13). Like humans but unlike mice, rabbits have abundant plasma CETP and exhibit hepatic apoB100 and intestinal apoB48 synthesis, and their lipoprotein profiles are LDL-rich(13). Availability of Watanabe heritable hyperlipidemic rabbits, a model for human familial hypercholesterolemia has provided many possibilities for the study of human lipoprotein metabolism and atherosclerosis. We envision that investigation of the possible role of EL in atherosclerosis using transgenic rabbits with overexpression and/or deletion of the EL gene should provide valuable insights into the pathogenesis of the disease. Towards this undertaking, it is essential to determine the rabbit endogenous EL expression and physiological functions in vivo. In the current study, we cloned rabbit EL cDNA, assembled/annotated the protein from the genomic structure of the gene, and investigated the tissue distribution of EL mRNA expression patterns in different tissues in rabbits. Our studies showed that EL is expressed in multiple tissues and mediates the plasma levels of HDLs in rabbits. EL was expressed in the lesions of aortic atherosclerosis of WHHL rabbits.
MATERIALS AND METHODS
Rabbits
Normal Japanese white (JW) rabbits and myocardial infarction-prone Watanabe heritable hyperlipidemic (WHHLMI) rabbits were used for the current study as described previously(14, 15). The tissues from different parts of rabbits were freshly collected for RNA and protein extraction, as well as in situ hybridization and immunohistochemical staining (see below). This study was approved by the Animal Care Committee of the University of Yamanashi and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Rabbit EL cDNA cloning and expression analysis
Total RNA was extracted from rabbit tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA). The first-strand cDNA was generated by the SuperScript III First-Strand Synthesis System (Invitrogen) with random primers following the manufacturer’s instructions. The cDNA served as a template for rabbit EL (rEL) cloning. Using human EL mRNA sequences (accession No: NM_006033), we performed BLAST search against the rabbit genome trace archive database (Oryctolagus cuniculus-WGS) provided by the National Center for Biotechnology Information. On the basis of the alignment results, we first assembled an electronic version of putative rEL mRNA sequence including 5′UTR, cds, and 3′UTR. To clone the rEL cDNA, we designed several pairs of primers for RT-PCR with PfuUltra high-fidelity DNA polymerase (Agilent Technologies, Inc., Santa Clara, CA). The amplified DNA was recovered from the agarose gel using QIAquick Gel Extraction kits (Qiagen, Valencia, CA) and cloned into pCR4Blunt-TOPO (Invitrogen). Then, several random clones from each PCR product were collected and sequenced with M13Forward and M13Reverse primers. The rEL mRNA sequences were finally assembled. For Northern blotting analysis, 10μg of total RNA from each tissue was fractionated on a 1.2% agarose gel and then transferred to nylon membranes. These membranes were hybridized with 32P-labeled rEL cDNA probe at 68°C for 16 h. After hybridization, the membranes were washed at 65°C in the presence of 0.1% SSC and 0.1% SDS. Visualization was achieved by exposure to BAS-2500 IP Reader (Fujifilm, Tokyo, Japan). The mRNA levels of the rEL were also evaluated by real-time RT-PCR using SYBR®Premix Ex Taq™ kits (Takara Bio Inc., Tokyo) according to the manufacturer’s instructions(14). The primers are listed in Table 1.
TABLE 1.
PRIMERS FOR REAL-TIME RT-PCR
| Gene | Sequence | Product (bp) | Tm (°C) | Accession No. |
|---|---|---|---|---|
| Rabbit EL | F: 5′-GAG CGG GAG CTC AGT ATC AG R: 5′-CAG CCG TCT CGA CAC TTG TA |
140 | 60.0 | NM_001195638 |
| Human EL | F: 5′-CTC GGG AAT GTC CAC TTG AT R: 5′-AGA GCC TCT TGT GGA TGT CG |
148 | 60.0 | NM_006033 |
| Rabbit GAPDH | F: 5′-ATC ACT GCC ACC CAG AAG AC R: 5′-GTG AGT TTC CCG TTC AGC TC |
146 | 58.3 | NM_001082253 |
| Human GAPDH | F: 5′-CAA TGA CCC CTT CAT TGA CCTC R: 5′-AGC ATC GCC CCA CTT GAT T |
172 | 60.9 | NM_002046 |
Generation of polyclonal antibody against rEL
A peptide corresponding to rEL #469~490 amino acids (KTSIAPGQELWFYKCRDGWRMK) was synthesized and used as an antigen to immunize rabbits for the generation of polyclonal Abs, which was performed by YenZym Antibodies, LLC (South San Francisco, CA). This rEL Ab can detect rEL proteins expressed by transfected 293 cells but showed no cross-reaction with human EL proteins (S-Fig. 1). Immuno-reactive rEL proteins in the post-heparin plasma and liver were investigated by Western blotting as described previously with some modifications(16, 17). The membrane was probed with the anti-rEL Ab (1:200) for 16 hours at 4°C and washed three times with wash buffer. Then, the membrane was incubated with Protein A-conjugated horseradish peroxidase (GE Healthcare, Piscataway, NJ) for 1 hour at room temperature for detecting rEL expression.
FISH mapping of rEL gene on G-banded rabbit chromosomes
Fluorescence in situ hybridization (FISH) analysis was performed on cultured rabbit fibroblasts to determine the chromosomal localization of the rEL gene as previously described(18). In brief, rabbit embryos were collected at day 13–14 of gestation and a piece of the skin was minced and digested using a solution of 0.25% trypsin and 0.25 mM EDTA for 30 min at 37°C. The cells were cultured in T75 flasks in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose (4.5 g/L) supplemented with 10% fetal bovine serum (Invitrogen). The fibroblasts between passages 2 and 5 were used for the current study. Mitotic chromosomes were prepared from rabbit fibroblasts as reported previously(19). The cells were treated with colcemid (0.1 μg/ml) for 1.5 h and then harvested by trypsinization. After treatment with hypotonic solution (75 mM KCl:1% Na citrate, 1:1) for 6–10 min at 37°C, the cells were fixed with acetic acid:methanol (1:3), dropped onto ice-cold glass slides, and air-dried. The slides were kept at −80°C until use. G-banding karyotyping was performed to identify each chromosome before FISH (20). Karyotyping was performed using the CytoVision imaging system (Applied Imaging Corp., San Jose, CA) based on the standard karyotype and nomenclature of Oryctolagus cuniculus(21).
