Abstract
An easier assessment model would be helpful for high-throughput screening of Aeromonas virulence. The previous study indicated the potential of Tetrahymena as a permissive model to examine virulence of Aeromonas hydrophila. Here our aim was to assess virulence of Aeromonas spp. using two model hosts, a zebrafish assay and Tetrahymena-Aeromonas co-culture, and to examine whether data from the Tetrahymena thermophila model reflects infections in the well-established animal model. First, virulence of 39 Aeromonas strains was assessed by determining the 50% lethal dose (LD50) in zebrafish. LD50 values ranging from 1.3×102 to 3.0×107 indicated that these strains represent a high to moderate degree of virulence and could be useful to assess virulence in the Tetrahymena model. In Tetrahymena-Aeromonas co-culture, we evaluated the virulence of Aeromonas by detecting relative survival of Aeromonas and Tetrahymena. An Aeromonas isolate was considered virulent when its relative survival was greater than 60%, while the Aeromonas isolate was considered avirulent if its relative survival was below 40%. When relative survival of T. thermophila was lower than 40% after co-culture with an Aeromonas isolate, the bacterial strain was regarded as virulent. In contrast, the strain was classified as avirulent if relative survival of T. thermophila was greater than 50%. Encouragingly, data from the 39 Aeromonas strains showed good correlation in zebrafish and Tetrahymena-Aeromonas co-culture models. The results provide sufficient data to demonstrate that Tetrahymena can be a comparable alternative to zebrafish for determining the virulence of Aeromonas isolates.
Introduction
Aeromonas is a Gram-negative bacterium that belongs to the family Aeromonadaceae [1]. Recently, the genus Aeromonas has received increased attention because it is not only an important disease-causing pathogen of fish and other cold blooded species, but also it is the etiological agent responsible for a variety of infectious complications in humans [2], [3]. As Aeromonas encompasses a diversity of strains or genotypes with varying virulence, it is often necessary to compare the degree of virulence of individual bacterial strains in order to assess the precise role and relative importance of various pathogenic mechanisms. To achieve this, a host must be chosen to evaluate and quantify the pathogenic potential of Aeromonas.
Intraperitoneal injection of mice with bacteria is useful in assessing the virulence of some bacteria, especially when combined with immune function modulation and evaluation of the dose that is lethal for 50% of the population (LD50) [4]. However, mice are not appropriate hosts for pathogens such as Aeromonas as these normally infect cold-blooded species living at low temperatures [5]. Recently, it has been reported that zebrafish is an excellent model for assessing the LD50 of Aeromonas hydrophila strains [6] and for examining host immune responses [7], but this model is not suitable for studying host-pathogen interactions at the molecular level [8]. In addition, using a vertebrate host to evaluate the virulence of bacteria is prohibitively time consuming, individually different, ethically problematic and expensive because large numbers of animals are required. In contrast, invertebrate model hosts represent a tempting alternative, as they not only address the defects mentioned above, but they are also associated with several important advantages, including simple immune systems, genetic tractability, and amenity to high-throughput experiments.
A number of different model systems, including amoeba [5], nematodes [9] and insects [10], have been introduced, and distinct model systems are more or less suitable for each particular pathogen. The Dictyostelium amoeba has been used as an alternative host to evaluate Aeromonas virulence [5]. However, Benghezal et al. [11] reported that Tetrahymena was more suitable than the amoeba model in a high-throughput screening study to identify inhibitors of Klebsiella pneumoniae virulence.
Tetrahymena thermophila is a free-living unicellular eukaryote that can be grown easily in cheap culture media over a wide range of temperature between 12°C to 41°C without the need for a CO2-enriched atmosphere. In addition, these cells are amenable to genetic analysis, most notably due to the small, fully sequenced genome, which allows for the elucidation of molecular interactions between host and pathogen [12], [13].
In the last decade, a lot of investigations have been performed on the interactions between Tetrahymena and certain bacteria, such as Yersinia pestis [14], [15], K. pneumoniae [11], Escherichia coli [16]–[18] and Vibrio fischeri [19]. A previous study in our laboratory described the interactions between A. hydrophila and T. thermophila, and indicated the potential of Tetrahymena as a permissive model to examine virulence of A. hydrophila [20].
The aim of this present study is to determine the correlation between the Tetrahymena–Aeromonas model system and the well-established animal model for assessing virulence of Aeromonas spp., in order to verify the reliability and accuracy of the former in evaluating bacterial virulence.
Results
The virulence of 39 Aeromonas strains was assessed by determining LD50 values in zebrafish, and LD50s of all strains in zebrafish ranged from 1.3×102 to 3.0×107 (Table 1). Most dying fish showed clinical signs typical of hemorrhagic septicemia. Colonies of the 39 Aeromonas strains were recovered from infected fish, and no evident external injuries were observed in the surviving fish. According to the degree of virulence described by Pu et al. [21], 18 strains were included in the virulent category (LD50s of 102−105) while the remaining 21 strains were classified as avirulent (LD50s >106).
Table 1. Virulence of 39 Aeromonas strains assessed in zebrafish and T. thermophila models.
| Strain | Aeromonasspecies | Source | Sequenceof gyrB | LD50 | Relative survivalOf Aeromonas (%) | Relative survival ofT. thermophila (%) |
| NJ-35 | A. hydrophila | Diseased Crucian carp | JX025789 | 1.3×102 | 82.0±3.5 | 16.2±1.4 |
| XY-16 | A. hydrophila | Diseased Crucian carp | JX025797 | 5.8×102 | 85.4±5.9 | 14.4±1.3 |
| NJ-34 | A. hydrophila | Diseased Crucian carp | JX025788 | 8.1×102 | 78.3±3.9 | 19.1±0.8 |
| XX-52 | A. hydrophila | Diseased Crucian carp | JX025794 | 1.3×103 | 75.6±1.3 | 17.1±1.4 |
| BSK-10 | A. hydrophila | Diseased Silver carp | JX413511 | 2.5×103 | 73.0±3.8 | 27.3±3.8 |
| CS-43 | A. hydrophila | Diseased Silver carp | JX025780 | 2.6×103 | 67.3±2.8 | 23.3±2.4 |
| XX-49 | A. hydrophila | Diseased Crucian carp | JX025793 | 6.3×103 | 74.6±3.0 | 26.0±5.6 |
| XX-58 | A. hydrophila | Diseased Silver carp | JX025795 | 7.5×103 | 61.1±6.6 | 23.3±3.0 |
| XX-22 | A. hydrophila | Diseased Common carp | JX025792 | 1.3×104 | 65.5±2.7 | 33.4±4.9 |
| NJ-1 | A. hydrophila | Healthy Crucian carp | JX025785 | 1.7×104 | 67.0±3.1 | 32.0±4.9 |
| XX-62 | A. hydrophila | Diseased Silver carp | JX025796 | 2.1×104 | 54.0±2.8 | 28.0±5.8 |
| NJ-24 | A. bestiarum | Water | JQ815386 | 3.9×104 | 62.2±5.3 | 31.7±3.3 |
| XX-14 | A. hydrophila | Diseased Silver carp | JX025791 | 4.9×104 | 66.2±4.8 | 29.0±2.7 |
| CS-2 | A. salmonicida | Water | JX025811 | 1.5×105 | 57.6±0.6 | 25.8±4.7 |
| XH-16 | A. veronii | Healthy Crucian carp | JX025900 | 3.4×105 | 59.6±6.7 | 37.0±5.4 |
| NJ-37 | A. hydrophila | Diseased Eel | JQ815377 | 4.3×105 | 63.1±5.1 | 42.0±4.9 |
| GY-6 | A. veronii | Healthy Common carp | JX025876 | 4.6×105 | 56.0±1.4 | 39.2±1.2 |
| J-1 | A. hydrophila | Diseased Crucian carp | JX401926 | 8.5×105 | 63.4±6.9 | 35.4±6.9 |
| NJ-28 | A. hydrophila | Diseased Crucian carp | JX025787 | 1.0×106 | 48.2±4.6 | 38.5±6.2 |
| NJ-16 | A. veronii | Water | JX025903 | 1.2×106 | 39.6±4.6 | 46.1±2.4 |
| XH-17 | A. veronii | Healthy Crucian carp | JX025899 | 1.2×106 | 38.6±5.7 | 51.0±2.4 |
| JH-19 | A. hydrophila | Healthy Chinese bream | JX025784 | 1.3×106 | 38.8±5.6 | 47.9±9.1 |
| NJ-3 | A. hydrophila | Water | JX025786 | 1.8×106 | 52.0±2.6 | 49.0±3.9 |
| XY-20 | A. veronii | Healthy Crucian carp | JX025951 | 2.6×106 | 27.1±4.7 | 58.6±7.4 |
| XY-7 | A. veronii | Healthy Grass carp | JX025956 | 2.9×106 | 44.6±5.5 | 51.2±6.3 |
| CS-51 | A. veronii | Water | JX025863 | 3.1×106 | 35.6±5.1 | 46.0±2.9 |
| JH-2 | A. veronii | Healthy Black carp | JX025890 | 4.8×106 | 31.7±2.7 | 61.7±5.3 |
| CS-16 | A. veronii | Healthy Chinese bream | JX025854 | 4.8×106 | 38.5±2.8 | 50.0±5.6 |
| NJ-7 | A. media | Water | JX025839 | 5.4×106 | 23.5±5.4 | 71.0±2.7 |
| XH-14 | A. veronii | Healthy Crucian carp | JX025901 | 5.4×106 | 14.2±2.2 | 68.3±4.2 |
| GY-13 | A. veronii | Healthy Common carp | JX025875 | 5.8×106 | 22.8±5.0 | 59.9±4.8 |
| CS-14 | A. veronii | Diseased Chines bream | JX025843 | 6.2×106 | 30.9±2.8 | 63.8±6.9 |
| GY-23 | A. hydrophila | Healthy Crucian carp | JX025782 | 6.7×106 | 34.5±5.0 | 57.0±5.2 |
| NJ-8 | A. media | Water | JX025838 | 9.8×106 | 10.8±1.8 | 81.2±5.5 |
| NJ-21 | A. media | Water | JX025837 | 1.1×107 | 19.5±3.1 | 72.0±5.2 |
| CS-34 | A. hydrophila | Healthy Crucian carp | JX025779 | 1.3×107 | 18.2±1.5 | 72.5±5.9 |
| JH-17 | A. hydrophila | Healthy Chinese bream | JX025783 | 2.0×107 | 31.0±1.2 | 62.0±7.2 |
| NJ-25 | A. media | Water | JX025836 | 2.6×107 | 22.2±0.8 | 76.7±5.1 |
| CS-15 | A. veronii | Diseased Chines bream | JX025842 | 3.0×107 | 13.4±1.1 | 77.6±4.1 |
Relative survival of each bacterium at 12 h is expressed as the number of Aeromonas CFU grown in the presence of T. thermophila relative to the number of Aeromonas CFU cultivated alone. Relative survival of T. thermophila at 12 h is expressed as the number of T. thermophila cells co-cultured with Aeromonas spp. relative to the number of T. thermophila cells cultured alone.
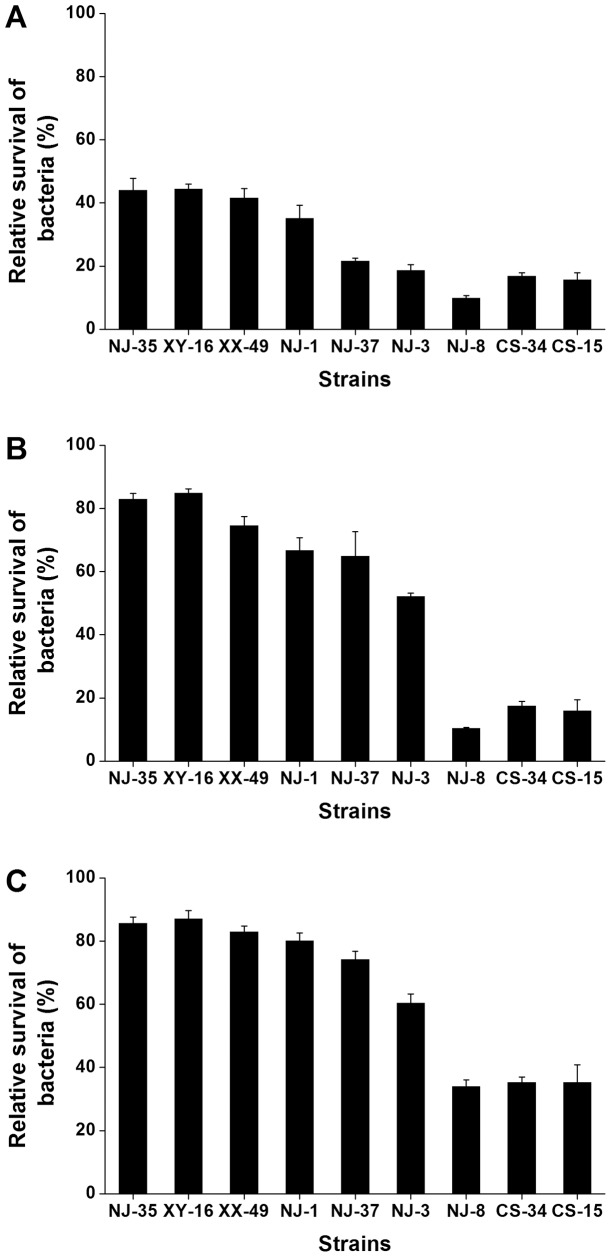
To obtain the most appropriate relative population size of bacteria to protozoa, we determined the relative survival of nine Aeromonas strains with differential virulence after co-culture with T. thermophila for 12 h in three proportions. As shown in Fig. 1, Aeromonas strains that had greater virulence also had higher relative survival after co-culture studies, which suggest that it is possible to use relative survival to evaluate bacterial virulence. Although T. thermophila could inhibit the growth of the Aeromonas strains, the extent of the relative survival of Aeromonas was very different when they were co-cultured in different proportions with T. thermophila. The relative survival of Aeromonas strains inoculated at 1×109 CFU/ml (Fig. 1C) was obviously greater than strains inoculated at the other two concentrations (Fig. 1A and Fig. 1B), and lowest relative survival was 34.0%. On the contrary, the Aeromonas strains inoculated at 1×108 CFU/ml had lower relative survival, and greatest relative survival was 44.5% (Fig. 1A). This result shows that the initial Aeromonas concentration of 5×108 CFU/ml gave the widest range of relative survival values (from 10.5% to 85.0%; Fig. 1B), and therefore was better for distinguishing Aeromonas strains of different virulence.