A BAC clone, LB1-130A10, which carried the rEL gene, was selected as the FISH probe for mapping the gene by screening rabbit BAC filter sets (LBNL-1 White Rabbit BAC Library from the BACPAC Resources, Oakland, CA) with the rEL cDNA sequences. The BAC DNA was isolated and purified using the QIAGEN Plasmid Midi Kit (QIAGEN) and labeled with biotin using BioNick DNA Labeling System (Invitrogen, Carlsbad, CA). G-banded metaphase slides that were karyotyped were treated with methanol/acetic acid fix solution, followed by incubation in 2xSSC at 73°C for 2 min and 10% phosphate buffered formalin at room temperature (RT) for 5 min. The slides were then dehydrated sequentially in 70%, 85%, and 100% ethanol at RT for 1 minute each. Approximately 10 μL of hybridization mix solution (50% formamide, 10% dextran sulfate and 2XSSC) containing 1 μg of labeled BAC DNA, 2 μg of sheared rabbit genomic DNA, and 5 μg of sheared salmon sperm DNA (Eppendorff, Hauppauge, NY) was denatured at 75°C for 5 min, re-annealed at 37°C for 30 min and then applied onto a destained slide. The slides were incubated with the hybridization mix solution at 75°C for 5 min before hybridization at 37°C overnight. After hybridization, the slides were washed with 0.4xSSC/0.3% NP40 at 73°C for 2 minutes and 2xSSC/0.1% NP40 at RT for 1 min. The FISH signals were visualized by avidin-fluorescein staining (Roche, Indianapolis, IN) and DAPI II counterstaining (Abbott Molecular, Des Plaines, IL), and analyzed using the CytoVision imaging system (Applied Imaging Corp., San Jose, CA). The rEL gene location was determined by comparing the FISH images with the corresponding karyotyped metaphase cells.
Preparation of rEL specific antisense oligonucleotides (ASOs)
On the basis of the rEL cDNA sequences that we cloned, we designed 90 sequences of 20-mer oligonucleotides by a linear “shot-gun” approach without targeting repeated sequences(22). We performed a rapid-throughput screen of the efficiency of 90 ASOs using cultured normal rabbit hepatocytes and evaluated rEL mRNA expression after treatment with 150 nM of each ASO for 24 h by real-time RT-PCR. We selected the top ten ASOs that showed the greatest potency in inhibiting rEL expression and further assessed their inhibitory activity at different concentrations (25, 50, 150, and 300 nM) (S-Fig. 2). On the basis of IC50 values, we tested four ASOs for their rEL inhibiting activity in JW rabbits in vivo. Finally, we obtained two rEL ASOs (designated as ISIS 440792 and ISIS 440825) that showed equal efficacies of inhibiting rEL expression of rabbit hepatocytes. Both ASOs had a uniform phosphorothioate backbone and a 20-base chimeric design with 2′-O-(methoxy)-ethyl (2′-MOE) modifications on the first five and last five bases(23). These modifications enhance their binding affinity to complementary sequences and their resistance to the action of nucleases. In the current study, we used ISIS 440825 for the injection experiment. A mismatch sequence (ISIS 141923), which has the same chemical composition as the rEL ASOs but without complementarity to any known rabbit gene sequences, was used as a control.
Analysis of plasma lipids and lipoproteins after ASO treatment
Male JW rabbits and WHHLMI rabbits (4-month-old) were intravenously injected with rEL ASO in saline solution (40 mg/kg), along with mismatched control, weekly for 6 weeks. Plasma total cholesterol, triglycerides, and HDL-cholesterol were determined using Wako assay kits (Wako Pure Chemical Industries, Osaka, Japan), and lipoproteins were analyzed using high-performance liquid chromatography (HPLC) (17) by Skylight Biotech, Inc. (Tokyo, Japan) at the end of the experiment. After 6 weeks of rEL ASO treatment, all rabbits were sacrificed and rEL expression in the liver was determined by real-time RT-PCR and Western blotting(24).
In situ hybridization and immunohistochemical staining of atherosclerotic lesions
Aortic atherosclerotic lesions of WHHL rabbits (7-month-old) and normal aorta of normal rabbits were quickly frozen in liquid nitrogen and then embedded in OCT compound solution. Serial frozen sections were used for in situ hybridization for rEL and immunohistochemical staining for macrophages and smooth muscle cells as described previously(24). The full protocol for in situ hybridization can be found in the Supplemental Methods.
Statistical analysis
All values are expressed as mean ± SEM and statistical significance was determined. Student’s t-test was used to compare the results. In all cases, statistical significance was set at P< 0.05.
RESULTS
Cloning and characterization of rEL cDNA
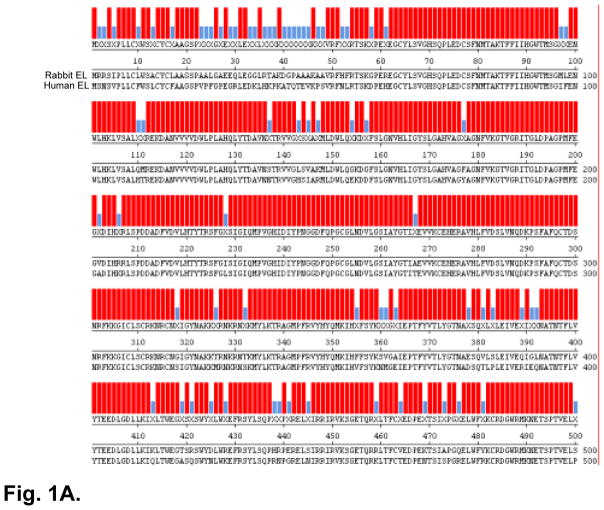
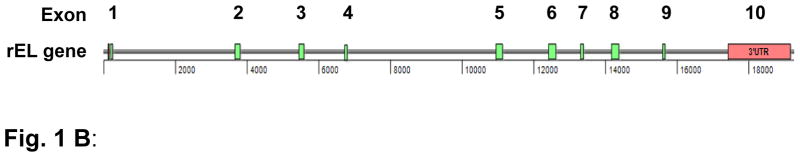
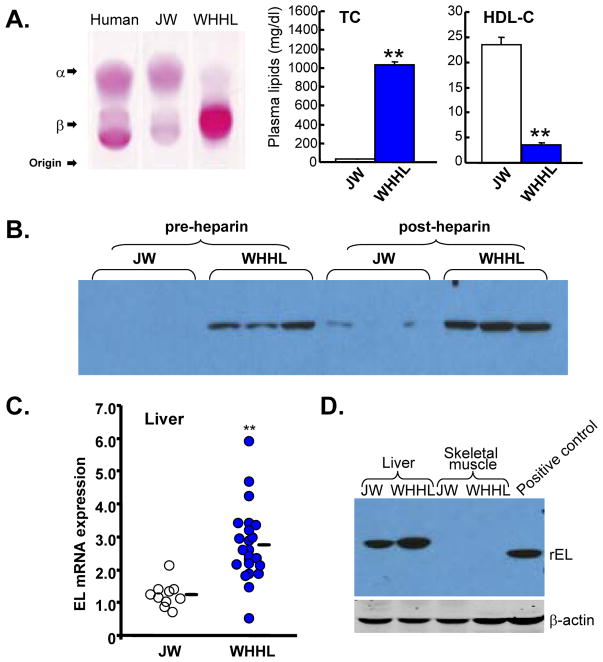
We successfully obtained the rEL cDNA sequences and submitted them to Genbank (accession No.: NM_001195638). The rEL cDNA contains an open reading frame of 1503 bp that codes for a 56.1 kDa protein. The amino acid sequence of rEL exhibits 85.4% identity with human EL (Fig. 1A). The predicted rEL protein size is similar to that of human EL(2). For example, the rEL contains a lipase domain (49-344 aa) and a PLAT/LH2 domain (347-483 aa) determined by Pfam family search (http://pfam.janelia.org/). The rEL enzymatically catalytic sites are S169, D193, and H274, which are conserved in human, mouse, and rat EL. 1-17 aa is the signal peptide predicted by online software (SIG-Pred: Signal Peptide Prediction). By the same strategy using the rEL cDNA sequences, we assembled the rEL gene by serial BLAST search. The rEL gene contains 10 exons, similarly to human and mouse EL gene (Fig. 1B). Our findings redefined the genomic structure of the rEL gene, which was recently predicted on the basis of the rabbit genome sequencing data (www.ncbi.nlm.nih.gov/gene/100342293). In addition, FISH analysis revealed that the rEL gene was located in the chromosome 9q14.3 15.1 region (Fig. 2), which is consistent with the predicted chromosomal location reported by the Broad Institute (www.ncbi.nlm.nih.gov/gene/100342293). Of note, the rabbit chromosome 9 is homologous to the human chromosome 18(25). The EL molecular features of human, rabbit, mouse, and rat are summarized in Table 2. In general, rEL possesses higher identity of both mRNA and protein sequences than mouse and rat EL.
Fig. 1.
A. Comparison of EL amino acid sequences between rabbit and human.
The resultant amino acid sequence of rabbit EL exhibits high identity (85.4%) to human EL. B: Rabbit EL gene structure (introns and exons). Rectangular boxes show the exons. Exons with green color are the coding region of rabbit EL mRNA. Pink color indicates 5′ and 3′ untranslated region (UTR) of rabbit EL mRNA.
Fig. 2.
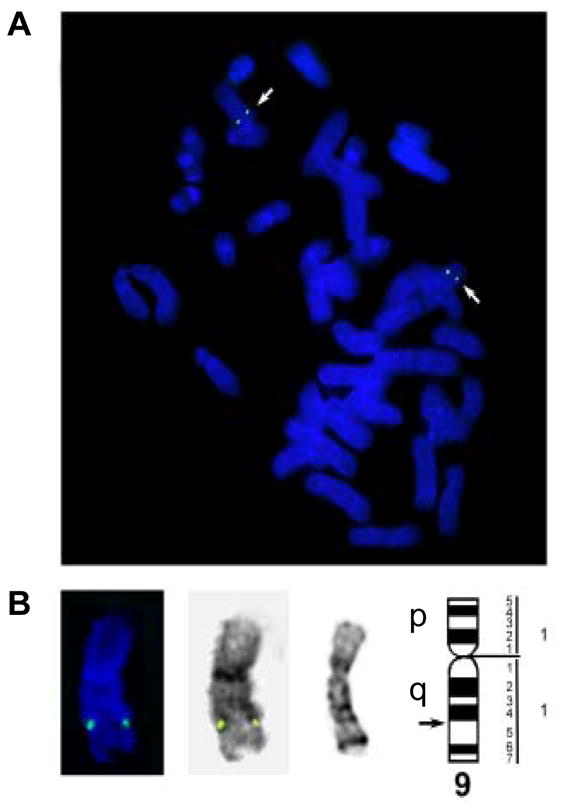
Localization of the rEL gene on chromosome 9.
A: A G-banded rabbit metaphase cell before FISH. Arrows indicate chromosomes 9. B: FISH on the same metaphase cell. Arrows indicate the rEL signals on chromosomes 9. C: Karyotype of the same cell. Arrows indicate chromosomes 9. D: Chromosomes 9 with the rEL signals from different metaphase cells. Far left: The rabbit chromosome 9 ideogram 20 (the arrow indicates the 9q14-q15 region where the rEL probe hybridized to); From left to right for each chromosome 9: G-banding before FISH, FISH and reversed DAPI banding after FISH of the same chromosome.