Figure 1. Relative survival of nine Aeromonas strains co-cultured with T. thermophila in three different proportions.
Relative survival of each bacterium at 12 h is expressed as the number of Aeromonas CFU in the co-culture with T. thermophila relative to the number of Aeromonas CFU grown alone. At the start of the experiment, T. thermophila was at 1×105 cells/ml while the Aeromonas spp. was at 1×108 CFU/ml (A), 5×108 CFU/ml (B) and 1×109 CFU/ml (C), respectively. Error bars represent standard deviations from four independent experiments, with each experiment being comprised of four individual measurements.
To further investigate the effect of T. thermophila on the growth of Aeromonas strains of differential virulence, 5,000∶1 co-cultures of Aeromonas (5×108 CFU/ml) and T. thermophila (1×105 cells/ml) were used and growth dynamics of the Aeromonas strains were measured after co-culturing for 12 h. Bacterial growth was determined by measuring absorbance at 450 nm every 2 h. The growth dynamics of each strain incubated in the presence of T. thermophila could be used to divide the bacterial isolates into three categories (Fig. 2). A. hydrophila NJ-35, XY-16 and XX-49 formed the first category and A. hydrophila XY-16, which had an LD50 of 5.8×102, was used as an example (Fig. 2A). In this category, co-culture with T. thermophila inhibited the growth of the Aeromonas strains but only very slightly, and these strains grew almost as well as control cultures grown in the absence of T. thermophila (Fig. 2A), indicating that to some extent they overcame predation by T. thermophila. The growth responses of strains in the second category can be illustrated using A. hydrophila NJ-1 (Fig. 1B), which had an LD50 of 1.7×104. In this second category, the populations of the Aeromonas strains (A. hydrophila NJ-1, NJ-37 and NJ-3) always increased during co-culture, but the growth rate was significantly lower than observed in control incubations where these bacteria were cultured alone. In the third category is A. hydrophila CS-34, A. veronii CS-15, and A. media NJ-8, an avirulent strain with an LD50 of 9.8×106 (Fig. 1C). The principal character of strains in this category is that the biomass of the Aeromonas cultured in the presence of T. thermophila either declined throughout the incubation period, or reached a peak before decreasing. These findings show that virulent Aeromonas strains were generally preyed upon less by T. thermophila compared to avirulent strains when cultured in the presence of T. thermophila. This indicates that the ability of an Aeromonas strain to resist T. thermophila-mediated phagocytosis is related to its virulence. This suggests that the virulence of Aeromonas strains can be assessed in the T. thermophila model by detecting relative survival of the bacteria when the co-culture is inoculated with population sizes of bacteria and protozoa at 5×108 CFU/ml and 1×105 cells/ml, respectively.
Figure 2. Growth of A. hydrophila XY-16 (A), A. hydrophila NJ-1 (B) and A. media NJ-8 (C) co-cultured in the presence or absence of T. thermophila.
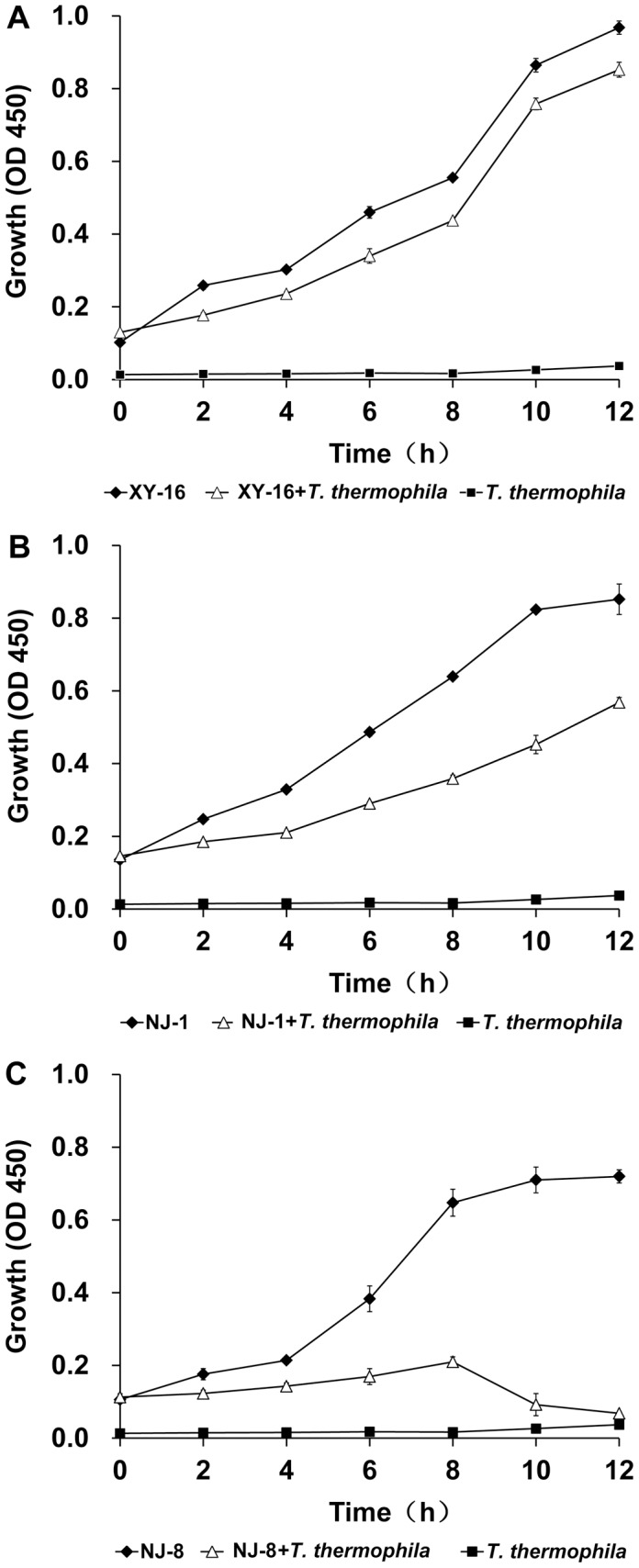
Bacterial growth was determined by measuring the absorbance at 450 nm (A450) during 12 h and (T. thermophila cells had only a negligible effect on A450). Data are expressed as the mean ± SD of four observations per time point.