TABLE 2.
COMPARISON OF HUMAN ELWITH RABBIT, MOUSE,AND RAT EL
| Species | Chromosome location | DNA (kb) | Exon number | RNA (kb) | Identity with human EL | Protein (kDa) | Identity with human EL |
|---|---|---|---|---|---|---|---|
| Human | 18q21.1 | 31 | 10 | 4.1 | - | 55 | - |
| Rabbit | 9q14.3 – 15.1 | 19 | 10 | 4.0 | 83.1% | 56 | 85.4% |
| Mouse | 18 E2 | 22 | 10 | 3.8 | 79.1% | 56 | 80.4% |
| Rat | 18q12.3 | 21 | 10 | 1.6 | 76.1% | 56 | 78.1% |
Because EL belongs to the lipase family, we also compared the rEL cDNA sequences with rabbit LPL and HL, both of which are important regulators in lipid metabolism(16, 17). Because rabbit LPL cDNA sequences were not available, we cloned the rabbit LPL cDNA and submitted it to Genbank (accession No.: NM_001177330) using the same method as we used for rEL cloning. We found that rEL protein was 49% homologous to rabbit LPL and 45% to that of rabbit HL.
Analysis of EL mRNA in tissues
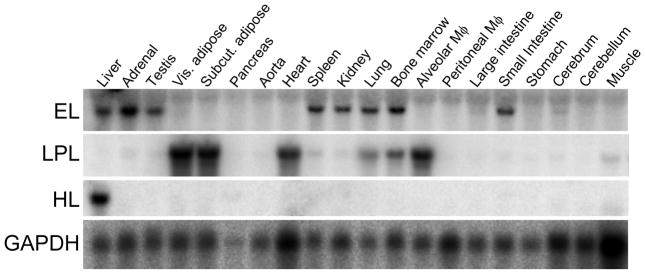
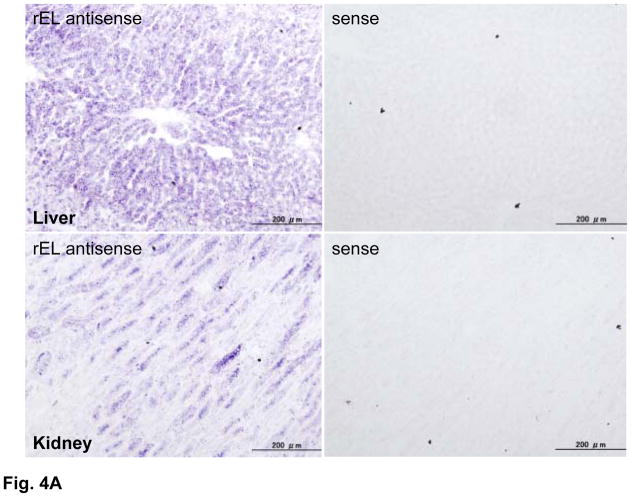
Using the rEL cDNA as a probe, we analyzed the tissue distribution of rEL mRNA by Northern blotting analysis. Among 20 tissues, high expression of rEL mRNA was demonstrated in the liver, adrenal, testis, spleen, kidney, lung, bone marrow, and small intestine (Fig. 3). This multiple expression pattern of rEL mRNA contrasted with the expression patterns of rabbit HL and LPL. Rabbit HL was exclusively expressed in the liver whereas rabbit LPL was mainly expressed in the adipose, heart, and macrophages (Fig. 3). To evaluate rEL expression in tissues, we also performed in situ hybridization of the rEL mRNA expression in the liver and kidney. As shown in Fig. 4A and B, hepatocytes expressed rEL mRNA while, in the kidney, tubular epithelial cells along with small vessels showed detectable rEL mRNA signals.
Fig. 3.
Northern blotting analysis of the rEL tissue expression. 20 tissues were freshly collected from a normal male JW rabbit. Northern blotting was performed as described in the Materials and Methods. Results are representative data derived from three independent analyses. GAPDH expression was used to show that equal amounts of RNA were loaded.
Fig. 4.
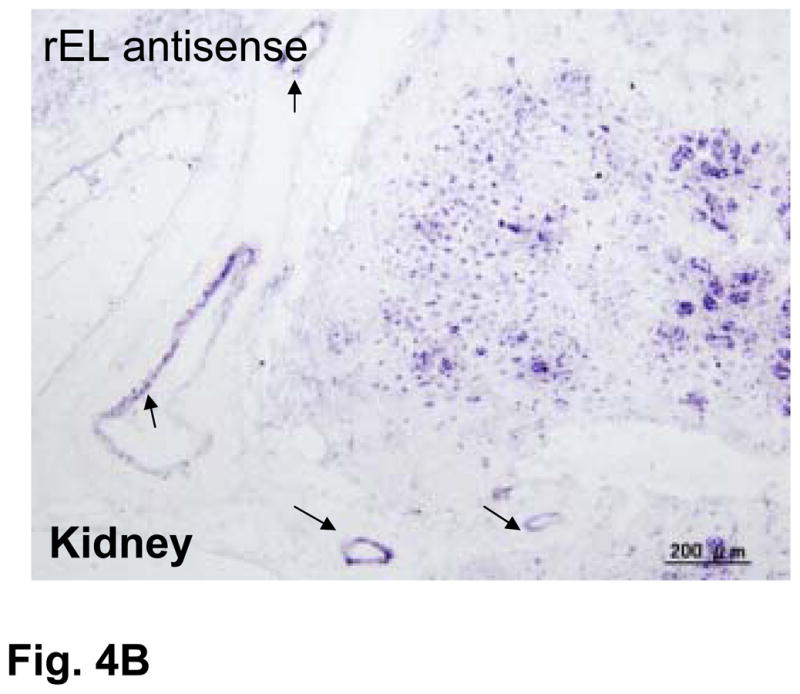
Demonstration of rEL mRNA expression in the liver (top) and kidney (bottom) by in situ hybridization (A). All tissues were hybridized with either rEL antisense or rEL sense (as a control) as described in the Materials and Methods. Arrows indicate small vessels of kidneys that shows rEL expression (B).
EL expression in WHHLMI rabbits
To study whether EL functions are associated with low levels of plasma HDL, we compared the EL expression of WHHLMI rabbits with that of chow-fed normal JW rabbits. WHHLMI rabbits are genetically deficient in LDL receptors(26) and are characterized by having high levels of plasma total cholesterol due to accumulation of LDLs, accompanied by a remarkable reduction of plasma HDL-C levels (Fig. 5A). We hypothesized that the low plasma levels of HDL-C exhibited in WHHLMI rabbits may be associated with increased expression of rEL. To examine this notion, we first compared the rEL proteins in both pre- and post-heparin plasma from WHHLMI with that from normal JW rabbits by Western blotting. As shown in Fig. 5B, rEL proteins in normal JW rabbits were detectable in both pre- and post-heparin plasma. In WHHLMI rabbits, both pre- and post-heparin plasma contained more rEL proteins than that in normal JW rabbits. Moreover, we found that the hepatic EL mRNA expression of WHHLMI rabbits were significantly higher than those of normal JW rabbits (Fig. 5C).
Fig. 5.
Analysis of normal and WHHL rabbit plasma lipoproteins and plasma lipids (A). Lipoprotein profile (2μl) was analyzed by1% agarose gel stained with Fat Red 7B for neutrolipids. Human plasma obtained from a healthy volunteer was used as a reference. Plasma total cholesterol and HDL-C were measured as described in the Materials and Methods. N=21 for JW rabbits and N=18 for WHHL rabbits. **P<0.001 vs. JW.
(B): Pre- and post-heparin plasma was obtained from two normal JW and WHHL rabbits and Western blotting was performed using polyclonal rEL Ab. Rabbit EL proteins expressed by transfected 293 cells were used as a positive control.
(C): Hepatic rEL expression was analyzed by real-time RT-PCR. **P<0.001 vs. JW. Each dot represents one sample from a rabbit.
Effect of EL antisense oligonucleotides on plasma HDLs
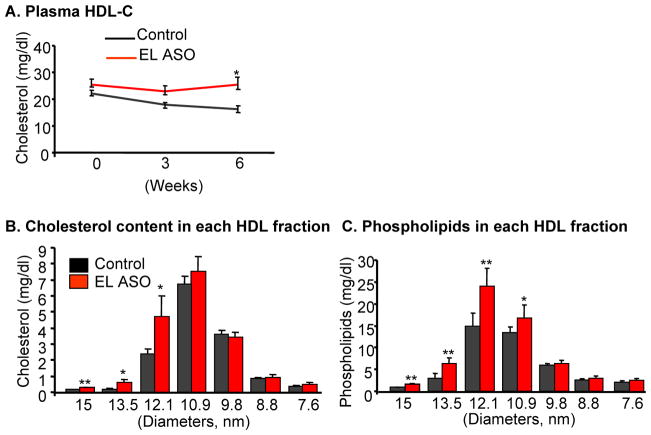
To evaluate whether rEL modulates the plasma HDL levels or HDL particles in vivo, we first intravenously injected rEL ASOs along with mismatched oligonucleotides (as a control) into normal chow-fed JW rabbits for 6 weeks. Rabbits injected with rEL ASOs did not show any changes in liver pathology and functions as serum levels of both alanine aminotransferase and aspartate aminotransferase were within normal ranges. Compared to control rabbits, rabbits injected with the rEL ASOs did not show significant changes in plasma total cholesterol and triglycerides (data not shown). However, rEL-injected rabbits showed a slight (not statistically significant) elevation of plasma HDL-C levels compared with control rabbits (Fig. 6A). In spite of these mild changes in HDL-C levels caused by rEL ASOs injection, large-sized (>12.1 nm) HDL cholesterol contents were significantly increased accompanied by a significant increase of phospholipids in these fractions in comparison with those of control rabbits, as shown by HPLC analysis (Fig. 6B and C). Examination by real-time RT-PCR and Western blotting revealed that rEL ASO treatment resulted in about 50% reduction of hepatic rEL expression (both mRNA and protein) compared with that of control rabbits (Fig. 6D). Furthermore, we injected rEL ASOs into WHHLMI rabbits, which had lower HDL-C levels than normal rabbits. As shown in S-Fig. 3, treatment with rEL ASOs did not lead to a significant change in plasma HDL-C levels of WHHLMI rabbits although a similar tendency was found in large-HDL fractions which were phospholipid-rich.
Fig. 6.
Effects of rEL antisense oligonucleotide injection on normal rabbits. Rabbits were intravenously injected with either rEL ASOs (ISIS 440825: 5′GCAGCTGACGGCCTGGCCCA3′) or mismatched control for 6 weeks. Plasma HDL-C (A), HDL fraction cholesterol (B), and HDL fraction phospholipid contents (C). HDL fractions according to their size were isolated by HPLC as described in the Materials and Methods and cholesterol and phospholipids of each fraction were measured. Hepatic EL expression was reduced as shown by real-time RT-PCR and Western blots (D). N=8 for control and 13 for ASO-injected group. *P<0.05 or **P<0.01 vs. control.