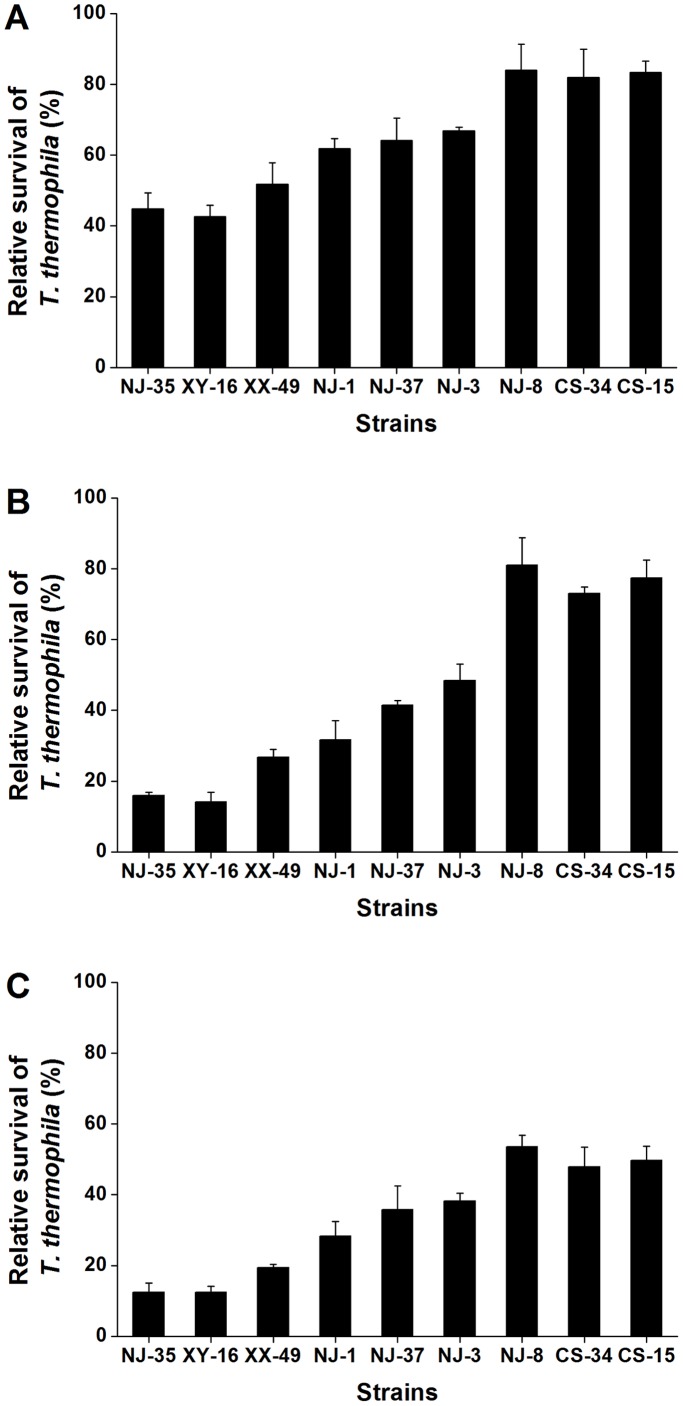
From a protozoan perspective, relative survival of T. thermophila after co-culture with nine Aeromonas strains was determined. There was a tendency for increased relative survival of T. thermophila as Aeromonas virulence decreased (Fig. 3), regardless of the relative proportion of protozoan cells in the co-culture. It suggests that relative survival of T. thermophila after co-culture is related negatively to bacterial virulence. However, relative survival of T. thermophila was related to the initial concentrations of bacteria, such that when T. thermophila was mixed with Aeromonas strains at an initial density of 1.5×109 CFU/ml, relative survival of T. thermophila was generally lower than 53.5% (Fig. 3C). In contrast, co-culture with Aeromonas strains at 2×108 CFU/ml had a much smaller impact on T. thermophila viability, as relative survival of T. thermophila was greater than 42.6% in each case (Fig. 3A). Nevertheless, relative survival of T. thermophila after co-culture with Aeromonas at 1×109 CFU/ml ranged from 14.2% to 81.0% (Fig. 3B). Therefore, an initial minimal population of size of 1×109 CFU/ml is able to provide better evidence at distinguishing Aeromonas strains of different virulence.
Figure 3. Relative survival of T.thermophila co-cultivated with Aeromonas strains in three different proportions.
Relative survival of T. thermophila at 12 h is expressed as the number of T. thermophila cells in the co-culture with Aeromonas spp. relative to the number of T. thermophila cells cultured alone. At the beginning of the experiment, T. thermophila was at 2×105 cells/ml while the Aeromonas spp. was at 2.0×108 CFU/ml (A), 1.0×109 CFU/ml (B) and 1.5×109 CFU/ml (C), respectively. Error bars represent SDs from a minimum of three independent experiments, with each experiment being comprised of six individual measurements.
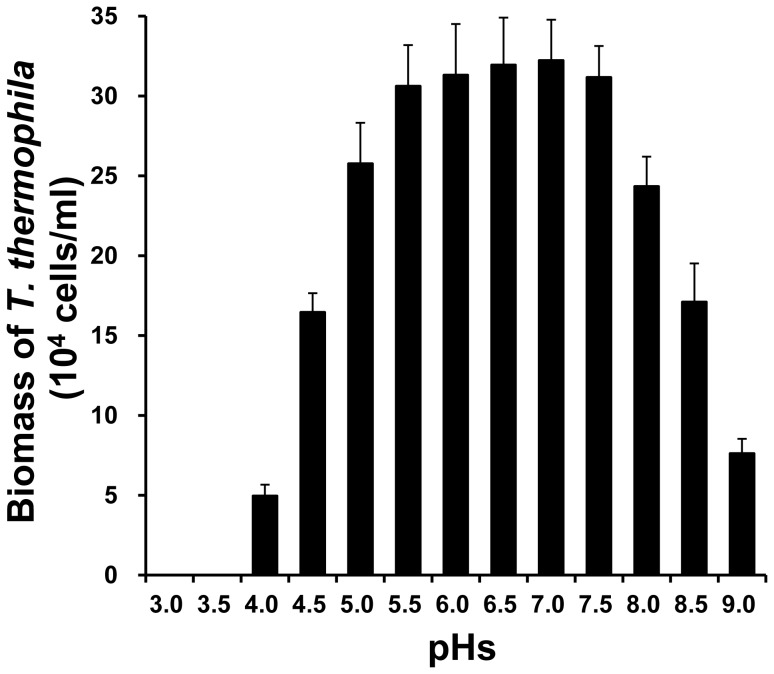
Considering that pH changes by Aeromonas might affect the viability of Tetrahymena, we measured the pH values of medium in which Aeromonas were grown for 12 h and evaluated the influence of extracellular pH on Tetrahymena survival. The pH values of medium were maintained in the range 6.4 to 6.7 at 12 h after Aeromonas were cultured. The number of viable Tetrahymena was drastically decreased at very low (less than 4.5) or very high (greater than 8.5) pH values, and even decreased to zero at pH3.0 and pH3.5. But no significant changes were seen at the pH ranges between 5.5 and 7.5(Fig. 4).The result ruled out the possibility that pH changes over co-culture may kill T. thermophila, thus indicating the changes of relative survival of T. thermophila by pH conditions could be neglected.
Figure 4. Effect of different pHs on the growth of T. thermophila.
Biomass of T. thermophila was measured after cultured in different pH SPP medium for 12 h. Data are expressed as the mean ± SD of six measurements per time point.
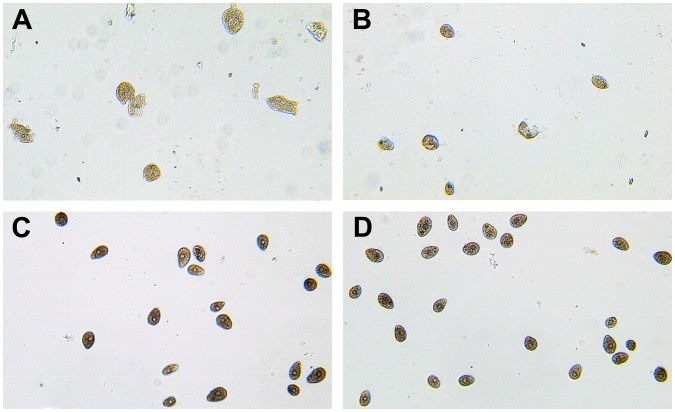
The effect of co-culture on growth of T. thermophila in a ratio of one cell to 5,000 Aeromonas cells was examined further. Biomass of T. thermophila cultured alone increased during the incubation and achieved a maximum concentration of 3.13×105 cells/ml at 12 h. The growth of T. thermophila co-cultivated with three representative Aeromonas strains, A. hydrophila XY-16, A. hydrophila NJ-1 and A. media NJ-8, can be seen in Fig. 5. Co-culture with A. hydrophila XY-16 caused survival of T. thermophila to reduce persistently during 12 h. The number of T. thermophila cells grown in the presence of A. hydrophila NJ-1 increased slightly before 4 h but then decreased, while the number of T. thermophila cells co-cultured with A. media NJ-8 increased throughout the incubation period but at a rate lower than the controls. These data show that virulent Aeromonas strains exert negative effects on the growth of T. thermophila and even decrease the survival of T. thermophila remarkably, while avirulent Aeromonas isolates decrease growth rate but do not affect total T. thermophila biomass. The reduced growth of T. thermophila in the presence of virulent Aeromonas coincided with the death of T. thermophila cells after 12 h culture. From Fig. 6A and Fig. 6B it is observed that T. thermophila cells co-cultured with virulent A. hydrophila XY-16 and NJ-1 became deformed and even lysed leading to cell death. In contrast, T. thermophila co-cultured with avirulent A. media NJ-8 grew well, were in tact (Fig. 6C) and looked similar to cells in the control group (Fig. 6D). The growth of T. thermophila during co-culture showed that the virulence of Aeromonas strains can be assessed based on relative survival of T. thermophila when the relative population size was 1×109 bacteria to 2×105 protozoan cells.
Figure 5. Growth of T. thermophila co-cultivated with Aeromonas isolates of different virulence.
Growth of T. thermophila co-cultured with A. hydrophila XY-16, A. hydrophila NJ-1 and A. media NJ-8 was measured for 12 h after initiating the co-culture. The control group was T. thermophila grown alone in sterile SPP medium. Data are expressed as the mean +SD of six measurements per time point.
Figure 6. Morphological changes of T. thermophila cells after co-cultivation with different Aeromonas strains.
T. thermophila was co-cultured with A. hydrophila XY-16 (A), A. hydrophila NJ-1 (B) and A. media NJ-8 (C) in SPP medium for 12 h before being observed under a light microscope. T. thermophila cultured in the absence of A. hydrophila (D) was used as a comparative control. Magnification × 100.
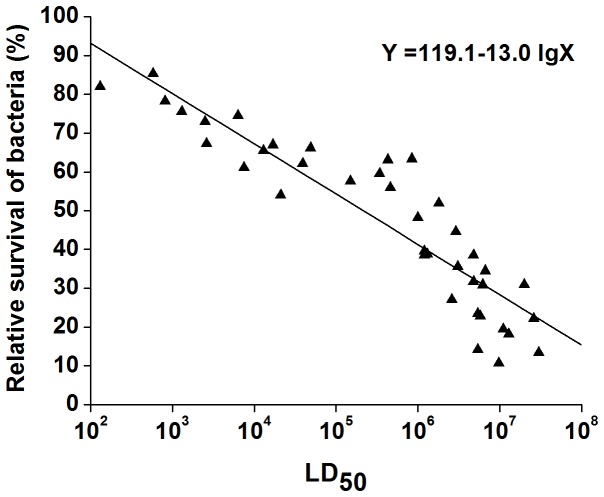
On the basis of these co-culture conditions, the virulence of all 39 Aeromonas strains was evaluated further in the T. thermophila model. The relative survival of the Aeromonas strains at 12 h was calculated to identify any correlation with LD50s determined in the zebrafish. In general, relative survival of the strains decreased as the LD50s increased. The Pearson correlation coefficient between relative survival of Aeromonas and the LD50s of these strains in zebrafish was −0.683 (P<0.01); thus, the correlation is categorized as moderate according to Munro [22]. The resulting regression equation to calculate relative survival is as follows:
Y = 119.1 - 13.0 lgX (X is the LD50 of the Aeromonas isolate and Y is relative survival of the Aeromonas isolate).
The standard deviation (SD) was 8.9, implying a prediction margin of ±SD. From the regression equation it is possible to predict that if the LD50 is above 1.0×106, then relative survival of the bacterium would be below 41.1%±8.9%. Relative survival values of the Aeromonas strains were all greater than 40% when the LD50s were lower than 1.0×106. Moreover, for most of the Aeromonas strains relative survival values were lower than 40% when the LD50s were above 1.0×106 (Fig. 7), except for three strains (NJ-28, NJ-3, XY-7). For avirulent Aeromonas isolates there was no relative survival value greater than 60%, while there was no relative survival value of virulent Aeromonas isolates less than 40%. This suggests that an Aeromonas isolate could be determined as virulent if relative Aeromonas survival was above 60% or, on the contrary, avirulent if relative survival was below 40%.
Figure 7. Linear regression model to predict relative survival of Aeromonas isolates from LD50 values.
Concentrations of Aeromonas and T. thermophila cells at the start of the experiment were 5×108 CFU/ml and 1×105 cells/ml, respectively. LD50s of 39 Aeromonas isolates in zebrafish were used as the abscissa and a trend line was added. Each experiment was repeated a minimum of four times. Data presented represent the mean of a four replicates.