EL expression in atherosclerotic lesions
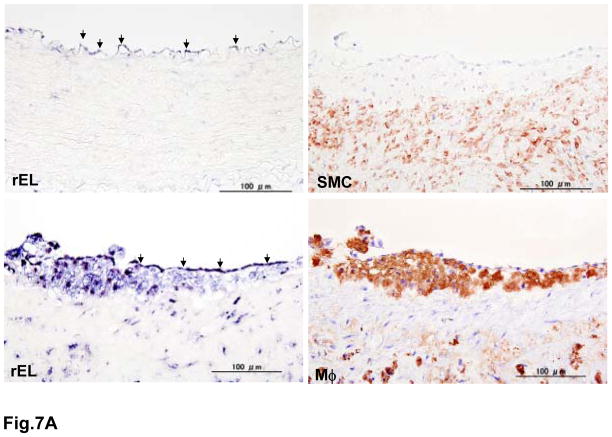
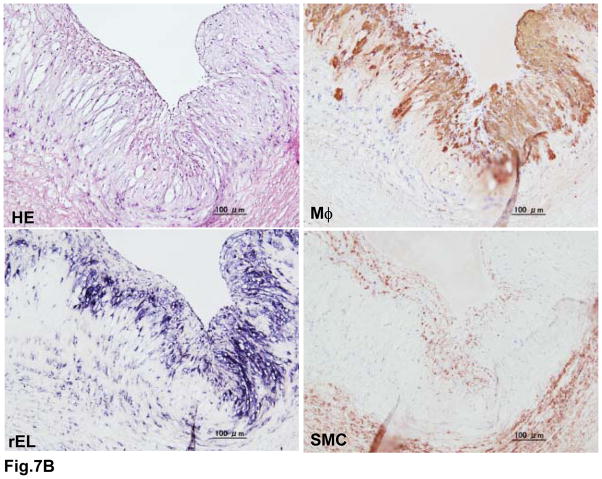
It has been reported that EL may be involved in the pathogenesis of atherosclerosis; therefore, we examined whether rEL was expressed in the lesions of rabbit atherosclerosis by in situ hybridization. For this purpose, we used 7-month-old WHHLMI rabbits because they developed aortic atherosclerosis. rEL mRNA was weakly demonstrated in the aortic endothelial cells of normal rabbits (Fig. 7A). In the aortic fatty-streak lesions, both endothelial cells and subendothelial macrophages showed abundant EL mRNA expression. In the advanced lesions, macrophages along with some smooth muscle cells showed rEL expression (Fig. 7B). rEL mRNA expression in the arterial wall was also quantified by a real-time RT-PCR method. Compared with the hepatic expression of rEL, aortic endothelial cells isolated from normal JW rabbit aortas expressed 4% of hepatic rEL whereas intimal lesions of aortas from WHHLMI rabbits expressed 8% of hepatic rEL.
Fig. 7.
Demonstration of rEL expression in the endothelial cells and lesions of aortic atherosclerosis of WHHLMI rabbits (A). Serial sections were used for both in situ hybridization and immunohistochemical staining as described in the Materials and Methods. In normal aorta, weak expression of rEL (indicated by arrowheads) was noted in the endothelial cells (top, left). In the aortic lesions, both endothelial cells (indicated by arrowheads) and macrophages (M ) (bottom) showed rEL expression but only a few smooth muscle cells (SMC) showed rEL expression (top, right). In advanced lesions of WHHLMI rabbits (B), many macrophages and some smooth muscle cells showed rEL expression.
DISCUSSION
In this study, we successfully cloned the rEL cDNA sequences and performed molecular characterization of the tissue expression of rEL in both normal and WHHLMI rabbits, an animal model appropriate for the study of human lipid metabolism and atherosclerosis(13, 27). We found that rEL exhibits high identity with human EL sequences compared with mouse and rat EL. The rEL is mapped to the chromosome 9, which is homologous to the human chromosome 18(25). In addition, rEL shows high homology with rabbit LPL and HL, although they are quite different in tissue expression pattern and enzymatic activity. It has been reported that the major substrate for EL is HDL phospholipids but that for LPL is TG-rich lipoproteins, whereas HL can hydrolyze TG-rich lipoproteins, HDLs, and LDLs(11). Although it is generally believed that EL is specifically expressed by vascular endothelial cells, EL expression has been reported in other tissues such as liver, lung, and kidney(1, 2). Consistent with this observation, our studies revealed that many tissues expressed rEL in rabbits, which is similar to the EL expression patterns in human tissues determined by real-time RT-PCR (Zhang J., et al., unpublished observations). Hepatocytes and renal epithelial cells along with endothelial cells can express EL mRNA, as shown by in situ hybridization. This multiple expression pattern of EL suggests that EL may exert diverse physiological functions in vivo. In tissues such as the liver, adrenal, and testis, EL may mediate the uptake of lipoproteins for the synthesis of cholesterol and hormones. EL can also hydrolyze the HDLs to release free fatty acids, which can provide energy for those expressing tissues(28). EL proteins are detected in pre-heparin plasma but EL contents are increased in WHHLMI rabbit plasma after injection with heparin, indicating that rEL conserved heparin-binding properties that can interact with heparan sulfate proteoglycans on the cellular surface(2).
To investigate whether EL mediates HDL metabolism in vivo, we adopted two strategies. First, we analyzed the rEL protein levels in plasma of WHHLMI rabbits, which have concomitant hypercholesterolemia and low HDL levels. Our results showed that hepatic mRNA expressions of rEL in WHHLMI rabbits were higher than those of normal JW rabbits, which was coincidently associated with low plasma HDL-C levels. Interestingly, rEL proteins of pre- and post-heparin plasma in WHHLMI rabbits were simultaneously increased compared with those of normal rabbits. It is currently unknown, however, whether increased rEL expression in WHHLMI rabbits is a cause of low HDL-C levels or a consequence of high LDL levels. It is possible that high expression of rEL is caused by extensive aortic atherosclerotic lesions (namely, chronic inflammation) because EL is highly regulated by inflammation(29). This phenomenon (high hepatic expression of EL) has not been observed in apoE KO and LDLr KO mice(11). In fact, hepatic EL expression of these KO mice was down-regulated while aortic EL was up-regulated(11, 30). It is noteworthy that many familial combined hyperlipidemic patients and homozygous familial hypercholesterolemic patients also have a reduction in HDL-C levels, (31) although the mechanisms for this phenomenon have not been completely elucidated. It will be interesting to investigate whether familial combined hyperlipidemic patients have high EL expression or whether increased EL expression is responsible for low HDL levels.