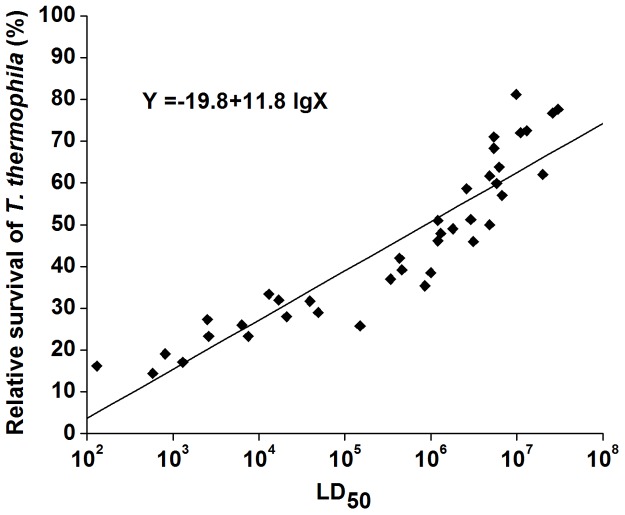
Relative survival of T. thermophila grown in the presence of the 39 Aeromonas strains increased as the LD50s of the Aeromonas strains increased (Fig. 8). This result indicates that there is a positive correlation between relative survival of T. thermophila and LD50 values in zebrafish since the Pearson correlation coefficient was 0.743 (P<0.01). The regression equation to calculate relative survival is as follows:
Figure 8. Linear regression model to predict relative survival of T. thermophila from LD50 values of the Aeromonas isolates.
Concentrations of Aeromonas and T. thermophila cells at the start of the experiment were 1×109 cells/ml and 2×105 cells/ml, respectively. LD50s of 39 Aeromonas isolates in zebrafish were used as the abscissa and a trend line was added. Each experiment was repeated a minimum of six times per time point. Data presented represent the mean of a four replicates.
Y = -19.8+11.8 lgX (X is the LD50 of the Aeromonas isolate and Y is relative survival of T. thermophila).
The SD was 7.9, implying a prediction margin of ±SD. Thus, from the regression equation it is estimated that if the LD50 is above 1.0×106, then relative survival of the bacteria would be greater than 51% ±7.9%.
The results show that almost all of the relative survival values of T. thermophila were lower than 40% when cultured in the presence of virulent Aeromonas isolates (LD50 values below 1.0×106), except for A. hydrophila NJ-37. On the contrary, relative survival of T. thermophila was above 40% when co-cultured with avirulent Aeromonas isolates (LD50s of greater than 1.0×106), except for A. hydrophila NJ-28. There was no relative survival value greater than 50% for T. thermophila co-cultured with a virulent Aeromonas isolate, while relative survival of T. thermophila was lower than 40% when co-cultured with avirulent isolates, except for A. hydrophila NJ28. Therefore, if relative survival of T. thermophila is greater than 50% after co-culture with an Aeromonas isolate, then this Aeromonas isolate is classified as avirulent. Conversely, the Aeromonas isolate is considered virulent if relative survival of T. thermophila in co-culture is less than 40%.
Discussion
Members of the genus Aeromonas are distributed widely in aquatic environments [23], [24]. Tetrahymena is a fresh-water protozoan that is ecologically highly successful. Thus, it is likely that these two organisms confront each other in the natural environment. Tetrahymena can predate other microorganisms and it uses phagocytosis to ingest and degrade these cells [25]; however, it seems that certain bacterial virulence factors have evolved to defend against phagocytosis by Tetrahymena [18], [26]. Therefore, it is not surprising that many bacterial virulence mechanisms are active against Tetrahymena and, for example, Ly and Muller [27] showed that hemolytic Listeria monocytogenes induces lysis of Tetrahymena pyriformis, while only a few protozoa underwent lysis in the presence of nonhemolytic Listeria innocua. Previous work by this group with A. hydrophila J-1 and NJ-4 showed that T. thermophila could distinguish virulent and avirulent A. hydrophila strains [20]. In this present study, we attempted to correlate data from an animal infection model with a less complex and more ethically acceptable Tetrahymena infection model.
Considering animal models remain the gold standard for bacterial virulence testing [28], we first determined LD50s of 39 Aeromonas strains in zebrafish. The LD50s ranged from 1.3×102 to 3.0×107, implying that these strains were differentially virulent and could therefore be used to study the Tetrahymena-Aeromonas infection model. Afterwards, nine Aeromonas strains of differential virulence were selected to determine the most appropriate experimental conditions useful for evaluating Aeromonas virulence in the Tetrahymena model. This present study demonstrates that the relative population sizes of the bacterium/protozoan play key roles in virulence interactions. When relative population sizes of bacteria to protozoan were sufficiently high (5×108 CFU/ml bacteria to 1×105 cell/ml protozoa or 1×109 CFU/ml bacteria to 2×105 cell/ml protozoa), it was more suited to distinguishing Aeromonas strains of different virulence.
High virulence Aeromonas strains could still grow in the presence of T. thermophila, while avirulent strains were largely phagocytozed. Moreover, T. thermophila grew well and cells were morphologically complete when co-cultivated with most avirulent Aeromonas strains; however, T. thermophila always became deformed and even lysed after co-culture with Aeromonas strains of high virulence. These results are in agreement with a previous report [20] and support the idea that resistance of bacteria to phagocytosis by T. thermophila correlates with virulence [11], [15]. Also, the virulence of an Aeromonas strain may be extrapolated from the ability of the protozoan to survive and grow in the presence of the bacterium.
To further support the idea that T. thermophila could be used reliably to evaluate Aeromonas virulence, the virulence of 39 Aeromonas strains was assessed in two ways using the T. thermophila model. From a bacterial perspective, an Aeromonas isolate could be determined as virulent when its relative survival in co-culture was above 60%, while the Aeromonas isolate was identified as avirulent when its relative survival was below 40%. This method is reliable for assessing Aeromonas virulence and discriminating virulent from avirulent strains. Unfortunately, we could not assess reliably the virulence of a few Aeromonas strains that had LD50 values between 1.0×105 and 1.0×106 because relative survival fluctuated from 60 to 40%. From a protozoan perspective, there were significant differences between the relative survival of T. thermophila co-cultured with virulent and avirulent Aeromonas strains. When relative survival of T. thermophila was lower than 40% in co-culture, the Aeromonas strain was regarded as virulent. In contrast, the strain was classified as avirulent if the relative survival of T. thermophila was greater than 50%. Hence, this implies that it is possible to evaluate the virulence of Aeromonas isolates by determining relative survival of co-cultured T. thermophila. Disappointingly, it was not possible to classify Aeromonas isolates as virulent or avirulent accurately when relative survival of T. thermophila was between 40 and 50%. This scenario involved Aeromonas strains with LD50s between 1.0×105 and 1.0×106, and is due mainly to the differences in virulence being only slight. Nevertheless, virulence assessments in the Tetrahymena-Aeromonas co-culture model using these two approaches generally complemented each other, and there existed good correlation between results obtained in the Tetrahymena model and in zebrafish, demonstrating the usefulness of this model system as a novel tool for investigating virulence determinants in Aeromonas spp.
The mechanisms that determine bacterial resistance to grazing protozoan are not known. Our previous study [20] showed that the extracellular products by Aeromonas might contribute to the death of T. thermophila. But the correlation of resistance with known virulence genes was not fully understood. Further work is required to enable understanding of interactions between the bacterial extracellular factors and host at the cellular and molecular level.
In conclusion, this is the first study to demonstrate that Tetrahymena can be used as an alternative host model for specifically determining virulence of Aeromonas strains. This protozoan model is an inexpensive and sensitive for such investigations, and can facilitate the systematic evaluation of virulence in hundreds or thousands of bacterial strains, and so is helpful for high-throughput screening of bacterial virulence factors. Moreover, as T. thermophila is amenable to genetic analysis and the entire genome has been published [29], [30], this system might also allow analysis of host resistance mechanisms.
Materials and Methods
Ethics Statement
Animal experiments were carried out according to animal welfare standards and approved by the Ethical Committee for Animal Experiments of Nanjing Agricultural University, China.
Strains, Media and Culture Conditions
A total of 39 Aeromonas strains were used in this study, including 20 A. hydrophila, 13 Aeromonas veronii, four Aeromonas media, one Aeromonas bestiarum, and one Aeromonas salmonicida (Table 1). All strains were identified by biochemical tests, polymerase chain reaction (PCR) amplification and sequencing of 16S rRNA and DNA gyrase subunit B (gyrB) genes. T. thermophila SB210 was obtained from Dr Miao Wei, Institute of Hydrobiology, Chinese Academy of Sciences.
Aeromonas strains were incubated in liquid Luria Bertani broth (LB) at 28°C. T. thermophila SB210 was grown axenically in SPP medium (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, 0.003% sequestrene) at 30°C.
LD50 Determinations
LD50 values of the 39 Aeromonas strains were determined in a zebrafish model. Eighty-day-old AB line inbred zebrafish weighing about 3g were bought from the Pearl River Fishery Research Institute, Chinese Academic of Fishery Science, and allowed to acclimate for at least 3 days before use. Single colonies of each bacterial strain were inoculated into LB and then overnight cultures were diluted 1∶100 in fresh LB. The inoculated cultures were incubated at 28°C and harvested during logarithmic phase of growth when the optical density at 600 nm reached 0.6. Harvested Aeromonas cells were washed twice in sterile phosphate-buffered saline (PBS), and then resuspended in PBS to appropriate concentrations. For each Aeromonas isolate, eight groups of ten fish were intraperitoneally injected with 0.02 ml of serially tenfold diluted bacterial suspensions containing 101–108 CFU. Another ten zebrafish (the control group) were injected intraperitoneally with 0.02 ml sterile PBS only. Zebrafish were placed in tanks, one for each bacterial infection group. Tanks were aerated and non-circulating, and were maintained at 28°C. The zebrafish were observed until one week post infection. Water was occasionally added to account for evaporation and any dead fish were removed from the tanks during the course of the experiments. For each determination, three separate experiments were performed in which ten fish were infected with each concentration of Aeromonas. Three replicate tanks of fish were used in the challenge tests. Survival data were analyzed by the method of Reed and Muench [31] for the calculation of LD50 values.
Effect of Co-culture on Aeromonas Growth
T. thermophila SB210 was cultured in sterile SPP medium at 30°C [18], [20] for 36 h until stationary phase of growth using an initial inoculum of 103 cells/ml. Cells in this culture were harvested by centrifugation at 2,000 g for 10 min at 15°C [20] with the consideration of keeping the normal morphology and athletic ability, washed twice with sterile SPP medium and adjusted to 1×105 cells/ml. This density of T. thermophila was selected on the basis of a preliminary sighting study, in which the starting density was screened from the fixed levels of 1×105 [11], 2×105 and 3×105 cells/ml as a density expected to show varying degrees of bacterial growth after co-cultured with Aeromonas strains of different virulence. Nine Aeromonas strains that had LD50s ranging between 1.3×102 and 3.0×107 in zebrafish were chosen by a stratified random sampling method [32] to be representative of differently virulent strains. Overnight cultures of these Aeromonas strains were diluted 100-fold in fresh LB and harvested by centrifugation at 4,000 g for 5 min after incubating at 28°C for 12 h. Harvested cultures were washed twice in sterile SPP medium and resuspended to 1×108, 5×108 and 1×109 CFU/ml, respectively. Five-hundred microliters of suspensions from these nine Aeromonas strains were mixed with equal volumes of T. thermophila SB210 cells. Then, 100 µl of these mixed cell suspensions were transferred into wells of 96-well plates; controls contained the bacterial strains at their respective concentrations mixed with an equal volume of SPP medium only and, similarly, T. thermophila SB210 cell suspensions supplemented with an equal volume of SPP medium. Sterile SPP medium was used as the blank well. Plates were incubated for 12 h at 28°C, and bacterial growth was determined every 2 h by measuring changes in absorbance of the suspensions at 450 nm. T. thermophila cells only accounted for negligible absorbance [11]. Evaporation during the incubation period was avoided by using a constant temperature-humidity incubator. Relative survival of bacteria (%) was expressed as the number of cells remaining in culture at 12 h relative to the number of cells present at the start of the co-culture. Each measurement was repeated in quadruplicate.
Effect of Co-culture on T. thermophila Viability
T. thermophila cells were harvested after being cultured at 30°C for 36 h, washed twice with sterile SPP medium and resuspended to 2×105 cells/ml. Nine Aeromonas strains, representative of differently virulent strains, were inoculated into LB medium and harvested after 12 h culture. The concentrations of these strains were adjusted to 2×108, 1×109 and 1.5×109 CFU/ml with sterile SPP medium. Five hundred microliters of each of the Aeromonas strains at the different concentrations were added to a same volume of T. thermophila cells in suspension in 2 ml Eppendorf tubes, and incubated at 28°C without shaking. To control for the effect of adding bacteria to T. thermophila cell suspensions, T. thermophila cells were separately incubated with an equal volume of sterile SPP medium. Every 2 h, 100 µl of each mixture was removed from the Eppendorf tubes, fixed with iodine solution and used to count the number of T. thermophila cells using a hemacytometer. The cellular morphology of T. thermophila cells incubated for 12 h was examined under a light microscope (Olympus cx21) after they had been fixed with iodine solution. Relative survival of T. thermophila (%) was expressed as the number of cells remaining in culture at 12 h relative to the number of cells present at the start of the co-culture. Each measurement was performed six times per time point and then mean values were calculated. Each experiment was repeated at least four times.
To evaluate the influence of pH changes by Aeromonas on T. thermophila viability, one milliliter of each of the nine Aeromonas strains with above different concentrations were cultured in the SPP medium with an initial pH value of 7.0 at 28°C for 12 h without shaking, and then pH values of SPP medium were measured by a pH meter (inoLab pH 7300, Germany). In addition, one milliliter of T. thermophila at an initial density of 2×105 cells/ml were cultured alone in the SPP medium with different pH values in the range 3.0 to 9.0 at 28°C for 12 h, and then T. thermophila were counted.
Assessment of Aeromonas Virulence using the T. thermophila Model
The virulence of the 39 Aeromonas strains for which LD50s had been determined in zebrafish was assessed using the Aeromonas-Tetrahymena infection model in two ways. First, cultures of the 39 Aeromonas strains were harvested, washed twice in SPP medium and adjusted to 5×108 CFU/ml, while T. thermophila cells were resuspended at 1×105 cells/ml. Second, the Aeromonas cultures were resuspended at 1×109 CFU/ml, while T. thermophila suspensions were adjusted to 2×105 cells/ml. Then the relative survival of T. thermophila was determined after co-culturing. The Pearson correlation coefficient (r) was used to quantify the relationship between the relative survival values of Aeromonas or T. thermophila and the LD50s of Aeromonas isolates in zebrafish. Additionally, linear regression models were used to predict the relative survival values from the LD50s of Aeromonas isolates in zebrafish. Each experiment was repeated at least four times.