Secondly, we screened 90 candidate rEL ASOs using cultured rabbit hepatocytes and obtained two potent ASOs for in vivo study. Although rEL ASO injection resulted in a 50% reduction of hepatic rEL expression, plasma HDL-C levels were not significantly improved in both normal and WHHLMI rabbits, suggesting that either inhibition of hepatic EL expression induced by rELASOs is not strong enough for lowering plasma HDL levels or reduced hepatic EL expression may be compensated by extra-hepatic EL expression since EL is expressed by multiple tissues as shown in Northern blotting results. Nevertheless, HDL particles in normal rabbits are indeed affected by rEL ASOs: large sized (>12 nm) phospholipid-rich HDLs are consistently increased, which supports the notion that the preferential substrate for EL is phospholipids of those large-sized HDLs(32). In spite of this, it remains to be determined whether these large-sized HDLs are more functional in terms of reverse cholesterol transport or beneficial for dyslipidemic patients. Recently, Hara et al. demonstrated that increased HDL particles in EL KO mice preserved anti-inflammatory and anti-atherosclerotic functions(33).
In addition to HDL metabolism, EL has been considered to be involved in the pathogenesis of atherosclerosis. Similar to lipoprotein lipase, both anti- and pro-atherogenic effects of EL through either catalytic or non-catalytic functions of EL have been proposed(11). As non-catalytic functions, EL may be atherogenic by forming a bridge between LDLs and arterial wall, thereby enhancing the retention of atherogenic lipoproteins. Using in situ hybridization, we demonstrated that rEL is widely expressed by vascular endothelial cells and macrophages in the lesions of WHHLMI rabbits, supporting the assertion that EL is involved in atherosclerosis(10). “Dual functions” of EL (anti-atherogenic and pro-atherogenic) may depend upon the site of EL expression, such as hepatic vs. vascular cells of EL(3). To examine these hypotheses and investigate underlying molecular mechanisms in vivo, we need to develop transgenic rabbits overexpressing EL in either liver or macrophages. Efforts are now being directed towards creating EL transgenic rabbits in our laboratory in order to evaluate the effects of EL on the development of atherosclerosis.
In summary, we have cloned rabbit EL cDNA and investigated the EL expression in both normal and WHHLMI rabbits. EL is expressed in multiple tissues and plays an important role in HDL metabolism.
Supplementary Material
Acknowledgments
We thank Watanabe, T., and Morimoto, M. for their help and support for this research. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (19790226, 71790514, 19390099, 21659078), National Institutes of Health (HL088391, HL068878, HL105114), a research grant for cardiovascular disease from the Ministry of Health, Labor and Welfare of Japan, and a research grant from AstraZeneca. YEC is an established investigator of the American Heart Association (0840025N).
The abbreviations
- ASOs
antisense oligonucleotides
- EL
endothelial lipase
- HL
hepatic lipase
- JW
Japanese white
- LPL
lipoprotein lipase
- WHHLMI
Watanabe heritable hyperlipidemic myocardial infarction-prone
References
- 1.Hirata K, Dichek HL, Cioffi JA, Choi SY, Leeper NJ, Quintana L, Kronmal GS, Cooper AD, Quertermous T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J Biol Chem. 1999;274:14170–5. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- 2.Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet. 1999;21:424–8. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- 3.Brown RJ, Rader DJ. Lipases as modulators of atherosclerosis in murine models. Curr Drug Targets. 2007;8:1307–19. doi: 10.2174/138945007783220614. [DOI] [PubMed] [Google Scholar]
- 4.Edmondson AC, Brown RJ, Kathiresan S, Cupples LA, Demissie S, Manning AK, Jensen MK, Rimm EB, Wang J, Rodrigues A, Bamba V, Khetarpal SA, Wolfe ML, Derohannessian S, Li M, Reilly MP, Aberle J, Evans D, Hegele RA, Rader DJ. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest. 2009;119:1042–50. doi: 10.1172/JCI37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T. Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003;111:347–55. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badellino KO, Wolfe ML, Reilly MP, Rader DJ. Endothelial lipase concentrations are increased in metabolic syndrome and associated with coronary atherosclerosis. PLoS Med. 2006;3:e22. doi: 10.1371/journal.pmed.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broedl UC, Maugeais C, Millar JS, Jin W, Moore RE, Fuki IV, Marchadier D, Glick JM, Rader DJ. Endothelial lipase promotes the catabolism of ApoB-containing lipoproteins. Circ Res. 2004;94:1554–61. doi: 10.1161/01.RES.0000130657.00222.39. [DOI] [PubMed] [Google Scholar]
- 8.Ishida T, Choi SY, Kundu RK, Spin J, Yamashita T, Hirata K, Kojima Y, Yokoyama M, Cooper AD, Quertermous T. Endothelial lipase modulates susceptibility to atherosclerosis in apolipoprotein-E-deficient mice. J Biol Chem. 2004;279:45085–92. doi: 10.1074/jbc.M406360200. [DOI] [PubMed] [Google Scholar]
- 9.Ko KW, Paul A, Ma K, Li L, Chan L. Endothelial lipase modulates HDL but has no effect on atherosclerosis development in apoE−/− and LDLR−/− mice. J Lipid Res. 2005;46:2586–94. doi: 10.1194/jlr.M500366-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Azumi H, Hirata K, Ishida T, Kojima Y, Rikitake Y, Takeuchi S, Inoue N, Kawashima S, Hayashi Y, Itoh H, Quertermous T, Yokoyama M. Immunohistochemical localization of endothelial cell-derived lipase in atherosclerotic human coronary arteries. Cardiovasc Res. 2003;58:647–54. doi: 10.1016/s0008-6363(03)00287-6. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda T, Ishida T, Rader DJ. Update on the role of endothelial lipase in high-density lipoprotein metabolism, reverse cholesterol transport, and atherosclerosis. Circ J. 2010;74:2263–70. doi: 10.1253/circj.cj-10-0934. [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Watanabe T. Cholesterol-fed and transgenic rabbits for the study of atherosclerosis. J Atheroscler Thromb. 2000;7:26–32. doi: 10.5551/jat1994.7.26. [DOI] [PubMed] [Google Scholar]