Statistical Analysis
Pearson correlation coefficients were calculated using the SPSS v20.0 statistics software. The linear regression model was fitted using the Origin 7.5 software. Error bars presented in the figures represent standard deviations of the means of multiple (>3) replicate experiments. A P-value <0.05 was considered to indicate a significant difference.
Funding Statement
The study was funded by National Nature Science Foundation (31072151) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ni XD, Wang N, Liu YJ, Lu CP (2010) Immunoproteomics of extracellular proteins of the Aeromonas hydrophila China vaccine strain J-1 reveal a highly immunoreactive outer membrane protein. FEMS Immunol Med Microbiol 58: 363–373. [DOI] [PubMed] [Google Scholar]
- 2. Bi ZX, Liu YJ, Lu CP (2007) Contribution of AhyR to virulence of Aeromonas hydrophila J-1. Res Vet Sci 83: 150–156. [DOI] [PubMed] [Google Scholar]
- 3. Janda JM, Abbott SL (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23: 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stelma Jr GN, Reyes AL, Peeler JT, Johnson CH, Spaulding PL (1992) Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus . Appl Environ Microbiol 58: 2776–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Froquet R, Cherix N, Burr SE, Frey J, Vilches S, et al. (2007) Alternative host model to evaluate Aeromonas virulence. Appl Environ Microbiol 73: 5657–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodriguez I, Novoa B, Figueras A (2008) Immune response of zebrafish (Danio rerio) against a newly isolated bacterial pathogen Aeromonas hydrophila . Fish Shellfish Immunol 25: 239–249. [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez I, Chamorro R, Novoa B, Figueras A (2009) beta-Glucan administration enhances disease resistance and some innate immune responses in zebrafish (Danio rerio). Fish Shellfish Immunol 27: 369–373. [DOI] [PubMed] [Google Scholar]
- 8. Pradel E, Ewbank JJ (2004) Genetic models in pathogenesis. Annu Rev Genet 38: 347–363. [DOI] [PubMed] [Google Scholar]
- 9. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, et al. (2001) A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 98: 10892–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miyazaki S, Matsumoto Y, Sekimizu K, Kaito C (2012) Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol Lett 326: 116–124. [DOI] [PubMed] [Google Scholar]
- 11. Benghezal M, Adam E, Lucas A, Burn C, Orchard MG, et al. (2007) Inhibitors of bacterial virulence identified in a surrogate host model. Cell Microbiol 9: 1336–1342. [DOI] [PubMed] [Google Scholar]
- 12. Orias E (1998) Mapping the germ-line and somatic genomes of a ciliated protozoan, Tetrahymena thermophila . Genome Res 8: 91–99. [DOI] [PubMed] [Google Scholar]
- 13. Orias E, Cervantes MD, Hamilton EP (2011) Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 162: 578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pushkareva VI (2003) Experimental evaluation of interaction between Yersinia pestis and soil infusoria and possibility of prolonged preservation of bacteria in the protozoan oocysts. Zh Mikrobiol Epidemiol Immunobiol 4: 40–44 (in Russian). [PubMed] [Google Scholar]
- 15. Breneva NV, Maramovich AS (2008) Modeling of interaction between Yersinia pestis and Tetrahymena pyriformis in experimental ecosystems. Zh Mikrobiol Epidemiol Immunobiol 5: 39–41 (in Russian). [PubMed] [Google Scholar]
- 16. Steinberg KM, Levin BR (2007) Grazing protozoa and the evolution of the Escherichia coli O157:H7 shiga toxin-encoding prophage. Proc Biol Sci 274: 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bukharin OV, Plotnikov AO, Nemtseva NV, Kovbyk LV (2008) Histone-antihistone interactions in the Escherichia coli-Tetrahymena pyriformis association. Mikrobiologiia 77: 219–225 (in Russian). [PubMed] [Google Scholar]
- 18. Lainhart W, Stolfa G, Koudelka GB (2009) Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila . J Bacteriol 191: 5116–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonnet JL, Bonnemoy F, Dusser M, Bohatier J (2008) Toxicity assessment of the herbicides sulcotrione and mesotrione toward two reference environmental microorganisms: Tetrahymena pyriformis and Vibrio fischeri . Arch Environ Contam Toxicol 55: 576–583. [DOI] [PubMed] [Google Scholar]
- 20. Li J, Zhang XL, Liu YJ, Lu CP (2011) Development of an Aeromonas hydrophila infection model using the protozoan Tetrahymena thermophila . FEMS Microbiol Lett 316: 160–168. [DOI] [PubMed] [Google Scholar]
- 21. Pu JY, Huang XX, Lu CP (2007) Virulence detection of Streptococcus suis type 2 in zebrafish. Sci Agric Sin 40: 2655–2658 (in Chinese). [Google Scholar]
- 22.Munro BH (1993) Selected nonparametric techniques. In: Munro BH, Page EB. Statistical methods for health care research. Philadelphia: Lippincott. 81–98.
- 23. Bin Kingombe CI, Huys G, Howald D, Luthi E, Swings J, et al. (2004) The usefulness of molecular techniques to assess the presence of Aeromonas spp. harboring virulence markers in foods. Int J Food Microbiol 94: 113–121. [DOI] [PubMed] [Google Scholar]
- 24. Ottaviani D, Santarelli S, Bacchiocchi S, Masini L, Ghittino C, et al. (2006) Occurrence and characterization of Aeromonas spp. in mussels from the Adriatic Sea. Food Microbiol 23: 418–422. [DOI] [PubMed] [Google Scholar]
- 25. Jacobs ME, DeSouza LV, Samaranayake H, Pearlman RE, Siu KW, et al. (2006) The Tetrahymena thermophila phagosome proteome. Eukaryot Cell 5: 1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Friman V-P, Hiltunen T, Laakso J, Kaitala V (2008) Prey resource availability drives evolution of predator prey interaction. Proc R Soc Lond B 275: 1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ly TM, Muller HE (1990) Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol 33: 51–54. [DOI] [PubMed] [Google Scholar]
- 28. Hayes SL, Rodgers MR, Lye DJ, Stelma GN, McKinstry CA, et al. (2007) Evaluating virulence of waterborne and clinical Aeromonas isolates using gene expression and mortality in neonatal mice followed by assessing cell culture’s ability to predict virulence based on transcriptional response. J Appl Microbiol 103: 811–820. [DOI] [PubMed] [Google Scholar]
- 29. Stover NA, Krieger CJ, Binkley G, Dong Q, Fisk DG, et al. (2006) Tetrahymena Genome Database (TGD): a new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res 34: D500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stover NA, Punia RS, Bowen MS, Dolins SB, Clark TG (2012) Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database (Oxford) 2012: bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reed LJ, Muench H (1938) A simple method of estimating fifty percent end points. Am J Hyg 27: 493–497. [Google Scholar]
- 32.Coehran WG (1977) Sampling techniques, 3rd edition. New York:John Wiley & Sons. 428p.