- 13.Fan J, Watanabe T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol Ther. 2003;99:261–282. doi: 10.1016/s0163-7258(03)00069-x. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Koike T, Ichikawa T, Hatakeyama K, Shiomi M, Zhang B, Kitajima S, Morimoto M, Watanabe T, Asada Y, Chen YE, Fan J. C-reactive protein in atherosclerotic lesions: its origin and pathophysiological significance. Am J Pathol. 2005;167:1139–48. doi: 10.1016/S0002-9440(10)61202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiomi M, Ito T, Yamada S, Kawashima S, Fan J. Development of an animal model for spontaneous myocardial infarction (WHHLMI rabbits) Arterioscler Thromb Vasc Biol. 2003;23:1239–44. doi: 10.1161/01.ATV.0000075947.28567.50. [DOI] [PubMed] [Google Scholar]
- 16.Fan J, Wang J, Bensadoun A, Lauer SJ, Dang Q, Mahley RW, Taylor JM. Overexpression of hepatic lipase in transgenic rabbits leads to a marked reduction of plasma high density lipoproteins and intermediate density lipoproteins. Proc Natl Acad Sci U S A. 1994;91:8724–8728. doi: 10.1073/pnas.91.18.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan J, Unoki H, Kojima N, Sun H, Shimoyamada H, Deng H, Okazaki M, Shikama H, Yamada N, Watanabe T. Overexpression of lipoprotein lipase in transgenic rabbits inhibits diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem. 2001;276:40071–40079. doi: 10.1074/jbc.M105456200. [DOI] [PubMed] [Google Scholar]
- 18.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–11. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chantry-Darmon C, Rogel-Gaillard C, Bertaud M, Urien C, Perrocheau M, Chardon P, Hayes H. 133 new gene localizations on the rabbit cytogenetic map. Cytogenet Genome Res. 2003;103:192–201. doi: 10.1159/000076310. [DOI] [PubMed] [Google Scholar]
- 20.Yerle M, Echard G, Gillois M. The high-resolution GTG-banding pattern of rabbit chromosomes. Cytogenet Cell Genet. 1987;45:5–9. doi: 10.1159/000132416. [DOI] [PubMed] [Google Scholar]
- 21.Standard karyotype of the laboratory rabbit, Oryctolagus cuniculus. Cytogenet Cell Genet. 1981;31:240–8. doi: 10.1159/000131653. [DOI] [PubMed] [Google Scholar]
- 22.Sohail M, Southern EM. Selecting optimal antisense reagents. Adv Drug Deliv Rev. 2000;44:23–34. doi: 10.1016/s0169-409x(00)00081-8. [DOI] [PubMed] [Google Scholar]
- 23.Altmann KH, Fabbro D, Dean NM, Geiger T, Monia BP, Muller M, Nicklin P. Second-generation antisense oligonucleotides: structure-activity relationships and the design of improved signal-transduction inhibitors. Biochem Soc Trans. 1996;24:630–7. doi: 10.1042/bst0240630. [DOI] [PubMed] [Google Scholar]
- 24.Koike T, Kitajima S, Yu Y, Nishijima K, Zhang J, Ozaki Y, Morimoto M, Watanabe T, Bhakdi S, Asada Y, Chen YE, Fan J. Human C-reactive protein does not promote atherosclerosis in transgenic rabbits. Circulation. 2009;120:2088–94. doi: 10.1161/CIRCULATIONAHA.109.872796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korstanje R, O’brien PC, Yang F, Rens W, Bosma AA, Van Lith HA, Van Zutphen LF, Ferguson-Smith MA. Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet Cell Genet. 1999;86:317–22. doi: 10.1159/000015325. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto T, Bishop RW, Brown MS, Goldstein JL, Russell DW. Deletion in cysteine-rich region of LDL receptor impedes transport to cell surface in WHHL rabbit. Science. 1986;232:1230–7. doi: 10.1126/science.3010466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiomi M, Ito T. The Watanabe heritable hyperlipidemic (WHHL) rabbit, its characteristics and history of development: a tribute to the late Dr. Yoshio Watanabe Atherosclerosis. 2009;207:1–7. doi: 10.1016/j.atherosclerosis.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 28.Strauss JG, Hayn M, Zechner R, Levak-Frank S, Frank S. Fatty acids liberated from high-density lipoprotein phospholipids by endothelial-derived lipase are incorporated into lipids in HepG2 cells. Biochem J. 2003;371:981–8. doi: 10.1042/BJ20021437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimokawa Y, Hirata K, Ishida T, Kojima Y, Inoue N, Quertermous T, Yokoyama M. Increased expression of endothelial lipase in rat models of hypertension. Cardiovasc Res. 2005;66:594–600. doi: 10.1016/j.cardiores.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Yu KC, David C, Kadambi S, Stahl A, Hirata K, Ishida T, Quertermous T, Cooper AD, Choi SY. Endothelial lipase is synthesized by hepatic and aorta endothelial cells and its expression is altered in apoE-deficient mice. J Lipid Res. 2004;45:1614–23. doi: 10.1194/jlr.M400069-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Hokanson JE, Krauss RM, Albers JJ, Austin MA, Brunzell JD. LDL physical and chemical properties in familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1995;15:452–9. doi: 10.1161/01.atv.15.4.452. [DOI] [PubMed] [Google Scholar]
- 32.Jin W, Millar JS, Broedl U, Glick JM, Rader DJ. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J Clin Invest. 2003;111:357–62. doi: 10.1172/JCI16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara T, Ishida T, Kojima Y, Tanaka H, Yasuda T, Shinohara M, Toh R, Hirata K. Targeted deletion of endothelial lipase increases HDL particles with anti-inflammatory properties both in vitro and in vivo. J Lipid Res. 2011;52:57–67. doi: 10.1194/jlr.M008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.